Abstract
Campylobacter species, especially Campylobacter jejuni and Campylobacter coli, are a major cause of human bacterial enteritis. Current detection in stools is done essentially by culture on selective and nonselective media with filtration. These methods were compared to 2 molecular biology methods, an in-house real-time PCR and a multiplex PCR named Seeplex Diarrhea ACE Detection, and 3 immunoenzymatic methods, Premier Campy, RidaScreen Campylobacter, and ImmunoCard Stat!Campy. Out of 242 stool specimens tested, 23 (9.5%) fulfilled the positivity criteria, i.e., they were positive by one or both culture methods or, in case of a negative culture, by a positive molecular method and a positive immunoenzymatic method. The striking feature of this study is the low sensitivity of culture, in the range of 60%, in contrast to immunoenzymatic and molecular tests.
INTRODUCTION
The incidence of Campylobacter-associated food poisoning has gradually increased, and the organism is now considered to be the leading cause of bacterial gastroenteritis worldwide (4). These infections can also lead to extraintestinal diseases and severe long-term complications (9). Campylobacter jejuni and Campylobacter coli are the most frequently isolated species in this context and in our experience account for 80% and 16%, respectively, of all the isolates received in our laboratory every year (2a). Campylobacter species are bacteria with a special culture requirement, i.e., a microaerobic environment. During stool processing, the bacteria may have long contact with a normal atmosphere, and in addition, the progressive decrease in oxygen tension when gas-generating kits are used may not favor adequate growth. Furthermore, selective media are commonly used, and the antibiotics incorporated may inhibit certain Campylobacter strains. Anecdotal data have shown that spiral or curved bacteria are sometimes observed on stool smears while Campylobacter growth does not occur. In a recent study, DNA sequences of the Campylobacter genome were detected by a metagenomic analysis, while the standard culture methods were negative (5). In a pilot study using real-time PCR as a diagnostic tool, we also detected more campylobacters than with culture, but without being able to confirm that true positives were detected, since, at that time, we did not use multiple detection methods to establish positivity when culture was negative. Some immunoenzymatic tests have already been commercialized for several years, such as the ProSpecT Campylobacter Microplate Assay (Remel) (8) and the RidaScreen Campylobacter (R-Biopharm, Darmstadt, Germany) evaluated in our study. In this study, we took advantage of the availability of several kits to compare Campylobacter detection by molecular methods (2 PCRs) and by 3 immunoenzymatic methods to the standard culture methods.
MATERIALS AND METHODS
Materials.
From 15 June to 30 October 2009, every stool specimen obtained from a symptomatic patient, i.e., a patient with a gastrointestinal illness, who was hospitalized for less than 48 h at Pellegrin Hospital (Bordeaux, France), was included. Stools were sent to the laboratory at room temperature without transport medium. The fresh, unpreserved stools were tested for culture within 4 h after arriving in the laboratory. The remaining part of the stool samples was then frozen at −80°C. The other tests were performed at the same time, once a week, after the samples were thawed.
Methods.
Two different PCRs were used for molecular diagnosis: (i) the in-house PCR routinely performed in the laboratory, which is a real-time fluorescence resonance energy transfer (FRET) PCR specific for C. jejuni and C. coli, targeting the gyrA gene, followed by a melting-curve analysis to differentiate C. jejuni and C. coli (3), and (ii) Seeplex Diarrhea-B1 ACE Detection (Seegene Inc., South Korea), which is a multiplex PCR based on dual-priming oligonucleotides (DPO) (10). The latter assay also permits the simultaneous amplification of target DNA of Salmonella spp. (Salmonella enterica and Salmonella bongori), Shigella spp. (Shigella flexneri, Shigella boydii, Shigella sonnei, and Shigella dysenteriae), Vibrio spp. (Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus), and Clostridium difficile toxin B.
The 3 different immunoenzymatic methods are all based on the use of specific monoclonal antibodies for a common antigen to C. jejuni and C. coli. The Premier Campy (Meridian Bioscience, Inc., Cincinnati, OH) and the RidaScreen Campylobacter test (R-Biopharm AG, Darmstadt, Germany) have an enzyme-linked immunosorbent assay (ELISA) format, while the ImmunocardStat!Campy (Meridian Bioscience) is an immunochromatographic rapid test.
Culture.
Two different culture methods were used: culture after a filtration step (6) and culture on selective medium without filtration. For the filtration method, a drop of a stool suspension prepared in a brucella broth was deposited on a 0.65-μm Millipore filter (Millipore, Billerica, MA), which was placed on Trypticase soy blood medium. Then, the medium was incubated at 37°C in a microaerobic atmosphere. For culture without filtration, a stool sample was directly inoculated on a Karmali agar plate (Oxoid, Basingstoke, Hampshire, United Kingdom) and incubated at 37°C in a microaerobic atmosphere. Colonies were observed 24 h and 48 h after culture on the selective medium and after 5 days when the filtration method was used. The colonies suspected to be Campylobacter species were confirmed based on motility, Gram staining, and oxidase activity. The hippurate test was used to differentiate C. jejuni from C. coli.
DNA extraction.
Genomic DNA was isolated using the QIAamp DNA stool minikit (Qiagen SA, Courtaboeuf, France) following the manufacturer's instructions.
Real-time PCR.
The PCR and hybridization reactions were performed in glass capillary tubes in a LightCycler thermocycler (Roche Diagnostics, Meylan, France). Each tube contained 7 μl of reaction mixture, including 0.7 μl of FastStart DNA Master Hybridization probe mixture (Roche Diagnostics), 3 mM MgCl2, 0.72 μM (each) forward and reverse primers, 0.2 μM each probe, and 0.7 μl of template DNA. Following initial denaturation at 95°C for 10 min, 50 amplification cycles (95°C for 6 s, 54°C for 12 s, and 72°C for 25 s) were performed, all with a temperature transition rate of 20°C/s. Fluorescence was measured at 640 nm after each cycle. This was followed by a melting program of 95°C for 60 s and 38°C for 50 s at a temperature transition rate of 20°C/s and 80°C for 0 s (hold time) at a rate of 0.1°C/s, with continuous monitoring of the fluorescence. The final step consisted of cooling at 20°C/s to 40°C with a 30-s hold (3).
Seeplex Diarrhea ACE Detection.
The reaction mixture was prepared as follows, For 1 reaction, the mixture contained 4 μl of 5× DB1 PM (primer mixture, containing the primer pairs for 5 pathogens and for the internal control and the template for the internal control), 3 μl of 8-Mop (8-methoxypsoralen) solution to prevent carryover contamination, 10 μl of 2× Multiplex Master Mix containing DNA polymerase, buffer with deoxynucleoside triphosphates (dNTPs) and MgCl2, and stabilizers. The reaction mixture tube was agitated by inverting it 5 times or by quick vortexing. Seventeen microliters of the reaction mixture was dispensed into 0.2-ml PCR tubes. Three microliters of each sample's nucleic acid was added to the reaction mixture tube in order to reach a total reaction volume of 20 μl. The tubes were placed in a preheated (94°C) thermal cycler. Amplifications were performed under the following conditions: 15 min at 94°C, followed by 40 cycles of 50 s at 94°C, 1.5 min at 60°C, and 1.5 min at 72°C, and finally 10 min at 72°C.
The detection step was performed using the ScreenTape System (Seegene Inc., South Korea). All of the reagents are included in multiple minigels, except the loading buffer, which contains the ladder and which is added to the PCR product at 6 μl of loading buffer for 2 μl of PCR product.
Premier Campy.
Fifty microliters of a well-mixed stool sample was transferred to the test tube containing 200 μl of sample diluent, and then the tube was vortexed for 15 s. One hundred microliters of the diluted stool sample was transferred to the microwell plate coated with specific monoclonal antibodies. After 60 min of incubation at room temperature, the microwell plate was washed with the washing buffer 5 times, and 2 drops of enzyme conjugate was added to each microwell and incubated for 30 min at room temperature. The microwell was washed 5 times before 2 drops of substrate was added and incubated for 10 min at room temperature. Then, 2 drops of stop solution was added, and the absorbance was read at 450 to 630 nm. According to the manufacturer, samples with a suspension at an optical density (OD) greater than 0.100 were considered positive.
RidaScreen Campylobacter.
One milliliter of sample dilution buffer was placed in a test tube, and 100 μl of liquid stools or 50 to 100 ng of solid stools was suspended and homogenized by vortexing. Two drops of the suspension was transferred to the well and incubated at room temperature for 60 min. The plate was washed 5 times, and 2 drops of the enzyme conjugate was added to the well and incubated at room temperature for 30 min; then, the plate was washed 5 times. Two drops of substrate was added to each well. The plate was incubated for 15 min at room temperature in the dark, and the reaction was stopped by adding 1 drop of stop reagent to each well. After the wells were mixed, the absorbance was measured at 450 nm. The cutoff corresponded to the manufacturer's recommendation. Samples with an OD 10% above the calculated cutoff were considered positive.
ImmunoCard Stat!Campy.
A small solid stool sample was suspended in 1,400 μl of diluent, or 50 μl of liquid stools was added to 1,400 μl of the sample diluent, depending on the stool consistency. The diluted specimen was vortexed for 15 s, and then 175 μl was transferred to the sampling port of the device. After 20 min of incubation at room temperature, the result was read and validated if the control line band was clearly visible. A positive result showed 2 bands, the control band and a test line band, whereas a negative result showed only the control band.
Definition of a Campylobacter-positive stool sample.
The following criteria were used to define a stool sample positive for Campylobacter: either one or both culture methods were positive, or in the case of a negative culture, a positive molecular method and an immunoenzymatic method were both positive.
RESULTS
Over the 4.5-month period, 242 cases fulfilled the inclusion criteria. Based on the case definition, 23 specimens were positive (9.5%): 16 were positive by culture, and 7 were culture negative but positive by both a molecular method and an immunoenzymatic method. A total of 37 specimens were positive by at least one method. The characteristics of these patients are presented in Table 1. The different combinations are presented in Table 2.
Table 1.
Distribution of the 23 positive cases studied according to age group and gender
| Age group | Total no. | No. male | No. female |
|---|---|---|---|
| Newborns | 1 | 0 | 1 |
| Infants | 3 | 2 | 1 |
| Children | 6 | 6 | 0 |
| Adults | 13 | 7 | 6 |
| Total | 23 | 15 | 8 |
Table 2.
Distribution of the positivity profiles of the cases using different techniques for detection of campylobacters
| No. of cases | Resulta |
||||||
|---|---|---|---|---|---|---|---|
| Culture with filtration | Culture without filtration | Real-time PCR | Seegene PCR | RidaScreen | Premier Campy | Immuno card Stat Campy | |
| Positive by culture (n = 16) | |||||||
| 7 | + | + | + | + | + | + | + |
| 3 | + | − | + | + | + | + | + |
| 1 | + | + | + | − | + | + | + |
| 1 | + | + | + | − | + | + | − |
| 3 | + | + | − | + | + | + | + |
| 1 | − | + | − | − | − | − | − |
| Positive by other tests (n = 7) | |||||||
| 6 | − | − | + | + | + | + | + |
| 1 | − | − | − | + | − | + | + |
| Positive by PCR only (n = 5) | |||||||
| 1 | − | − | + | + | − | − | − |
| 2 | − | − | − | + | − | − | − |
| 2 | − | − | + | − | − | − | − |
| Positive by Ag tests only (n = 9) | |||||||
| 3 | − | − | − | − | − | + | + |
| 3 | − | − | − | − | − | − | + |
| 2 | − | − | − | − | + | + | + |
| 1 | − | − | − | − | + | − | + |
+, positive; −, negative.
All methods were positive in only 7 cases. Culture using filtration allowed the detection of 3 more cases than the selective medium. In contrast, the selective medium permitted the detection of a strain negative by the other methods; it was indeed Arcobacter butzleri. Of the positive culture specimens, very few were missed by Seegene PCR (three), the in-house PCR (four), ImmunoCard (two), and the ELISAs (one).
In 6 out of 7 samples that fulfilled the positivity criteria when culture was negative, all of the methods were positive.
Of the 14 other cases where at least one technique was positive, 9 were positive using immunoenzymatic methods only (6 samples with at least 2 immunoenzymatic methods and 3 samples with 1), and 5 were positive using molecular methods only (1 sample with the 2 molecular methods).
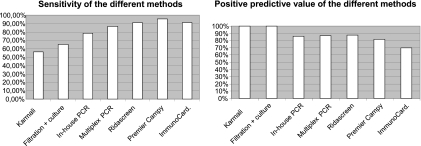
The specificities and negative predictive values (NPV) of the different methods were all in the range of 95 to 100%. The sensitivities and positive predictive values (PPV) of the different methods are presented in Fig. 1. Molecular methods gave 3 false-positive results each and 3 and 5 false-negative results, respectively, with the Seegene PCR and the in-house PCR. The different immunoenzymatic methods gave 2 false-negative results and 3 false-positive results with RidaScreen Campylobacter, 1 false-negative result and 5 false-positive results with Premier Campy, and 2 false-negative results and 9 false-positive results with ImmunocardStat!Campy.
Fig. 1.
Sensitivities and positive predictive values of the different techniques used for Campylobacter detection.
The Campylobacter species identified in 16 positive culture cases were 10 C. jejuni, 4 C. coli, 1 Campylobacter sp., and 1 A. butzleri, while they were 4 C. jejuni and 2 C. coli for the 6 identified by real-time PCR only (Table 3). The immunoenzymatic test did not allow differentiation between C. jejuni and C. coli.
Table 3.
Species identification of the Campylobacter strains and related bacteria detected in the study
| Species | No. identified by culture | No. identified by real-time PCRa | No. identified by culture and real-time PCR | Total |
|---|---|---|---|---|
| C. jejuni | 10 | 12 | 8 | 14 |
| C. coli | 4 | 6 | 4 | 6 |
| A. butzleri | 1 | 0 | 0 | 1 |
| Campylobacter sp. | 1 | 0 | 0 | 1 |
| Total | 16 | 18 | 12 | 22 |
RT-PCR, reverse transcription-PCR.
As stated previously, the Seegene multiplex PCR is also able to detect other pathogenic intestinal bacteria. In this study, we focused only on S. enterica detection: 14 samples were positive both by culture and by the Seegene multiplex PCR, 8 were positive only by the multiplex PCR, and 5 were positive only by culture. There was no sample positive at the same time for S. enterica and for Campylobacter sp.
DISCUSSION
The main result of this study is the lack of sensitivity of culture methods for detecting Campylobacter species in stools. Indeed, if we consider C. jejuni and C. coli, the filtration method could detect only 15 cases (65%) and selective media 13 cases (54.5%). This is in line with previous observations made in our laboratory using real-time PCR, when apparently almost 1/3 of the campylobacters (7/23) were missed by culture, but this study is the first to clearly point out the limited sensitivity of culture.
The study took advantage of the recent availability of diagnostic methods other than culture. The definition of a case positive for Campylobacter when culture is not available is a matter of debate, as for any microorganism in this context. Due to the numerous methods compared in this study, we were able to consider a sample positive when positive results were found simultaneously for at least one molecular method and an immunoenzymatic method. Given that these methods look either at a specific DNA target or at a specific antigen of Campylobacter, we are confident that this is a reasonable choice. When culture is not efficient, another strategy could be to use a molecular method as the reference method. However, we did not take this approach because of the limits frequently highlighted for PCR specificity.
It is striking that in 6 cases out of 7 falling into the category of positive samples, all methods were positive except culture, indicating the weakness of the method. However, the interest of culture is that it allows precise identification of bacteria and testing of their susceptibilities to antibiotics, which are both important elements. Furthermore, all Campylobacter spp. and related bacteria present, e.g., Arcobacter sp. and Helicobacter sp., have a chance to grow (2), provided the medium is kept long enough, which was not the case in this study on selective medium. Another explanation could be the use of only one selective medium in this study while several are commercially available worldwide. However, this selective medium was found to be the best in a previous study performed in our laboratory. The relatively high number of samples positive only by immunoenzymatic methods (n = 9) is also interesting. In 3 cases, the rapid test (ImmunocardStat!Campy) was the only positive method; for the other 6 cases, 2 immunoenzymatic methods were positive, and we can expect that they are based on a different target because they are from different manufacturers. Such results deserve further study to determine if there are cross-reactions with antigens from other bacteria or if they are true Campylobacter antigens. Four samples were positive with one of the 2 molecular methods: 2 with the real-time PCR and 2 with the multiplex PCR. At this time, these 4 results can only be considered false-positive results due to a lack of specificity of the technique. For one sample, both molecular methods were positive whereas the other methods were negative. Other tests were repeated for verification. In the past, a few studies evaluated the accuracy of immunoenzymatic tests but not of PCRs. The RidaScreen Campylobacter tested in this study was used by Tissari and Rautelin on 1,050 stool samples in comparison to culture on a selective medium (7). They found that 98 samples were positive for Campylobacter by culture and 952 were negative (including 46 yielding other enteropathogens). Only 68 out of the 98 positive samples were also positive with the RidaScreen Campylobacter, giving a sensitivity of 69%, and 75 out of 952 negative samples were positive with the RidaScreen Campylobacter. As the authors did not perform other diagnostic tests, they could not verify if they were true positives and concluded there was a lack of specificity of the test (92%). Another test, the ProSpecT Campylobacter Microplate Assay, available for 10 years in the United States and now available in Europe, has also been evaluated only against culture methods, with varying results. For Tolcin et al., the test showed a sensitivity of 96% and a specificity of 99% (8), but for Dediste et al. the sensitivity was only 91.2%, and 25/1,104 supposed false positives (specificity, 97.7%) could not be confirmed by another method (1). Besides sensitivity, specificity, NPV, and PPV, it is interesting to highlight the added values of some of the new tests. The advantage of the multiplex Seegene PCR is its ability to detect other bacteria at the origin of intestinal infections. The main advantage of the ImmunoCardStat!Campy is its rapidity, with the results being obtained in less than 30 min, as well as its convenience in comparison to ELISAs and PCRs.
In conclusion, this study highlights the limits of culture methods in detecting campylobacters in stools. Currently, the best accuracy is obtained by ELISAs, followed by molecular methods. However, the rapid detection by ImmunoCardStat!Campy is attractive, and further studies should be performed to specify the conditions of its use. We may have to change the guidelines for Campylobacter detection in the future.
Footnotes
Published ahead of print on 5 January 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Dediste A., et al. 2003. Evaluation of the ProSpecT Microplate Assay for detection of Campylobacter: a routine laboratory perspective. Clin. Microbiol. Infect. 9:1085–1090 [DOI] [PubMed] [Google Scholar]
- 2. Lastovica A., Allos B. M. 2008. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and Campylobacter coli, p. 123–149 In Nachamkin I., Szymanski C. M., Blaser M. J. (ed.), Campylobacter, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 2a. Mégraud F., Labadi L., Camou C., Lehours P. 2009. A molecular strategy to identify campylobacters and related species. Data from the Campylobacter National Reference Center in France, p. 246 Proceedings of the CHRO, Niigata, Japan [Google Scholar]
- 3. Ménard A., Dachet F., Prouzet-Mauleon V., Oleastro M., Megraud F. 2005. Development of a real-time fluorescence resonance energy transfer PCR to identify the main pathogenic Campylobacter spp. Clin. Microbiol. Infect. 11:281–287 [DOI] [PubMed] [Google Scholar]
- 4. Moore J. E., et al. 2005. Campylobacter. Vet. Res. 36:351–382 [DOI] [PubMed] [Google Scholar]
- 5. Nakamura S., et al. 2008. Metagenomic diagnosis of bacterial infections. Emerg. Infect. Dis. 14:1784–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steele T. W., McDermott S. N. 1984. The use of membrane filters applied directly to the surface of agar plates for the isolation of Campylobacter jejuni from feces. Pathology 16:263–265 [DOI] [PubMed] [Google Scholar]
- 7. Tissari P., Rautelin H. 2007. Evaluation of an enzyme immunoassay-based stool antigen test to detect Campylobacter jejuni and Campylobacter coli. Diagn. Microbiol. Infect. Dis. 58:171–175 [DOI] [PubMed] [Google Scholar]
- 8. Tolcin R., et al. 2000. Evaluation of the Alexon-trend ProSpecT Campylobacter microplate assay. J. Clin. Microbiol. 38:3853–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wassenaar T. M., Blaser M. J. 1999. Pathophysiology of Campylobacter jejuni infections of humans. Microbes Infect. 1:1023–1033 [DOI] [PubMed] [Google Scholar]
- 10. Woo H. Y., et al. 2009. Dual-priming oligonucleotide-based multiplex PCR for the detection of Helicobacter pylori and determination of clarithromycin resistance with gastric biopsy specimens. Helicobacter 14:22–28 [DOI] [PubMed] [Google Scholar]