Abstract
Upon starvation, a dense population of rod-shaped Myxococcus xanthus bacteria coordinate their movements to construct mounds in which some of the cells differentiate to spherical spores. During this process of fruiting body formation, short-range C-signaling between cells regulates their movements and the expression of genes important for sporulation. C-signaling activates FruA, a transcription factor that binds cooperatively with another transcription factor, MrpC2, upstream of the fmgA and fmgBC promoters, activating transcription. We have found that a third C-signal-dependent gene, herein named fmgD, is subject to combinatorial control by FruA and MrpC2. The two proteins appear to bind cooperatively upstream of the fmgD promoter and activate transcription. FruA binds proximal to the fmgD promoter, as in the fmgBC promoter region, whereas MrpC2 binds proximal to the fmgA promoter. A novel feature of the fmgD promoter region is the presence of a second MrpC2 binding site partially overlapping the promoter and therefore likely to mediate repression. The downstream MrpC2 site appears to overlap the FruA site, so the two transcription factors may compete for binding, which in both cases appears to be cooperative with MrpC2 at the upstream site. We propose that binding of MrpC2 to the downstream site represses fmgD transcription until C-signaling causes the concentration of active FruA to increase sufficiently to outcompete the downstream MrpC2 for cooperative binding with the upstream MrpC2. This would explain why fmgD transcription begins later during development and is more dependent on C-signaling than transcription of fmgA and fmgBC.
Myxococcus xanthus is a Gram-negative soil bacterium that undergoes multicellular development when starved, providing an attractive model to study signaling and gene regulatory mechanisms (49). Cells glide in swarms over solid surfaces, seeking prey bacteria on which to feed (3). When nutrients become limited, cells alter their movements, including the frequency with which they reverse their direction of gliding (15). Approximately 105 cells coordinate their movements to construct a mound-shaped fruiting body. A majority of the cells undergo programmed cell death during the developmental process, but some cells in the fruiting body differentiate into dormant, stress-resistant, spherical spores (30). The mature fruiting body is a dense mound of spores capable of germinating in response to nutrients and producing a swarm of cells ready to feed, grow, and divide.
Three signals governing M. xanthus fruiting body development are fairly well understood. Starvation triggers a stringent response that leads to the production of intracellular (p)ppGpp and induction of early developmental genes (11, 41). The second signal, A-signal, is a mixture of amino acids and peptides produced extracellularly by the activity of secreted proteases (24, 34). A-signaling appears to allow cells to measure their density and, if sufficient, proceed into development by beginning to form mounds and expressing A-signal-dependent genes (23, 25). The third signal is also extracellular, but unlike the A-signal, which is diffusible, the C-signal is a short-range signal involving CsgA (40), a protein associated with the outer membrane, where it is cleaved by a secreted protease to a 17-kDa form that appears to act as the signal (18, 27, 35), although a receptor remains to be identified. C-signaling is necessary for cells to build large, stable mounds, apparently because cells become aligned in the outer domain of a nascent fruiting body as they move in circles (16, 37) and efficient C-signaling between aligned cells (19) causes some to form spores, which are swept to the interior of the fruiting body by continued movement of cells in the outer domain (37). Many studies support a model in which an increasing level of C-signaling governs mound formation followed by sporulation within mounds to produce mature fruiting bodies (reviewed in references 13, 39, and 42). C-signaling is also necessary for normal developmental gene expression after the early mound stage (20). Genes that are normally expressed shortly after early mound formation exhibited reduced expression in a csgA mutant incapable of C-signaling, and genes that are normally expressed later failed to be expressed. For these two classes of genes, their differential timing of expression and level of dependence on C-signaling is likely related but is not understood.
Recently, an understanding of the regulation of genes in the early class that depends only in part on C-signaling has been achieved. Two transcription factors bind cooperatively upstream of the fmgA and fmgBC promoters and activate transcription (28, 29). The fmg designation stands for “FruA- and MrpC2-regulated gene.” FruA is similar to response regulators of two-component signal transduction systems and has been proposed to be phosphorylated in response to C-signal (6, 33), but a cognate histidine kinase has not been identified, and some evidence suggests that FruA might function without being phosphorylated (28). MrpC is similar to the cyclic AMP receptor protein (CRP) family (43), and it is inhibited during growth by phosphorylation (31, 32). Compared with the unphosphorylated form, the phosphorylated form of MrpC binds weakly to DNA. Starvation inhibits phosphorylation of MrpC, and some of the unphosphorylated form may be cleaved to MrpC2, lacking the 25 N-terminal residues (46). MrpC2 binds better than MrpC to sites upstream of the mrpC and fruA promoters, activating transcription so that the concentrations of MrpC, MrpC2, and FruA increase in starving cells (32, 46). MrpC also binds to the mazF promoter region and activates expression of MazF, a toxic endoribonuclease that causes programmed cell death during development (30). Paradoxically, MrpC acts as an antitoxin by binding to MazF and inhibiting its activity. Presumably, this allows some cells in the population to escape programmed cell death and eventually form spores inside fruiting bodies. Binding of MrpC2 to MazF and to the mazF promoter region remains to be tested, as does binding of MrpC to the fmgA and fmgBC promoter regions. MrpC2 binds cooperatively with FruA upstream of the fmgA and fmgBC promoters but in different arrangements; MrpC2 binds proximal to the fmgA promoter, and FruA binds proximal to the fmgBC promoter (28, 29). Cooperative binding by the two transcription factors has been proposed to integrate starvation signaling and cell death through MrpC and MrpC2 with positional information via C-signaling through FruA, determining spatiotemporal gene expression and cell fate (28).
Here, we report that MrpC2 and FruA combinatorially regulate a gene in the late class that depends completely on C-signaling. The gene was identified by an insertion of the transposon Tn5 lac at the Ω4403 locus, because it created a transcriptional fusion to lacZ, and the M. xanthus strain exhibited developmentally regulated β-galactosidase activity (22). Developmental lacZ expression was abolished in a csgA mutant incapable of C-signaling (20). The gene at the Ω4403 locus (MXAN1501) (9) is predicted to code for a subtilisin-type serine protease, but the Tn5 lac insertion in the coding region caused no discernible growth or developmental defect (8, 22). DNA downstream of position −80 (relative to the transcriptional start site) was shown to be sufficient for C-signal-dependent activation of the promoter (8), and mutational analysis identified three positive cis-regulatory sequences upstream of the promoter (47). Two of the sequences, a 5-bp element (consensus sequence GAACA) at positions −63 to −59 and a C box (consensus sequence CAYYCCY, in which Y means C or T) at positions −52 to −46, are also present upstream of the fmgA and fmgBC promoters in regions important for promoter activity that are bound by MrpC2 and FruA (7, 28, 29, 50-52). Therefore, we tested whether MrpC2 and FruA bind upstream of the MXAN1501 promoter. The two proteins appear to bind cooperatively to DNA with FruA proximal to the promoter, as in the fmgBC promoter region, and this combination of proteins activates transcription, based on the effects of mutations upstream of the promoter that impair both DNA binding in vitro and promoter activity in vivo. Hence, we name the MXAN1501 gene fmgD (for FruA- and MrpC2-regulated gene D). Importantly, the fmgD promoter region contains a second MrpC2 binding site that partially overlaps the promoter and the FruA binding site. We propose that cooperative binding of MrpC2 to the two sites accounts for the differential timing and dependence on C-signaling of fmgD compared with those of fmgA and fmgBC.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 10 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm with DE3, a λ prophage carrying the T7 RNA polymerase gene | Novagen |
| SMhisMrpC2 | BL21(DE3) containing pET16b/His10-MrpC2 | 28 |
| SMFruAhis | BL21(DE3) containing pET11km/FruA-His6 | 28 |
| M. xanthus | ||
| DK1622 | Wild type | 14 |
| DK5285 | fruA::Tn5 lac Ω4491 | 21 |
| MPL1-6, -7, -8 | sglA1 attB::pMF100 | This work |
| MPL2-2, -7, -9 | sglA1 fruA::TnV Ω786 attB::pMF100 | This work |
| MPV4037-1, -5, -8 | attB::pPV04037 | This work |
| MPV4441-1, -2, -3 | attB::pPV04441 | This work |
| Plasmids | ||
| pET16b/His10-MrpC2 | pET16b with a gene encoding His10-MrpC2 under the control of a T7 RNA polymerase promoter | 32 |
| pET11km/FruA-His6 | pET11km with a gene encoding FruA-His6 under the control of a T7 RNA polymerase promoter | S. Inouye |
| pREG1727 | Apr Kmr P1-inc attP ′lacZ | 8 |
| pMF100 | pREG1727 with fmgD DNA from positions −80 to +382 | 8 |
| pGEM7Zf | Aprlacα | Promega |
| pMF0100 | pGEM7Zf with fmgD DNA from positions −80 to +382 | 8 |
| pPV74 | pMF0100 with a G-to-T mutation at position −74 | 47 |
| pPV4037 | pMF0100 with a TTGA-to-GGTC mutation at positions −40 to −37 | This work |
| pPV04037 | pREG1727 with a 537-bp XhoI-BamHI fragment from pPV4037 | This work |
| pPV4441 | pMF0100 with a CGGG-to-ATTT mutation at positions −44 to −41 | This work |
| pPV04441 | pREG1727 with a 537-bp XhoI-BamHI fragment from pPV4441 | This work |
Growth and development.
M. xanthus strains were grown at 32°C in CTT (1% Casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH 7.6]) medium (12) or on CTT agar (1.5%) plates. When required, 40 μg kanamycin sulfate per ml was added. Fruiting body development was performed on TPM (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH 7.6]) agar (1.5%) plates as described previously (22).
Site-directed mutagenesis and construction of plasmids.
The QuikChange site-directed mutagenesis kit (Stratagene) was used to create mutations in pMF0100 using primer pairs LK714 and LK715 or LK716 and LK717 (Table 2). Sequencing of the M. xanthus DNA portion of the plasmid was performed to ensure the presence of only the desired mutation. The BamHI-XhoI fragment bearing the mutant fmgD promoter region was gel purified and subcloned into BamHI-XhoI-digested pREG1727. Escherichia coli DH5α served as the host for plasmid construction and propagation.
TABLE 2.
Primers used in this study
| Primer | Sequence | Descriptiona or reference |
|---|---|---|
| LK714 | CACGGACCGCCGTCTCATCCCTCATTTTTGATTCATGAATAAGCCG | −67 forward with a CGGG-to-ATTT mutation at −44 to −41 |
| LK715 | CGGCTTATTCATGAATCAAAAATGAGGGATGAGACGGCGGTCCGTG | −22 reverse, complement of LK714 |
| LK716 | GCCGTCTCATCCCTCCGGGGGTCTTCATGAATAAGCCG | −59 forward with a TTGA-to-GGTC mutation at −40 to −37 |
| LK717 | CGGCTTATTCATGAAGACCCCCGGAGGGATGAGACGGC | −22 reverse, complement of LK716 |
| LK1359 | CAAAAACGGCTTATTCATGAATC | −16 reverse |
| LK1376 | CGCCATGGCATGTTCAATCA | −85 forward |
| LK1407 | CTCGAGCAGCTGAAGCTGG | MCS of pPV74 plus −80 to −78 forward |
| LK1861 | CCTTGAGCGCGATGGAGATA | 52 |
| LK1862 | CTCGGCGGCCTCATCGAC | 52 |
| LK2104 | TGGCATGTTCAATCACGGACCGCCGTCT | −80 forward |
| LK2105 | AGACGGCGGTCCGTGATTGAACATGCCA | −53 reverse, complement of LK2104 |
| LK2106 | TCACGGACCGCCGTCTCATCCCTCCGGG | −68 forward |
| LK2107 | CCCGGAGGGATGAGACGGCGGTCCGTGA | −41 reverse, complement of LK2106 |
| LK2108 | CGTCTCATCCCTCCGGGTTGATTCATGA | −57 forward |
| LK2109 | TCATGAATCAACCCGGAGGGATGAGACG | −30 reverse, complement of LK2108 |
| LK2304 | TGGCAGTGGAAATCACGGACCGCCGTCT | −80 forward with a TGTTC-to-GTGGA mutation at −75 to −71 |
| LK2305 | AGACGGCGGTCCGTGATTTCCACTGCCA | −53 reverse, complement of LK2304 |
| LK2306 | TGGCATGTTCAATCACGTCAATCCGTCT | −80 forward with a GACCG-to-TCAAT mutation at −63 to −59 |
| LK2307 | AGACGGATTGACGTGATTGAACATGCCA | −53 reverse, complement of LK2306 |
| LK2308 | TGGCAGTGGAAATCACGTCAATCCGTCT | −80 forward with a TGTTC-to-GTGGA mutation at −75 to −71 and a GACCG-to-TCAAT mutation at −63 to −59 |
| LK2309 | AGACGGATTGACGTGATTTCCACTGCCA | −53 reverse, complement of LK2308 |
| LK2446 | CATCCCTCCGGGTTGATTCATGAATAAGCCGTTTTTG | −52 forward |
| LK2447 | CAAAAACGGCTTATTCATGAATCAACCCGGAGGGATG | −16 reverse, complement of LK2446 |
| LK2453 | CAAAAACGGCTTATTCATGAAGA | −16 reverse for PCR with pPV4037 as template |
The number is relative to the start site of fmgD transcription, and the orientation (forward or reverse) is relative to the direction of fmgD transcription.
Construction of M. xanthus strains and determination of lacZ expression during development.
Strains containing a plasmid integrated at the Mx8 phage attachment site, attB, were constructed by electroporation (17). Transformants were selected on CTT agar plates containing kanamycin sulfate and screened on TPM agar plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside/ml, in order to avoid rare transformants with unusual developmental lacZ expression (47). Three transformants were chosen for further analysis, and β-galactosidase activity was measured as described previously (22).
ChIP.
M. xanthus strains DK1622 and DK5285 were used for ChIP as described previously (28, 52), except Dynabeads protein G (Invitrogen) (100 μl/ml of cell extract) was used instead of protein A Sepharose beads for preclearing and immunoprecipitation. The primers used for PCR of the fmgD promoter region were LK1359 and LK1376, and the primers used for PCR of the rpoC coding region were LK1861 and LK1862 (Table 2).
Preparation of His10-MrpC2 and FruA-His6.
Recombinant proteins were expressed in E. coli and purified as described previously (28, 32).
Preparation of DNA fragments.
DNA fragments from the fmgD promoter region were generated by PCR using wild-type or mutant plasmid (Table 1) as the template and the oligonucleotide primers listed in Table 2. For electrophoretic mobility shift assays (EMSAs), 32P-labeled DNA was synthesized by PCR after labeling the primers with [γ-32P]ATP using T4 polynucleotide kinase (New England BioLabs), and the DNA fragment was purified after 15% PAGE (38). Alternatively, complementary primers were labeled with 32P as just described, mixed, boiled for 10 min, and placed at room temperature for 3 h, and then the double-stranded DNA fragment was purified as just described.
EMSAs.
EMSAs were performed as described previously (52), except that binding reaction mixtures were incubated at 25°C for 15 min.
RESULTS
Expression of fmgD depends on fruA.
To determine whether FruA is involved in regulation of fmgD, expression from an fmgD-lacZ transcriptional fusion was measured during development of wild-type and fruA mutant M. xanthus strains. The fmgD-lacZ fusion was integrated at the Mx8 phage attachment site in the M. xanthus chromosome via site-specific recombination as described previously (8). Expression from the fmgD promoter was abolished in the fruA mutant (Fig. 1), indicating that FruA directly or indirectly regulates transcription of fmgD.
FIG. 1.
Expression of fmgD-lacZ during development. β-galactosidase-specific activity during development was measured for lacZ fused to fmgD (positions −80 to +382) in the wild type (⧫) or in a fruA mutant (▪). The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Points show the averages of results from three transformants, and error bars depict one standard deviation of the data.
MrpC and FruA associate with the fmgD promoter region in vivo.
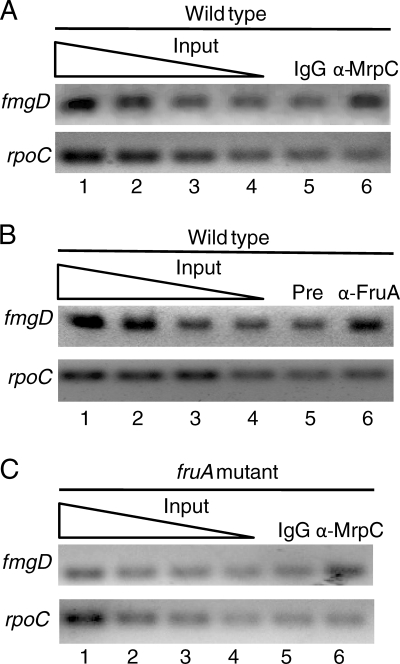
To determine whether MrpC (and/or MrpC2) and FruA are associated with the fmgD promoter region in vivo, we performed ChIP assays with polyclonal antibodies against MrpC that also recognize MrpC2 and with polyclonal antibodies against FruA. Wild-type M. xanthus cells were collected after 18 h of development and subjected to ChIP with affinity-purified immunoglobulin G (IgG) antibodies against MrpC (or, as a control, with total IgG from nonimmunized rabbits) or with antiserum against FruA (or, as a control, preimmune antiserum). DNA recovered after ChIP was analyzed by PCR with primers designed to amplify the fmgD promoter region or, as a control, the rpoC coding region. The PCR analysis showed that the fmgD promoter region, but not the rpoC coding region, was reproducibly enriched by ChIP with the anti-MrpC antibodies relative to the IgG control (Fig. 2 A, lanes 5 and 6) and with the anti-FruA antibodies relative to the preimmune control (Fig. 2B, lanes 5 and 6). These results indicate that MrpC and/or MrpC2 and FruA are present in the vicinity of the fmgD promoter at 18 h into development, when expression of fmgD-lacZ was observed (Fig. 1).
FIG. 2.
Association of MrpC and/or MrpC2 and FruA with the fmgD promoter region during development. ChIP analysis of M. xanthus at 18 h into development. Cells were treated with formaldehyde and lysed. Cross-linked chromatin was immunoprecipitated with antibodies. DNA was amplified with primers for the fmgD promoter region (positions −85 to −16 relative to the start site of transcription) or for the rpoC coding region (positions +1780 to +1905 relative to the predicted translation start) as a control. A 2-fold dilution series of input DNA purified from 0.025, 0.0125, 0.00625, or 0.003125% of the total cellular extract prior to immunoprecipitation was used as a template in parallel PCRs to show that the PCR conditions were in the linear range of amplification for each primer set. (A) Wild-type strain DK1622 with affinity-purified IgG antibodies against MrpC (α-MrpC), or, as a control, total IgG (IgG) from nonimmunized rabbits. (B) Wild-type strain DK1622 with antiserum against FruA (α-FruA) or, as a control, preimmune antiserum (Pre). (C) fruA mutant strain DK5285 antibodies, as described for panel A.
Association of MrpC and/or MrpC2 with the fmgA and fmgBC promoter regions was dependent on FruA, presumably due to cooperative binding of the two proteins just upstream of the promoters (28, 29). To determine whether association of MrpC and/or MrpC2 with the fmgD promoter region depends on FruA, ChIP assays were performed on a fruA mutant at 18 h into development. Interestingly, the fmgD promoter region was reproducibly enriched by ChIP with the anti-MrpC antibodies relative to the IgG control (Fig. 2C, lanes 5 and 6). We conclude that MrpC and/or MrpC2 do not require FruA to associate with the fmgD promoter region, in contrast to the fmgA and fmgBC promoter regions.
MrpC2 and FruA bind cooperatively to the fmgD promoter region.
Previous studies showed that MrpC2 and FruA bind cooperatively just upstream of the fmgA and fmgBC promoter regions (28, 29). To determine whether this is the case for the fmgD promoter region, EMSAs were performed with a DNA fragment spanning from positions −80 to −16 and purified His10-MrpC2 and FruA-His6. His10-MrpC2 produced two shifted species (Fig. 3, lane 2), suggesting that the fragment includes two binding sites (see below). We reasoned that the more abundant, lower species was a mixture of His10-MrpC2 bound singly to one site, and the upper species was two molecules of His10-MrpC2 bound to the DNA fragment (i.e., both sites occupied). FruA-His6 produced a single shifted complex (Fig. 3, lane 3), indicative of a single binding site. The combination of proteins produced two shifted species (Fig. 3, lane 4). The total amount of shifted species was greater than expected for additive binding, a pattern shown previously to be indicative of cooperative binding (28). The upper species is presumably a mixture of DNA fragments with two sites occupied (one His10-MrpC2 and one FruA-His6, or two His10-MrpC2), and the lower species is presumably a mixture of DNA fragments with one site occupied. Much more of the lower species is formed by the combination of His10-MrpC2 and FruA-His6 than by either protein alone, which might reflect initial cooperative binding of the two proteins followed by dissociation of FruA-His6 (whose binding is weaker), as discussed previously for fmgA and fmgBC (28, 29). Occasionally, binding of both proteins may be lost during the electrophoresis, giving rise to the observed smear between the lower species and the unbound probe DNA fragment.
FIG. 3.
Binding of MrpC2 and FruA to the fmgD promoter region. EMSAs with 32P-labeled fmgD DNA (2 nM) spanning from positions −80 to −16 and no protein, His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM) as indicated, were electrophoresed on an 8% polyacrylamide gel. The two arrowheads indicate the shifted species produced by the combination of proteins.
MrpC2 binds to sequences important for fmgD promoter activity.
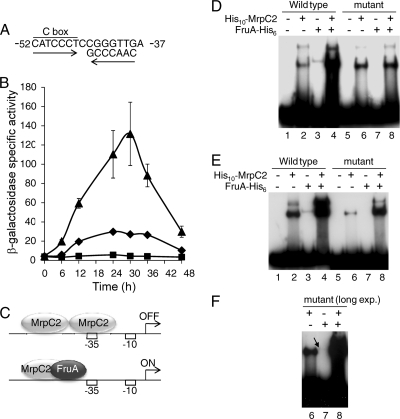
Mutational analysis of the fmgD promoter region previously identified three positive cis-regulatory sequences, a 10-bp element at positions −79 to −70, a 5-bp element at positions −63 to −59, and a C box at positions −52 to −46 (Fig. 4 A) (47). We note that within the 10-bp element, there is a sequence matching the 5-bp element consensus sequence (GAACA), on the other strand at positions −71 to −75. Sequences similar to the 5-bp element consensus sequence and the C box (consensus sequence CAYYCCY, in which Y means C or T) are present in the promoter regions of other C-signal-dependent genes (7, 50, 51) and have been shown to be important for the binding of MrpC2 and FruA in the fmgA and fmgBC promoter regions (28, 29, 52). To localize MrpC2 and FruA binding in the fmgD promoter region, three oligonucleotide pairs were used as probes in EMSAs. Each oligonucleotide pair forms a 28-bp duplex that was tested for binding of His10-MrpC2, FruA-His6, and the combination of proteins. His10-MrpC2 produced a shifted complex only with DNA from positions −80 to −53 (Fig. 4A, lane 2), whereas FruA-His6 produced a shifted complex only with DNA from positions −57 to −30 (Fig. 4A, lane 11). No enhancement of complex formation was observed with the combination of proteins, apparently because none of the 28-bp duplexes allows binding of both proteins. We conclude that MrpC2 binds upstream of FruA in the fmgD promoter region.
FIG. 4.
MrpC2 binds to sequences important for fmgD promoter activity. (A) Sequences important for fmgD promoter activity and binding of MrpC2 and FruA to different probes. The top part shows the sequence of the fmgD promoter region with three positive cis regulatory elements labeled. A short segment of the other strand and divergent arrows highlight the inverted 5-bp elements. The location and ends of three oligonucleotide pairs used as probes (1 nM) are depicted with downward arrows pointing to the corresponding EMSAs (lanes 1 to 12). No protein, His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM) were added as indicated, and reactions were electrophoresed on an 8% polyacrylamide gel. The black arrowhead points to a shifted complex produced by His10-MrpC2, and the white arrowhead points to a shifted complex produced by FruA-His6. Lanes 13 to 18 show EMSAs with mutant −80 to −53 oligonucleotide pairs in which TGTTC at positions −75 to −71 was changed to GTGGA (lanes 13 and 14), GACCG at positions −63 to −59 was changed to TCAAT (lanes 15 and 16), or both changes were made (lanes 17 and 18). The light gray spots in lanes 16 to 18 are artifacts of X-ray film processing, not shifted complexes. (B) Effect of a 1-bp change at position −74 on binding of MrpC2 and FruA. EMSAs were performed as described above except with wild-type or mutant fmgD DNA (2 nM) spanning from positions −80 to −16. Exposure to film was longer in this experiment than in the experiment shown in Fig. 3, so the two shifted species in lane 4 are unresolved. Intervening lanes were deleted from the image. (C) A longer exposure of the experiment described for panel B allowed detection of a faint, shifted complex in lane 6 (arrowhead).
To determine whether MrpC2 recognizes the inverted 5-bp elements at positions −63 to −59 and at −71 to −75, we measured binding of His10-MrpC2 to three mutant versions of the DNA spanning from positions −80 to −53. A 5-bp change in either 5-bp element, or simultaneous 5-bp changes in both elements, eliminated binding (Fig. 4A, lanes 14, 16, and 18). These results indicate that MrpC2 binds to the inverted 5-bp elements, presumably as a dimer (31, 32). We refer to this as the “upstream” MrpC2 binding site.
The mutation in the 5-bp element at positions −63 to −59 (GACCG to TCAAT) that eliminated binding of His10-MrpC2 in vitro (Fig. 4A, lane 16) was shown previously to abolish fmgD promoter activity in vivo (47). The effect on fmgD promoter activity of the 5-bp change at positions −71 to −75 has not been measured; however, a 1-bp change at positions −74 (G to T) abolished promoter activity (47). Therefore, binding of His10-MrpC2 and FruA-His6 to a DNA fragment (−80 to −16) bearing the 1-bp change was measured in EMSAs. Binding of His10-MrpC2 was greatly reduced (Fig. 4B, lane 6), although a single, faint, shifted complex was detected in a long exposure (Fig. 4C, lane 6). Binding of FruA-His6 was unchanged (Fig. 4B, lane 7). The combination of proteins exhibited little or no enhancement of binding (Fig. 4B, lane 8). Taken together, the data suggest that binding of MrpC2 to the upstream site is important for cooperative binding of FruA and for fmgD promoter activity.
Localization of a second MrpC2 binding site and of the FruA binding site.
His10-MrpC2 produced two shifted species with the DNA segment from positions −80 to −16 (Fig. 3, lane 2), but the 28-bp duplex from positions −80 to −53 produced only a single shifted complex, and neither of the other 28-bp duplexes (−68 to −41 and −57 to −30) produced a shifted complex (Fig. 4A), suggesting that DNA between positions −30 and −16 might be important for binding of a second molecule of MrpC2. To test this idea and to further localize the site of FruA binding, additional DNA segments were used as probes in EMSAs. A segment from positions −80 to −30 produced a single shifted complex with His10-MrpC2 (Fig. 5 A, lane 10), consistent with the idea that binding of a second MrpC2 molecule requires DNA between positions −30 and −16 (Fig. 5A, lane 2). FruA-His6 also produced a single shifted complex with DNA from positions −80 to −30 (Fig. 5A, lane 11), as expected since FruA bound the 28-bp duplex from positions −57 to −30 (Fig. 4A, lane 11). The combination of His10-MrpC2 and FruA-His6 exhibited enhanced binding (relative to either protein alone) to DNA from positions −80 to −30 (Fig. 5A, lane 12), similar to that observed with the DNA fragment from positions −80 to −16 (Fig. 5A, lane 4). These results provide additional evidence that MrpC2 at the upstream site can bind cooperatively with FruA and indicate that binding of MrpC2 to a second site, which we refer to as the “downstream” site below, requires DNA between positions −30 and −16.
FIG. 5.
Localization of a second MrpC2 binding site and of the FruA binding site. (A) EMSAs with 32P-labeled DNA fragments (2 nM) as indicated and no protein, His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM) as indicated, were electrophoresed on an 8% polyacrylamide gel. (B) A longer exposure of the experiment described for panel A allowed detection of faint, shifted complexes with DNA from positions −68 to −16 (lanes 6 to 8). (C) EMSAs were performed as described for panel A with DNA from positions −52 to −16. The arrowhead points to a faint, shifted complex produced by His10-MrpC2.
Binding of MrpC2 to the downstream site does not promote cooperative binding of FruA. A fragment from positions −68 to −16, lacking the upstream MrpC2 site, produced single, faint, shifted complexes with His10-MrpC2 or FruA-His6 but no enhancement of binding by the combination of proteins (Fig. 5A and B, lanes 6 to 8). The pattern is much like that observed for the DNA fragment from positions −80 to −16 bearing the 1-bp change at position −74 (Fig. 4C, lanes 6 to 8). We infer that in both cases, MrpC2 bound to the downstream site and did not bind cooperatively with FruA. We note that FruA-His6 alone bound the fragment from positions −80 to −16 more strongly than the fragment from positions −68 to −16 (Fig. 5A, compare lanes 3 and 7), indicating that DNA upstream of position −68 enhances FruA binding.
A fragment from positions −52 to −16 showed weak binding by His10-MrpC2 (Fig. 5C, lane 2), presumably to the downstream site, and showed no binding by FruA-His6 (Fig. 5C, lane 3). Since FruA bound the 28-bp duplex from positions −57 to −30 (Fig. 4A, lane 11), DNA between positions −57 and −52 is important for FruA binding.
FruA acts positively and the downstream MrpC2 binding site likely acts negatively to regulate fmgD promoter activity.
Within the region bound by FruA (−57 to −30) is a C box sequence at positions −52 to −46 (Fig. 4A) that was shown previously to function as a positive regulatory element for fmgD promoter activity (47). We noticed an imperfect match to the C box consensus sequence at positions −38 to −44 on the other DNA strand (Fig. 6 A) and hypothesized that FruA binds to these inverted repeats as a dimer, since FruA-His6 shifted the 28-bp duplex from positions −57 to −30 to about the same position as a presumed dimer of His10-MrpC2 (32) shifted the 28-bp duplex from positions −80 to −53 (Fig. 4A). Monomers of the two transcription factors are predicted to be similar in size. The sequence from positions −44 to −37 had not been subjected to mutational analysis. Therefore, we tested the effects of two 4-bp changes in this region on fmgD promoter activity. Strikingly, a 4-bp change from CGGG to ATTT at positions −44 to −41 abolished fmgD-lacZ expression, and a 4-bp change from TTGA to GGTC at positions −40 to −37 increased expression about 4-fold (Fig. 6B). These results suggested adjacent or partially overlapping activator and repressor binding sites. We reasoned that binding of MrpC2 to the downstream site might repress transcription, and binding of MrpC2 to the upstream site cooperatively with FruA might activate transcription (Fig. 6C).
FIG. 6.
FruA acts positively and the downstream MrpC2 binding site likely acts negatively to regulate fmgD promoter activity. (A) Sequences within the region bound by FruA. Perfect and imperfect matches to the C box consensus sequence (CAYYCCY [Y means T or C]) are indicated by arrows. (B) Effects of two mutations on fmgD promoter activity. β-Galactosidase-specific activity during development was measured for lacZ fused to fmgD (positions −80 to +382) with no mutation (⧫), a 4-bp mutation at positions −44 to −41 (▪), or a 4-bp mutation at positions −40 to −37 (▴). The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Points show the averages of results from three transformants, and error bars depict one standard deviation of the data for mutant promoter regions. The error bars are too small to be seen in the case of the mutation at positions −44 to −41. For the wild-type promoter region, one transformant was measured in this experiment. (C) Model for regulation of the fmgD promoter. Early in development, MrpC2 binds cooperatively to the upstream and downstream sites, preventing transcription (top). Later in development, C-signaling activates FruA, which binds cooperatively with MrpC2 bound to the upstream site, activating transcription (bottom). (D) Effect of a 4-bp change at positions −44 to −41 on binding of MrpC2 and FruA. EMSAs were performed as described in the Fig. 3 legend, with wild-type or mutant fmgD DNA (2 nM) spanning from positions −80 to −16. A shorter exposure of the result for wild-type DNA (lanes 1 to 4) is shown in Fig. 3. A longer exposure is shown here for comparison with mutant DNA (lanes 5 to 8). (E) Effect of a 4-bp change at positions −40 to −37. EMSAs were performed as described for panel D. (F) A longer exposure of the experiment described for panel E allowed detection of a faint, shifted complex in lane 7 (arrow).
To test the model depicted in Fig. 6C, binding of His10-MrpC2 and FruA-His6 to DNA fragments (−80 to −16) bearing the 4-bp changes was measured in EMSAs. In agreement with the model, the mutation at positions −44 to −41 eliminated binding of FruA-His6 alone, and no enhancement of binding was observed in combination with His10-MrpC2 (Fig. 6D). This result, together with the finding that fmgD-lacZ expression was abolished by the mutation at positions −44 to −41, provides evidence that FruA is a direct activator of fmgD transcription.
The EMSA result for the mutation at positions −40 to −37 was more complex but also consistent with the model. Binding of His10-MrpC2 alone was reduced, and only one shifted complex was observed (Fig. 6E, lane 6), even in a long exposure (Fig. 6F, lane 6), suggesting that the mutation eliminated His10-MrpC2 binding to the downstream site. The mutation also weakened binding of FruA-His6 alone; a very faint shifted complex was observed in a long exposure (Fig. 6F, lane 7). The effects of the mutation on binding of both proteins suggest that the downstream MrpC2 site partially overlaps the FruA site. Importantly, the combination of proteins produced a pattern indicative of cooperative binding (Fig. 6E, lane 8). Despite weakened binding of FruA alone, it can still bind cooperatively with MrpC2 bound to the upstream site. We infer that the 4-fold increase in fmgD-lacZ expression brought about by the mutation at positions −40 to −37 is likely due to loss of MrpC2 binding to the downstream site, consistent with the notion that MrpC2 represses transcription when bound to this site (Fig. 6C). Because the mutation at positions −40 to −37 is adjacent to the promoter −35 region, we cannot exclude the possibility that the mutation enhances RNA polymerase binding that is productive for transcription.
DISCUSSION
The main finding of this work is that fmgD, like fmgA and fmgBC, is subject to combinatorial control by MrpC2 and FruA. The two transcription factors appear to bind cooperatively in all three promoter regions, but the arrangements of binding sites differ. FruA binds upstream of MrpC2 in the fmgA promoter region, whereas MrpC2 binds upstream of FruA in the fmgBC promoter region. In terms of cooperative binding that activates transcription, the fmgD promoter region resembles the fmgBC promoter region, with MrpC2 binding to an upstream site and FruA binding downstream, adjacent to the promoter. However, our results show that MrpC2 also binds to a downstream site that overlaps the fmgD promoter, likely repressing transcription when bound at this position. The downstream MrpC2 site appears to overlap the FruA site, so the two transcription factors may compete for binding, resulting in repression when MrpC2 is bound (Fig. 6C, top) and in activation when FruA is bound (Fig. 6C, bottom). This model would explain why fmgD transcription begins later during development and exhibits greater dependence on C-signaling than fmgA and fmgBC transcription, since C-signaling activates FruA, and a higher concentration of active FruA might be needed to outcompete MrpC2 for binding to its downstream site in the fmgD promoter region.
While fmgD transcription exhibits greater dependence on C-signaling than fmgA and fmgBC transcription, all three promoters depend absolutely on FruA (29, 52) (Fig. 1). This implies that FruA is active to some extent in the absence of C-signaling. In a csgA mutant incapable of C-signaling, developmental expression of fmgA and fmgBC is reduced 2-fold to 4-fold (5, 7, 20). Apparently, the concentration of active FruA is high enough to permit some cooperative binding with MrpC2, partially activating the fmgA and fmgBC promoters. In contrast, developmental expression of fmgD is abolished in a csgA mutant (20). We infer that the concentration of active FruA is insufficient to permit cooperative binding with MrpC2 bound to the upstream site in the fmgD promoter region, perhaps due to MrpC2 bound to the downstream site. A csgA mutant accumulates MrpC and MrpC2 normally (data not shown).
Consistent with the notion that the fmgD promoter region differs from the fmgA and fmgBC promoter regions, ChIP analysis revealed MrpC and/or MrpC2 associated with the fmgD promoter region even in a fruA mutant (Fig. 2C). In contrast, association of MrpC and/or MrpC2 with the fmgA and fmgBC promoter regions required FruA (28, 29). We infer that MrpC and/or MrpC2 can occupy the fmgD promoter region in the absence of FruA due to the presence of a higher-affinity binding site (i.e., the upstream MrpC2 site) and/or due to the presence of two binding sites (to which MrpC2 binds cooperatively; see below).
Comparison of MrpC2 and FruA binding to different DNA fragments suggests that MrpC2 binding to the upstream site in the fmgD promoter region is cooperative with either a second molecule of MrpC2 binding downstream or with FruA binding downstream. A 1-bp change at position −74 in the upstream MrpC2 binding site nearly abolished binding of His10-MrpC2 in vitro (Fig. 4B). The small amount of binding detected in a long exposure likely reflects weak binding of MrpC2 to the downstream site (Fig. 4C). In agreement with this interpretation, His10-MrpC2 bound very weakly to DNA fragments lacking the upstream site (Fig. 5). FruA-His6 bound to DNA with the 1-bp change at position −74, but in combination with His10-MrpC2 there was little or no enhancement of binding (Fig. 4B), and a similar result was seen with a DNA fragment from positions −68 to −16 lacking the upstream MrpC2 site (Fig. 5B). Likewise, a DNA fragment lacking the FruA site due to a mutation at positions −44 to −41 showed little or no enhancement of binding by the combination of proteins (Fig. 6D), and DNA fragments lacking the downstream MrpC2 site due to truncation (Fig. 5A, lane 10) or due to a mutation at positions −40 to −37 (Fig. 6E, lane 6) exhibited considerably less binding by His10-MrpC2 than a DNA fragment from positions 80 to −16 that contains both MrpC2 sites (Fig. 5A, lane 2, and 6E, lane 2). Taken together, the results suggest that MrpC2 binds cooperatively to upstream and downstream sites in the fmgD promoter region, and MrpC2 binds cooperatively to the upstream site, with FruA binding downstream (Fig. 6C). The combination of FruA and MrpC2 produced much more shifted species than either protein alone, indicative of strong cooperative binding.
The sites bound by MrpC2 and FruA in the fmgD promoter region contain sequences that match consensus binding sequences. The upstream MrpC2 site contains the sequence ATGTT(N8)GACCG from positions −76 to −59, and the downstream MrpC2 site contains the sequence GGGTT(N8)AATAA from positions −43 to −26, which match the GTGTC(N8)GACAC consensus sequence (32) at six and five out of 10 positions, respectively. The FruA site contains the sequence TCTCA(N6)CGGG from positions −55 to −41, which matches the consensus sequence for binding of the FruA DNA-binding domain, GGG(C/T)(A/G)(N4-6)(C/T)GGG (48), at six out of nine positions.
Binding of both MrpC2 to its upstream site and FruA to its site appears to be required for fmgD promoter activity. Mutations at positions −63 to −59 or at position −74 that impair binding of MrpC2 to its upstream site (Fig. 4), as well as a mutation at positions −44 to −41 that impairs FruA binding (Fig. 6D), abolish developmental fmgD-lacZ expression (47) (Fig. 6B). Hence, fmgD, like fmgA and fmgBC, is subject to combinatorial control by MrpC2 and FruA. As noted previously (28), the dev operon also appears to utilize this mechanism (S. Mittal, P. Viswanathan, A. Campbell, and L. Kroos, unpublished data), and expression of this operon is crucial for sporulation (4, 45). Also as noted previously (28, 29), at promoters utilizing this mechanism, MrpC2 and FruA occupy a location typical for class I activators, which contact the C-terminal domain of the α subunits of RNA polymerase (1), and both MrpC2 and FruA might make such contacts at each promoter, based on studies of activator pairs at both synthetic (26, 44) and natural (2) promoters.
Our work with fmgD reveals complexity in combinatorial control by MrpC2 and FruA. Our previous work with fmgA and fmgBC revealed flexibility in the arrangement of MrpC2 and FruA binding sites (28, 29). The new feature uncovered here is the presence of a second MrpC2 binding site that we propose mediates repression of fmgD transcription until the concentration of active FruA increases enough to overcome the repression. According to our model, the added complexity delays fmgD expression relative to that of fmgA and fmgBC and makes fmgD expression more dependent on C-signaling (since C-signaling activates FruA). Since cell alignment in the outer domain of the nascent fruiting body is believed to promote C-signaling (reviewed in references 13, 39, and 42), our model predicts that fmgD expression would be localized to the fruiting body outer domain, as has been observed (36). We have speculated that the role of MrpC and MrpC2 in combinatorial control of fmg genes is to signal persistent starvation and prevent MazF-mediated programmed cell death of cells destined to form spores (28). Our work with fmgD suggests that MrpC2 also fine-tunes expression of particular fmg genes via more than one binding site in the promoter region.
Acknowledgments
We are grateful to Sumiko Inouye for providing plasmids, protocols, and antibodies and to Sheenu Mittal for providing purified His10-MrpC2 and FruA-His6 and technical advice.
This research was supported by NSF grant MCB-0744343 and by the Michigan Agricultural Experiment Station.
This article is dedicated to the memory of Jun-seok Lee, whose courage and generosity continue to inspire those he knew.
Footnotes
Published ahead of print on 21 January 2011.
REFERENCES
- 1.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 2.Beatty, C. M., D. F. Browning, S. J. Busby, and A. J. Wolfe. 2003. Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J. Bacteriol. 185:5148-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berleman, J. E., and J. R. Kirby. 2009. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 33:942-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boysen, A., E. Ellehauge, B. Julien, and L. Sogaard-Andersen. 2002. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J. Bacteriol. 184:1540-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandner, J. P., and L. Kroos. 1998. Identification of the Ω4400 regulatory region, a developmental promoter of Myxococcus xanthus. J. Bacteriol. 180:1995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellehauge, E., M. Norregaard-Madsen, and L. Sogaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal coordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30:807-817. [DOI] [PubMed] [Google Scholar]
- 7.Fisseha, M., D. Biran, and L. Kroos. 1999. Identification of the Ω4499 regulatory region controlling developmental expression of a Myxococcus xanthus cytochrome P-450 system. J. Bacteriol. 181:5467-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisseha, M., M. Gloudemans, R. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman, B. S., et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nature Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser, D., M. Robinson, and L. Kroos. 2010. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb. Perspect. Biol. 2:a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser, D., and R. Welch. 2004. Dynamics of fruiting body morphogenesis. J. Bacteriol. 186:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashefi, K., and P. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 18.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S. K., and D. Kaiser. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926-928. [DOI] [PubMed] [Google Scholar]
- 20.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 21.Kroos, L., A. Kuspa, and D. Kaiser. 1990. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J. Bacteriol. 172:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 23.Kuspa, A., L. Kroos, and D. Kaiser. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267-276. [DOI] [PubMed] [Google Scholar]
- 24.Kuspa, A., L. Plamann, and D. Kaiser. 1992. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 174:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuspa, A., L. Plamann, and D. Kaiser. 1992. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 174:7360-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langdon, R. C., and A. Hochschild. 1999. A genetic method for dissecting the mechanism of transcriptional activator synergy by identical activators. Proc. Natl. Acad. Sci. U. S. A. 96:12673-12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobedanz, S., and L. Sogaard-Andersen. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17:2151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittal, S., and L. Kroos. 2009. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc. Natl. Acad. Sci. U. S. A. 106:1965-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittal, S., and L. Kroos. 2009. Combinatorial regulation by a novel arrangement of FruA and MrpC2 transcription factors during Myxococcus xanthus development. J. Bacteriol. 191:2753-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nariya, H., and M. Inouye. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55-66. [DOI] [PubMed] [Google Scholar]
- 31.Nariya, H., and S. Inouye. 2005. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol. Microbiol. 58:367-379. [DOI] [PubMed] [Google Scholar]
- 32.Nariya, H., and S. Inouye. 2006. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol. Microbiol. 60:1205-1217. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 34.Plamann, L., A. Kuspa, and D. Kaiser. 1992. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J. Bacteriol. 174:3311-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rolbetzki, A., M. Ammon, V. Jakovljevic, A. Konovalova, and L. Sogaard-Andersen. 2008. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus. Dev. Cell 15:627-634. [DOI] [PubMed] [Google Scholar]
- 36.Sager, B., and D. Kaiser. 1993. Spatial restriction of cellular differentiation. Genes Dev. 7:1645-1653. [DOI] [PubMed] [Google Scholar]
- 37.Sager, B., and D. Kaiser. 1993. Two cell-density domains within the Myxococcus xanthus fruiting body. Proc. Natl. Acad. Sci. U. S. A. 90:3690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 40.Shimkets, L. J., R. E. Gill, and D. Kaiser. 1983. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc. Natl. Acad. Sci. U. S. A. 80:1406-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singer, M., and D. Kaiser. 1995. Ectopic production of guanosine penta-and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9:1633-1644. [DOI] [PubMed] [Google Scholar]
- 42.Sogaard-Andersen, L., et al. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48:1-8. [DOI] [PubMed] [Google Scholar]
- 43.Sun, H., and W. Shi. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J. Bacteriol. 183:4786-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tebbutt, J., V. A. Rhodius, C. L. Webster, and S. J. Busby. 2002. Architectural requirements for optimal activation by tandem CRP molecules at a class I CRP-dependent promoter. FEMS Microbiol. Lett. 210:55-60. [DOI] [PubMed] [Google Scholar]
- 45.Thony-Meyer, L., and D. Kaiser. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol. 175:7450-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueki, T., and S. Inouye. 2003. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:8782-8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viswanathan, P., and L. Kroos. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 185:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viswanathan, P., T. Ueki, S. Inouye, and L. Kroos. 2007. Combinatorial regulation of genes essential for Myxococcus xanthus development involves a response regulator and a LysR-type regulator. Proc. Natl. Acad. Sci. U. S. A. 104:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitworth, D. E. (ed.). 2008. Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 50.Yoder, D., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4400 promoter region provides insight into developmental gene regulation by C signaling. J. Bacteriol. 186:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoder, D., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J. Bacteriol. 186:3766-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoder-Himes, D., and L. Kroos. 2006. Regulation of the Myxococcus xanthus C-signal-dependent Ω4400 promoter by the essential developmental protein FruA. J. Bacteriol. 188:5167-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]