Abstract
Sixty-four of 85 (75.3%) smear-negative respiratory (n = 78) and nonrespiratory (n = 7) samples with positive cultures of Mycobacterium tuberculosis complex (MTC) were detected by the GeneXpert system using the Xpert MTB/RIF assay (GX). In addition, GX found rpoB mutations in all six of the rifampin-resistant strains detected. The test was negative in 20 culture-negative and 20 nontuberculous culture-positive samples (100% specificity). GX offers high potential for the diagnosis of tuberculosis due to its capacity for direct detection of MTC, its rapidity, and its simplicity.
Tuberculosis (TB) remains one of the biggest health problems in developing and industrialized countries and is associated with high rates of morbidity and mortality (19). The emergence and spread of Mycobacterium tuberculosis strains resistant to multiple drugs represent a serious threat to TB control worldwide (18). Early diagnosis of active TB and detection of multidrug-resistant (MDR) strains are essential to interrupt transmission. Traditionally, acid-fast bacillus (AFB) smear microscopy has been the initial method for diagnosis, due to its speed, simplicity, and low cost. However, its low sensitivity (45 to 80% of positive cultures), as well as the fact that a significant percentage (17%) of bacillus transmission is due to smear-negative pulmonary cases, limits the usefulness of this technique (1). On the other hand, due to the intrinsic slow growth of these microorganisms, mycobacterial culture (the gold standard method for TB diagnosis) takes several weeks to provide microbiological confirmation. Therefore, other techniques are needed to reduce turnaround time to diagnosis (13, 16).
In recent years, direct-detection methodologies, most of them based on nucleic acid amplification, have appeared as potentially useful tools for the rapid diagnosis of TB (7–9, 11–14). Guidelines for their use have been defined and recently updated (6).
A real-time automated integrated system, the GeneXpert system using the Xpert MTB/RIF assay (Cepheid, Sunnyvale, CA) (GX), has recently been developed and evaluated (2, 3, 10). GX integrates DNA extraction, genomic amplification (heminested PCR), semiquantitative detection of M. tuberculosis complex (MTC), and rifampin (RIF) resistance determination in a single cartridge, thus reducing labor time and cross-contamination risk.
The aim of the present study was to evaluate the effectiveness of GX for direct detection of MTC and RIF resistance in smear-negative clinical respiratory and nonrespiratory samples.
One hundred twenty-five smear-negative clinical samples from 122 patients, collected over a 10-year period, were retrospectively studied; 85 samples had MTC-positive cultures, 20 samples (all respiratory) had isolates of nontuberculous mycobacteria (5 rapidly growing mycobacteria, 4 Mycobacterium avium-intracellulare complex isolates, 3 Mycobacterium kansasii isolates, and 8 others), and 20 samples had negative mycobacterial cultures (Table 1). Nonsterile clinical samples were pretreated according to the conventional N-acetyl-l-cysteine-NaOH digestion-decontamination procedure, and sterile specimens were processed directly. Afterwards, 1 ml of these samples was directly frozen at −80°C, and the remaining volume was used for the performance of the following tests: (i) microscopic examination for acid-fast organisms (auramine-rhodamine and Ziehl-Neelsen stains) and (ii) mycobacterial culture using Lowenstein-Jensen and Bactec MGIT 960 (Becton Dickinson, Towson, MD) as solid and liquid media, respectively. The GX assay was performed according to manufacturer's instructions, with 1 ml of the frozen samples. Briefly, the GX assay consists of the inactivation of the sample with NaOH and isopropanol (sample reagent [SR]), at a 1:2 ratio, for 15 min; afterwards, the mixture is introduced into the Xpert MTB/RIF cartridge and then loaded into the Xpert instrument for DNA extraction and amplification of a 192-bp segment of the rpoB gene. The detection consists of the hybridization of the amplicon with five overlapping probes complementary to the rpoB “core” region (81 bp) determining the RIF resistance (9, 15, 17). The cartridge also includes spores of Bacillus globigii as an internal control of the sample processing and the real-time PCR assay.
Table 1.
Sources and distribution of 125 smear-negative clinical samples tested by GX
| Specimen type (total no. of samples) | No. of samples with indicated result by culturea |
||
|---|---|---|---|
| Positive for MTC | Positive for NTMb | Negative | |
| Respiratory specimens | |||
| Sputum (92) | 72 | 19 | 1 |
| Bronchial aspirate (12) | 5 | 1 | 6 |
| Pulmonary biopsy (1) | 1 | ||
| Nonrespiratory specimens | |||
| Pleural fluid (4) | 2 | 2 | |
| Gastric aspirate (5) | 2 | 3 | |
| Urine (2) | 2 | ||
| Stool (1) | 1 | ||
| Cerebrospinal fluid (3) | 3 | ||
| Ascitic fluid (2) | 2 | ||
| Lymph node aspirate (1) | 1 | ||
| Skin biopsy (1) | 1 | ||
| Mammary abscess (1) | 1 | ||
Mycobacterial growth in liquid and/or solid media.
NTM, nontuberculous mycobacteria.
Every MTC isolate was also tested for susceptibility against antituberculous first-line drugs by using the BACTEC 460 radiometric system (Becton Dickinson, Towson, MD). The critical concentration of RIF used was 2 μg/ml. Isolates showing resistance to RIF were further analyzed by sequencing of an internal region of the rpoB gene, which included the rifampin resistance-determining region (RRDR), in order to identify mutations associated with phenotypic resistance (17).
Despite the fact that all samples had been stored frozen (−80°C) for several years (1 to 10 years), GX detected the DNA of MTC, in a total time of 2 h, in 64 of 85 (75.3%) clinical samples with negative smears and positive cultures of MTC. Percentages of GX positivity, according to time of storage of MTC culture-positive respiratory samples, were studied in order to determine whether the prolonged freezing of the specimens had an impact on the result of this technique. No statistical difference in terms of sensitivity was found, i.e., 80.6% (29/36) in samples stored frozen for 5 to 10 years, versus 76.2% (32/42) in those stored frozen for less than 5 years. These results reflect the robustness of GX even under demanding conditions such as the long-term storage of the samples.
As expected, a higher sensitivity (61 of 78 [78.2%]) was found for respiratory specimens (Table 2). However, the actual effectiveness of GX in nonrespiratory samples is difficult to establish due to the low number of samples studied (n = 7), of which three were GX positive (one urine, one stool, and one gastric aspirate sample) and four negative (two pleural fluid, one urine, and one gastric aspirate sample).
Table 2.
GX results for the detection of M. tuberculosis complex in 125 smear-negative clinical samples
| GX resulta | No. of MTC culture-positive specimens |
No. of MTC culture-negative specimens |
Total | Predictive valued (%) | |||
|---|---|---|---|---|---|---|---|
| Respiratory | Nonrespiratory | Positive for NTMb, respiratory | Negative for mycobacteriac |
||||
| Respiratory | Nonrespiratory | ||||||
| Positive | 61 | 3 | 0 | 0 | 0 | 64 | 100 |
| Negative | 17 | 4 | 20 | 9 | 10 | 60 | 65 |
The GX sensitivity was 75.3% (95% confidence interval [95% CI], 64.5 to 83.7%), and the specificity was 100% (95% CI, 88.8 to 100%).
NTM, nontuberculous mycobacteria.
One respiratory culture-negative sample had an invalid result using GX.
The positive predictive value is given for the positive GX results, and the negative predictive value is given for the negative GX results.
Seven of the 85 samples (8.2%) with MTC-positive culture had a RIF-resistant isolate also resistant to isoniazid (MDR strains). GX detected DNA of MTC in six of them and found rpoB mutations in all six GX-positive samples (lack of hybridization of one of the rpoB probes). Mutations in these strains were confirmed by sequencing; all showed a Ser531Leu shift. Although GX can detect the nucleotide substitutions, a susceptibility test is also needed to confirm phenotypic resistance. Nevertheless, mutations in rpoB and phenotypic resistance to RIF are highly correlated in 95 to 98% of cases (17); therefore, GX's high sensitivity for detecting these genotypic variations means that the technique has a high predictive value for the rapid diagnosis of multidrug resistance. Consequently, GX may assist clinicians to provide proper initial treatment of patients with potential MDR TB and thus minimize its spread. As regards the samples with negative MTC culture, no positive results were obtained by GX, showing 100% specificity (Table 2). Only one sample (a gastric aspirate with negative culture for mycobacteria) showed an invalid GX result. In this case, the Bacillus globigii spore internal control was negative, suggesting that the sample was not properly processed for DNA extraction in the cartridge or that it contained PCR inhibitors.
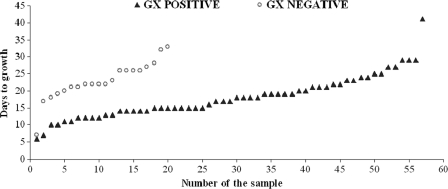
The impact of the bacterial load of the sample (inoculum size) on the GX result was also evaluated. Since the GX assay gives a semiquantitative result when MTC is detected, an initial correlation between this GX result and the time to growth in the liquid medium was established. An inverse association between the median days to growth and the semiquantitative GX result was found: only one sample had a high semiquantitative result (average cycle threshold [CT] value of 15.4), with a median time to growth in liquid medium of 13 days; 8 samples had a medium result (average CT value of 19.6), with a median of 10 days; 26 samples had a low result (average CT value of 25.9), with a median of 15 days; and 29 samples showed a very low result (average CT value of 31.1), with a median of 20 days. Afterwards, the correlation between the time to growth in liquid medium and the qualitative result (positive/negative) of GX was determined. The median time required to grow in liquid medium for GX-positive samples with MTC isolates was statistically lower than that for GX-negative samples (17 versus 22 days; P = 0.003) (Fig. 1). All of these data confirmed the expected association between the inoculum size present in the sample and the result of the technique. However, the global data obtained in the present study indicate that GX has a high sensitivity, since all samples analyzed had a low mycobacterial load.
Fig. 1.
Time to growth of Mycobacterium tuberculosis complex in liquid medium according to GX qualitative result (positive result, n = 57; negative result, n = 20). Eight samples (seven GX positive and one GX negative) were excluded due to negative growth in liquid medium (n = 4), bacterial contamination of the sample (n = 3), or lack of liquid culture (n = 1).
The relatively high cost of GX is an important issue that TB control programs should consider prior to implementation of this assay. Its clinical and epidemiological advantages should be weighed against the resources available in each setting.
In summary, the GX technique has demonstrated a high capacity for detecting MTC and for predicting multidrug resistance in smear-negative clinical samples. Moreover, its rapidity, simplicity, and low laboriousness make the technique a good candidate for routine use in many clinical laboratories whenever the clinical criteria for its application are met (4–6).
Acknowledgments
This study was supported by Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III-FEDER, Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008).
We are grateful to IZASA, S.A., for providing us with the GX reagents.
R. Moure and L. Muñoz received a grant from the Institut d'Investigació Biomèdica de Bellvitge (IDIBELL).
Footnotes
Published ahead of print on 29 December 2010.
REFERENCES
- 1. Behr M. A., et al. 1999. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet 353:444–449 [DOI] [PubMed] [Google Scholar]
- 2. Blakemore R., et al. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boehme C. C., et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Catanzaro A. 1997. What is the appropriate use of the rapid diagnostic tests for tuberculosis? Monaldi Arch. Chest Dis. 52:27–32 [PubMed] [Google Scholar]
- 5. Catanzaro A., et al. 2000. The role of clinical suspicion in evaluating a new diagnostic test for active tuberculosis: results of a multicenter prospective trial. JAMA 283:639–645 [DOI] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention 2009. Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb. Mortal. Wkly. Rep. 58:7–10 [PubMed] [Google Scholar]
- 7. D'Amato R. F., Miller A. 1998. Rapid diagnosis of pulmonary tuberculosis using Roche AMPLICOR Mycobacterium tuberculosis PCR test. Methods Mol. Biol. 92:203–214 [DOI] [PubMed] [Google Scholar]
- 8. Dinnes J., et al. 2007. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol. Assess. 11:1–196 [DOI] [PubMed] [Google Scholar]
- 9. El-Hajj H. H., Marras S. A., Tyagi S., Kramer F. R., Alland D. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 39:4131–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Helb D., et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hillemann D., Rusch-Gerdes S., Richter E. 2006. Application of the Genotype MTBDR assay directly on sputum specimens. Int. J. Tuberc. Lung Dis. 10:1057–1059 [PubMed] [Google Scholar]
- 12. Lemaitre N., et al. 2004. Comparison of the real-time PCR method and the Gen-Probe amplified Mycobacterium tuberculosis direct test for detection of Mycobacterium tuberculosis in pulmonary and nonpulmonary specimens. J. Clin. Microbiol. 42:4307–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moore D. F., Guzman J. A., Mikhail L. T. 2005. Reduction in turnaround time for laboratory diagnosis of pulmonary tuberculosis by routine use of a nucleic acid amplification test. Diagn. Microbiol. Infect. Dis. 52:247–254 [DOI] [PubMed] [Google Scholar]
- 14. Musial C. E., Tice L. S., Stockman L., Roberts G. D. 1988. Identification of mycobacteria from culture by using the Gen-Probe Rapid Diagnostic System for Mycobacterium avium complex and Mycobacterium tuberculosis complex. J. Clin. Microbiol. 26:2120–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piatek A. S., et al. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359–363 [DOI] [PubMed] [Google Scholar]
- 16. Taegtmeyer M., et al. 2008. The clinical impact of nucleic acid amplification tests on the diagnosis and management of tuberculosis in a British hospital. Thorax 63:317–321 [DOI] [PubMed] [Google Scholar]
- 17. Telenti A., et al. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–650 [DOI] [PubMed] [Google Scholar]
- 18. WHO 2008. Anti-tuberculosis drug resistance in the world. Fourth global report. The WHO/IUATLD global project on anti-tuberculosis drug resistance surveillance. World Health Organization, Geneva, Switzerland [Google Scholar]
- 19. WHO 2010. Global tuberculosis control: a short update to the 2009 report. World Health Organization, Geneva, Switzerland [Google Scholar]