Abstract
Blastocystis anaerobic parasites are widespread worldwide in the digestive tract of many animal species, including humans. Epidemiological Blastocystis studies are often limited by the poor sensitivity of standard parasitological assays for its detection. This report presents a highly sensitive real-time quantitative PCR (qPCR) assay developed to detect Blastocystis parasites in stool samples. The assay targets a partial sequence of the Blastocystis small ribosomal subunit (SSU) rRNA gene, allowing subtyping (ST) of Blastocystis isolates by direct sequencing of qPCR products. This qPCR method was assessed in a prospective study of 186 patients belonging to two cohorts—a group of 94 immunocompromised patients presenting hematological malignancies and a control group of 92 nonimmunocompromised patients. Direct-light microscopy and xenic in vitro stool culture analysis showed only 29% and 52% sensitivity, respectively, compared to our qPCR assay. Of the 27 (14.5%) Blastocystis-positive patients, 8 (4%) experienced digestive symptoms. No correlation was found between symptomatic patients and immune status, parasite load, or parasite subtypes, although subtyping of all isolates revealed a high (63.0%) prevalence of ST4. Two unexpected avian subtypes were found, i.e., ST6 and ST7, which are frequently isolated in Asia but rarely present in Western countries. In conclusion, this qPCR proved by far the most sensitive of the tested methods and allowed subtype determination by direct sequencing of qPCR products. New diagnostic tools such as the qPCR are essential for evaluating the clinical relevance of Blastocystis subtypes and their role in acute or chronic digestive disorders.
INTRODUCTION
Blastocystis anaerobic parasites are commonly found in the digestive tract in species of several animal groups. It is the most prevalent protozoon found in human fecal samples and has widespread geographic distribution (38). The role of Blastocystis as a human pathogen remains unclear, but studies have associated Blastocystis with acute or chronic digestive disorders (19, 26). Knowledge about its life cycle is equally limited, and various morphological forms, including the cyst and vacuolar forms that are commonly found in feces, have previously been described. The cyst represents the smallest (2 to 5 μm in diameter) form of the organism and is responsible for the environmental dissemination of the parasite. The results of in vivo experiments previously performed with mice have suggested that cysts represent the infectious stage (18, 35, 37), but the most common form detected during parasitological examination of feces is the vacuolar form (2 to 200 μm in diameter) (41, 44). The vacuolar form also represents the major parasitic stage observed in in vitro cultures of Blastocystis parasites.
Recent molecular studies revealed high genetic diversity among Blastocystis strains, identifying 10 different subtypes (ST1 to ST10) that are defined by the sequence of a 600-bp region of the gene encoding the 18S rRNA of the small ribosomal subunit (SSU rRNA) (20, 27, 33). Subtype distribution differs among hosts such as mammals and birds, but recent observations indicate that numerous subtypes previously considered “zoonotic” are also found in humans (1, 20, 38). Studies of zookeepers suggest direct transmission of Blastocystis from animals to humans, while cyst detection in Scottish and Malaysian sewage evidences waterborne transmission (21, 29, 37). Subtypes ST1 to ST9 have been recovered from human fecal samples, with ST3 as the predominant ST followed by ST1, ST2, and ST4. Subtypes ST5 to ST9 are rarely found in human feces (29). Some subtypes have a particular distribution, such as avian subtypes ST6 and ST7, which are more frequently found in Asia and the Middle East (38). Prevalence levels also differ between areas and are higher in developing countries, reaching 60% in Indonesian children (22). Blastocystis species are also widely observed in developed countries, including the United States (23%), France (3%), and the United Kingdom (3.9%) (2, 23, 36). However, prevalence data are largely dependent on the methods used for detection. Direct-light microscopy (DLM) of fecal smears and formol-ether concentration techniques (FECT) greatly underestimate the prevalence of Blastocystis parasites compared to short-term xenic in vitro cultures (XIVC) (14, 30, 36, 45). Nevertheless, XIVC is a time-consuming diagnostic method, and some subtypes present a slow growth rate under culture conditions (30). Several nonquantitative PCR (non-qPCR) assays targeting the SSU rRNA gene to detect and discriminate between Blastocystis isolates directly from stool samples have previously been developed, and Stensvold et al. demonstrated that these molecular methods are more sensitive than XIVC (30). However, nonquantitative PCR approaches do not establish whether there is a correlation between parasite load and clinical features. There is debate over the relationships between parasite abundance and symptoms in blastocystosis. Stenzel and Boreham associated symptomatic patients with a parasite count of over 5 per ×400 field by DLM, whereas Leder et al. found no correlation (13, 34). However, microscopy diagnosis frequently misses cysts, thus underestimating parasite load. In this context, only a qPCR approach could provide clear answers (14, 25, 36, 40). There has been only one recently reported development of a real-time PCR assay targeting a 152-bp fragment of an unknown region of the Blastocystis genome (10). This PCR protocol was able to detect the three major subtypes found in humans, i.e., ST1, ST3, and ST4, and the lower limit of detection was 760 Blastocystis parasites per 100 mg of stools, but it was applied to stool samples from only 3 patients. Thus, there is a real need for highly sensitive quantitative tools validated for all subtypes in order to gain insights into the physiopathology of Blastocystis infections.
Blastocystosis is generally associated with nonspecific symptoms such as diarrhea and/or abdominal pain (3, 13). There are reports of acute gastroenteritis and cutaneous disorders in some cases (8, 15), while other studies have indicated that blastocystosis may be correlated to chronic symptoms. There is evidence that Blastocystis species are associated with irritable bowel syndrome (IBS) (7, 42). Numerous studies have tackled the pathogenic ambivalence of Blastocystis. A majority of these data focused on parasite factors in an attempt to correlate parasite density and/or subtype with pathogenic power, with few studies addressing host factors. A high prevalence of Blastocystis in HIV patients was previously found to be associated with clinical relevance in severely immunocompromised subjects (3, 9). There are case reports of blastocystosis from other immunocompromised patients, and one study focused on patients suffering from hematological malignancies (HM), but it was limited to symptomatic patients (24, 39).
This paper reports development of a highly sensitive and real-time qPCR assay targeting a region of the Blastocystis small subunit rRNA (SSU rRNA) gene to detect Blastocystis parasites in stool samples. Direct sequencing of PCR products was then used to discriminate between subtypes. This qPCR method was validated in a prospective epidemiological study on two cohorts—one group of patients with hematological malignancies and one control group of nonimmunocompromised patients.
MATERIALS AND METHODS
Clinical specimens.
Stool samples from 186 patients were collected at the Diagnostic Parasitology laboratory at the Clermont-Ferrand Hospital complex (France), in a prospective study performed from September 2009 to April 2010. Among the 186 patients, 94 (52 males and 42 females; mean age, 55.8 ± 14.4 years) were hospitalized for HM. For HM patients, blood cell counts of polymorphonuclear neutrophils and lymphocytes were recorded on the day corresponding to stool sampling. The control group, which included 92 nonimmunocompromised patients, had an age range (mean age, 59.4 ± 17.4 years) and a sex ratio (50 males and 42 females) effectively equivalent to those of the HM group. All stool samples were analyzed within 12 h by direct-light microscopy (DLM) and xenic in vitro culture. Aliquots of each stool were also stored at −20°C before subsequent DNA extraction for qPCR assays.
XIVC.
A 500-mg aliquot of each stool sample was cultured at 37°C in Jones medium (supplemented with 10% horse serum) in an anaerobic chamber (11). Antibiotics (100 IU/ml penicillin and 100 μg/ml streptomycin; Invitrogen, France) were added as previously described (28). Cultures were examined using direct-light microscopy after 72 h.
DLM.
Both fecal smears and XIVC were examined using an Olympus BX 40F4 microscope under low-power (magnification of ×10) and high-power (magnification of ×40) lenses. Parasite density in fecal smears was evaluated as the number of vacuolar forms per field (magnification, ×40). Positive samples were classified into 3 categories: under 5 parasites per field (“<5”), 5 to 10 parasites per field (“5 to 10”), and over 10 parasites per field (“>10”).
Axenic in vitro culture.
An axenic culture of Blastocystis subtype 7 (ST7) was maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% horse serum (Invitrogen, France) and cultured in an anaerobic chamber at 37°C. The strain was obtained from a stool sample from a Singapore patient and was kindly provided by K. Tan (National University of Singapore).
DNA extraction.
Total DNA from 200 mg of each stool sample was extracted with a DNA stool minikit (Qiagen, France) and eluted in a final volume of 200 μl according to the manufacturer's recommendations. DNA extracts were stored at −20°C until qPCR analysis.
DNA samples for qPCR controls.
DNA controls representing all subtypes found in humans were used to validate our qPCR method. DNA samples from published isolates of ST1 (LS1 strain), ST2 (LS9 strain), ST3 (LS22 strain), and ST4 (LS21 strain) were kindly provided by E. Viscogliosi (Institut Pasteur, Lille, France) (28). DNA samples from rare subtypes (ST5, ST6, ST8, and ST9) were kindly provided by C. Rune Stensvold (Statens Serum Institut, Copenhagen, Denmark). For ST7, we used DNA extract from our axenic culture. We checked the absence of aspecific amplification due to the main human intestinal parasites by the use of DNA from Giardia intestinalis, Entamoeba histolytica-Entamoeba dispar, Entamoeba coli, and Endolimax nana.
Primer design.
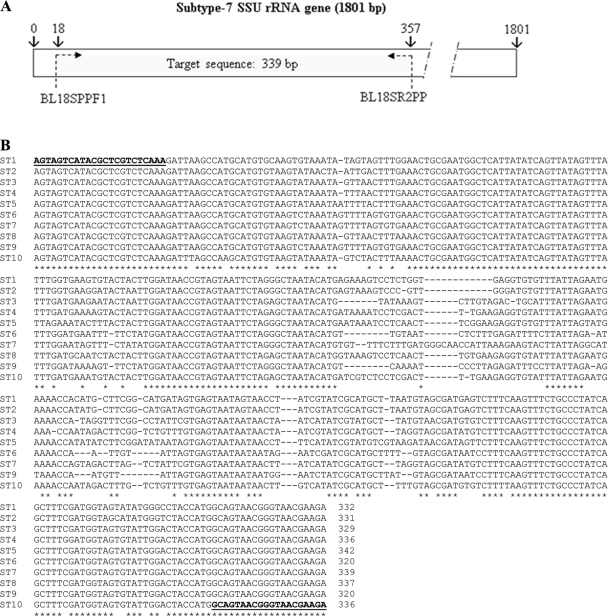
An alignment of the SSU rRNA subtype gene sequences from ST1 to ST10 was performed using ClustalW2 software (http://www.ebi.ac.uk/Tools/clustalw2/index.html) to determine a target sequence for sequencing-enabled subtyping. A set of primers was selected in conserved regions flanking a nonconserved region of the SSU rRNA gene for the 10 subtypes by the use of Primer3 software (http://frodo.wi.mit.edu/primer3/). Primer pair BL18SPPF1 (5′-AGTAGTCATACGCTCGTCTCAAA-3′) and BL18SR2PP (5′-TCTTCGTTACCCGTTACTGC-3′) amplified a DNA fragment of 320 to 342 bp, depending on the subtype. Target sequence identity among the 10 subtypes ranged from 82 to 95%.
qPCR assay.
qPCR amplifications were performed using a Rotor-Gene 6000 system (Corbett Life Science, France) and a 20-μl reaction volume containing 2 μl of the reagent from an LC-FastStart DNA Master SYBR green kit (Roche Diagnostics, France), 3.65 mM MgCl2, 0.5 μM each primer (BL18SPPF1 and BL18SR2PP), and 2 μl of DNA extract (equivalent to 2 mg of stool). After denaturing at 95°C for 5 min was performed, 45 cycles were run with 5 s of denaturation at 95°C, 10 s of annealing at 68°C, and 15 s of extension at 72°C. To establish the coefficient of correlation of the PCR assay, 10-fold dilutions of genomic DNA (total DNA extract from a sample containing 107 Blastocystis parasites/ml) from an axenic in vitro culture of subtype 7 (ST7) were realized. Each dilution was run in 4 replicate experiments. For data analysis, the melting curve and cycle threshold (CT) values were selected as the evaluation parameters. The readout of the reaction with melting temperatures of 78°C to 85°C, a dF/dT fluorescence value above 2, and a CT value below the CT of the detection limit was used to validate a positive reaction. All clinical sample extracts were controlled for PCR inhibitors: 1.5 μl of the DNA extracts was mixed with 0.5 μl of extract from positive controls and analyzed by qPCR. Samples with a CT value greater than the CT value of the positive control alone (2-fold diluted) were subjected to repeat testing in a qPCR after a 10-fold dilution.
Lower limit of detection and repeatability and reproducibility assays.
To establish external standard curves for the quantification of Blastocystis and to determine the limit of detection of our qPCR assay, we ran calibration range analysis of Blastocystis (ST7 from the axenic in vitro culture) in Blastocystis-negative stool samples. Ten-fold dilutions of Blastocystis consisting of 107 to 101 vacuolar forms per gram of stools were realized. Three range points (106, 104, and 102 Blastocystis/g) were tested 8 times in the same experiment to validate repeatability. The same samples were tested once a day for 5 days to determine reproducibility. To analyze the reproducibility and repeatability of the qPCR assay, both intra- and interassay coefficients of variation (CV) were assessed. CV values were calculated as the standard deviation “σ” divided by the mean “μ” (CV = σ/μ).
Sensitivity and specificity of the qPCRs.
The sensitivity and specificity of the qPCRs were calculated for all 186 fecal samples from the clinical study and compared to XIVC and DLM results. Sensitivity was determined as the number of true positives divided by the sum of true positives and false negatives. Specificity was determined as the number of true negatives divided by the sum of true negatives and false positives.
Blastocystis subtyping.
Subtyping was performed by direct qPCR product sequencing after purification with a Nucleospin extract II kit (Macherey-Nagel, France). Sequencing was performed by Eurofins MWG Operon (Germany) using the same primers as those used for PCR (BL18SPPF1 and BL18SR2PP). Sequences were analyzed using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/). Subtypes were assigned according to the recent classification method by Stensvold et al. for a query coverage > 98% with exact match or identity > 98% (30).
Other pathogens.
Other parasites were detected by direct-light microscopy of fecal smears, and stool samples were concentrated using merthiolate iodine formaldehyde (MIF) and Parasitochrome Plus (Bailenger method; Biomedical diagnostics, France). Positive stool samples from symptomatic patients were screened to detect viral and bacterial pathogens. Major viruses responsible for gastroenteritidis were screened, including Norovirus strains (by using the Rida Quick Norovirus test [R-Biopharm, France]) and Rotavirus and Adenovirus strains (by using the Rida Quick Rotavirus/Adenovirus Combi test [R-Biopharm, France]). Conventional bacteriological media were used to detect Salmonella, Shigella, Campylobacter, and Clostridium difficile species.
Statistical analysis.
Except for the variables needing a two-tailed Fischer exact test, the data were analyzed using Epi-Info version 6.02 software and the χ2 test.
RESULTS
Real-time qPCR development.
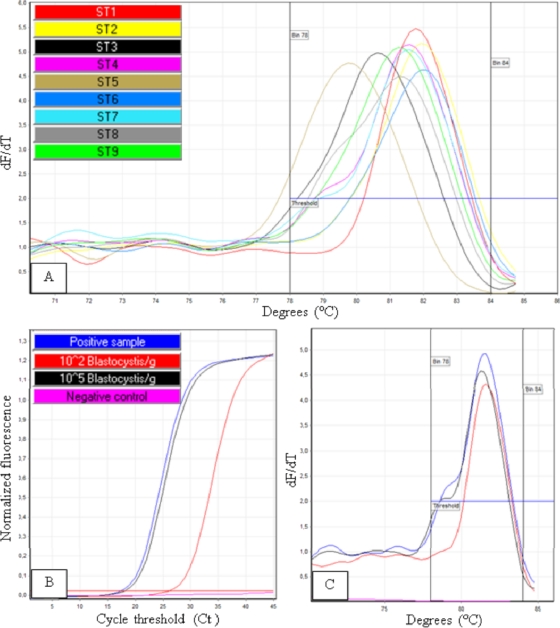
A qPCR assay targeting a region of the Blastocystis SSU rRNA gene was developed to detect and quantify subtypes ST1 to ST10 directly from human stool specimens. The amplified DNA fragments were 320 to 342 bp in size, depending on subtype (Fig. 1). As shown in Fig. 2A, DNAs from the 9 subtypes (ST1 to ST9) found in human feces were successfully amplified, with melting temperatures between 78°C and 85°C, depending on subtype. The criteria used to define a positive reaction were a melting temperature of 78°C to 85°C and a dF/dT fluorescence value above 2 (Fig. 2B and C). A good negative regression was observed in comparisons of 10-fold dilutions of Blastocystis ST7 DNA extract and CT values (R2 = 0.99). PCR efficiency was 0.87. With an extraction range of live parasites in stool samples from 107 to 10 Blastocystis parasites/g, it was possible to detect as few as 102 Blastocystis parasites/g of stools. The standard curve from the extraction range was then used to quantify parasite load in the stool samples.
Fig. 1.
(A) Localization of the target sequence in the SSU rRNA gene for subtype 7. (B) ClustalW alignment of the target sequence (SSU rRNA) from subtype ST1 to ST10. Primers (BL18SPPF1 and BL18SPPR2) are indicated in bold and underlined. The target sequence length ranges from 320 to 342 bp, and sequence identity among the 10 subtypes ranges from 82% to 95%.
Fig. 2.
(A) Subtype DNA samples from ST1 to ST9 were subjected to the qPCR assay. Melting peaks for these subtypes were between 78°C and 84°C. (B and C) Readouts of the reaction with a melting temperature of 78°C to 84°C, dF/dT fluorescence above 2, and CT value below the CT of the limit of detection were used to indicate a positive result.
Both intra- and interassay CV values were assessed to analyze the reproducibility and repeatability of the qPCR assay. Eight replicates of 106, 104, and 102 Blastocystis/g were analyzed in a single run. The CT intra-assay CV values for the replicate experiments were 0.47%, 0.55%, and 0.47% for the 106, 104, and 102 concentrations, respectively. In addition, 5 runs of the 3 concentrations were performed at different days. The CT interassay CV values for the range points 106, 104, and 102 Blastocystis/g were 0.82%, 0.50%, and 1.06%, respectively.
Prospective study with two patient cohorts.
Stool samples from 186 patients were collected in a prospective study performed from September 2009 to April 2010. Among the 186 patients, 94 presented hematological malignancies (HM) and 92 were nonimmunocompromised (control group). The qPCR assay gave an overall prevalence of 27 (14.5%) Blastocystis-positive patients among the 186. The mean of the ages of Blastocystis-positive patients was 57.2 ± 16.4 years (57.7 ± 16.0 years in Blastocystis-negative patients), with a female-to-male gender ratio of 1.45 (0.82 in Blastocystis-negative patients). The prevalence values for the 2 cohorts were 16% (15 of 94) in the HM group and 13% (12 of 92) in the control group. For each Blastocystis-positive sample, parasite density was quantified using qPCR. Parasite counts ranged from <102 to >107 Blastocystis/g of stool sample (Table 1). Among the 27 Blastocystis-positive patients, two were coinfected by Giardia intestinalis. Of the 25 patients infected only with Blastocystis parasites, 8 (4 from the HM group and 4 controls) experienced overt symptoms. Bacteriology and virology assay results were negative for these 8 patients, 7 of whom had diarrhea episodes of liquid (n = 3), soft (n = 1), mucous (n = 1), or normal (n = 2) stools on the day of sampling. One patient had normal stools associated with constipation. Of the three Blastocystis-positive patients with diarrhea, one had weight loss, two had abdominal pain, and one also had anorexia. Three Blastocystis-positive patients were on antibiotherapy. No correlation was found between parasite count and clinical expression of symptoms.
Table 1.
Clinical and biological data from Blastocystis-positive patients
| Patient no. | Age (yr) | Sexa | Hematological malignancies | Blood cell count (cells/mm3) |
Symptom(s) | Antibiotherapy | Stool | Other pathogen | Result by Blastocystis diagnostic method: |
STg | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphonuclear neutrophilsb | Lymphocytesc | DLMd | XIVCe | qPCRf | |||||||||
| 4 | 47 | F | Acute myeloid leukemia 2 | 480 | 500 | Diarrhea, abdominal pain | No | Liquid | G. intestinalis | >10 | P | 4.85 × 104 | 4 |
| 21 | 57 | F | Myeloma | 2,040 | 40 | /i | No | Hard | / | N | P | 7.15 × 105 | 4 |
| 25 | 60 | M | Acute myeloid leukemia 2 | 1,690 | 650 | / | No | Soft | / | 5–10 | P | >107 | 7 |
| 28h | 54 | F | Marginal-zone lymphoma | 3,090 | 1,640 | Diarrhea, wt loss | No | Liquid | / | N | N | 7.51 × 105 | 4 |
| 30 | 71 | F | High-grade lymphoma | 2,520 | 1,810 | / | No | Soft | / | N | N | 2.42 × 102 | 4 |
| 34h | 30 | F | Pelvic mesenchymal tumor | 4,470 | 440 | Diarrhea, abdominal pain, anorexia | No | Soft | / | N | P | >107 | 3 |
| 35h | 67 | M | Acute myeloid leukemia | 140 | 920 | Diarrhea | No | Liquid | / | N | N | <102 | 3 |
| 44 | 74 | M | Follicle center lymphoma | 8,730 | 640 | / | yes | Normal | / | N | N | >107 | 4 |
| 60h | 78 | F | Acute myeloid leukemia 2 | 1,960 | 80 | Constipation | yes | Normal | / | N | N | 4.62 × 104 | 4 |
| 80 | 58 | F | Aplasia | 0 | 1,230 | / | No | Normal | / | N | N | >107 | 4 |
| 81 | 68 | M | Non-Hodgkin's lymphoma | 4,670 | 4,080 | / | No | Soft | / | N | P | 1.07 × 106 | 4 |
| 89 | 72 | M | Acute myeloid leukemia | 7,240 | 700 | / | No | Normal | / | N | N | 1.79 × 102 | 4 |
| 91 | 35 | F | Acute leukemia | 0 | 0 | / | yes | Normal | / | <5 | P | 1.18 × 106 | 3 |
| 92 | 53 | F | Uterine lymphoma | 6,650 | 640 | / | No | Normal | / | 5–10 | P | 7.17 × 105 | 6 |
| 93 | 33 | F | Acute myeloid leukemia 4 | 1,190 | 560 | / | No | Hard | / | N | N | <102 | 4 |
| T13 | 59 | F | / | NDj | ND | / | No | Soft | / | <5 | N | 1.56 × 106 | 4 |
| T17 | 39 | F | / | ND | ND | / | No | Hard | G. intestinalis | <5 | P | >107 | 1 |
| T18 | 32 | F | / | ND | ND | / | No | Soft | / | N | N | 1.68 × 102 | 2 |
| T25 | 46 | M | / | ND | ND | / | No | Soft | / | N | P | 2.22 × 106 | 3 |
| T28 | 89 | F | / | ND | ND | / | No | Soft | / | N | N | 9.82 × 102 | 7 |
| T63 | 50 | M | / | ND | ND | / | No | Soft | / | N | P | >107 | 4 |
| T64 | 45 | M | / | ND | ND | / | No | Soft | / | >10 | P | >107 | 4 |
| T77h | 71 | M | / | ND | ND | Diarrhea | Yes | Normal | / | >10 | P | >107 | 7 |
| T79h | 56 | M | / | ND | ND | Diarrhea | Yes | Mucous | / | N | N | <102 | 4 |
| T85h | 82 | F | / | ND | ND | Diarrhea, abdominal pain | No | Normal | / | N | N | 5.79 × 106 | 4 |
| T87h | 76 | M | / | ND | ND | Diarrhea | No | Liquid | / | N | N | <102 | 4 |
| T90 | 43 | F | / | ND | ND | / | No | Hard | / | N | P | 9.64 × 106 | 4 |
M, male; F, female (F).
Normal values, 1,500 to 4,000 cells/mm3.
Normal values, 2,000 to 4,000 cells/mm3.
Parasite count per field (×400) by direct-light microscopy (DLM).
Results of xenic in vitro culture (XVIC). P, positive; N, negative.
Parasite count per gram of stool by qPCR.
ST, subtype.
Patient presenting with blastocystosis.
/, absence.
ND, not determined.
All the PCR-positive samples were then sequenced to discriminate between subtypes and determine subtype prevalences in both the HM and control groups (Table 2). Among the 27 Blastocystis-positive samples, 15 were from the HM group and 12 were from the control group. Six different subtypes (ST1, -2, -3, -4, -6, and -7) were determined by direct sequencing of qPCR products. ST3, ST4, and ST7 were identified in both groups, with ST4 being the most predominant (17/27), followed by ST3 (4/27) and ST7 (3/27). In contrast, ST6 was found only in the HM group (1/15), while ST1 and ST2 were found only in the nonimmunocompromised group (1/12 for both). Considering symptomatic patients with no other pathogens identified (8/27; Table 2), ST4 still proved the most dominant (5/8), followed by ST3 (2/8) and ST7 (1/8).
Table 2.
Subtype distribution of Blastocystis isolates collected from both the immunocompromised hematological malignancy group and nonimmunocompromised control group patients as determined using the qPCR assay
| Group | % of isolates of indicated subtype (no. of isolates) |
|||||
|---|---|---|---|---|---|---|
| ST1 | ST2 | ST3 | ST4 | ST6 | ST7 | |
| HMa | 20.0 (3) | 66.7 (10) | 6.7 (1) | 6.7 (1) | ||
| Control | 8.3 (1) | 8.3 (1) | 8.3 (1) | 58.3 (7) | 16.7 (2) | |
| Total | 3.7 (1) | 3.7 (1) | 14.8 (4) | 63.0 (17) | 3.7 (1) | 11.1 (3) |
HM, hematological malignancy.
Comparison of real-time qPCR with microscopy and culture methods.
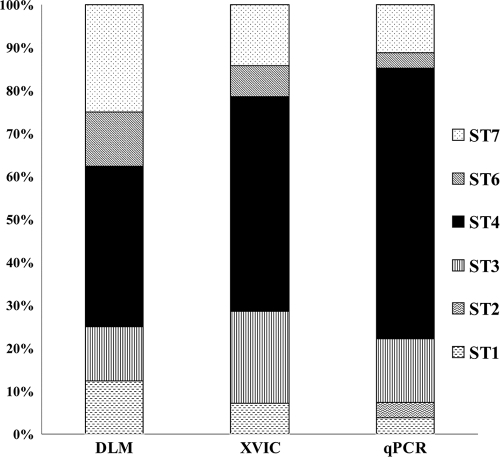
We compared our qPCR assay with conventional diagnostic methods, including direct-light microscopy (DLM) and xenic in vitro culture (XIVC) of fresh stool samples. Only 8 Blastocystis-positive samples were detected by DLM and 14 by XIVC (Table 1), whereas the qPCR assay detected 27 Blastocystis-positive samples (confirmed by sequencing of amplified products), making it by far the most sensitive method. The qPCR assay was then used as the “gold standard” to calculate the sensitivity of both XIVC (52%) and DLM (29%). The limit of detection of qPCR was established as 102 Blastocystis parasites/g of stools, but we were unable to determine the limit of detection for XIVC. Indeed, XIVC did not detect any stool containing less than 4.85 × 104 Blastocystis parasites/g of stools, but it also failed to return a positive result for four samples containing more parasites than this threshold, i.e., one containing 7.51 × 105 Blastocystis parasites/g of stools, one containing 5.79 × 106 Blastocystis parasites/g, and two others containing >107Blastocystis parasites/g as determined by qPCR (patients 28, T85, and 44 and 80, respectively; see Table 1). We identified 8 false-positive samples using our qPCR by sequencing the PCR product (specificity = 95.0%), whereas the specificities of both the DLM diagnostic method and the XIVC diagnostic method were 100%. Cross-comparison between diagnostic methods revealed the impact of each method on subtype distribution. Indeed, the prevalence of ST4 was 38% by the DLM method, rising to 50% with XVIC and 63% with our qPCR diagnostic method (Fig. 3).
Fig. 3.
Distribution of the Blastocystis subtypes from our prospective study according to diagnostic method used. Eight samples tested positive with DLM versus 14 with XIVC and 27 with qPCR. The prevalence of subtype 4 determined with DLM was just 38%, increasing to 50% with XVIC and to 63% with qPCR.
DISCUSSION
Highly sensitive tools are needed to study the epidemiological features of blastocystosis. Until now, short-term xenic in vitro culture (XIVC) has been widely considered the gold standard method. Several studies have shown that XIVC is more efficient than DLM or formol-ether concentration (FECT) in detection of Blastocystis parasites in stool samples (14, 40, 45). Our study confirmed the poor (57%) sensitivity of DLM compared to XIV (previously reported by Leelayoova et al. and Termmathurapoj et al.; 14, 40). The superiority of XIVC is likely due to multiplication of the parasites in culture and to the detection of small forms such as cysts after excystation. Cysts are common in stools, with a prevalence rate of up to 28.5% in stool samples from Blastocystis-positive patients, but escape diagnosis due to their small size, resulting in significant underestimation of prevalence (14, 25, 36, 40). In our study, the overall prevalence according to DLM was only 4%, a result similar to the 3% found by Pinel et al. in a study performed with 2,581 patients in another region of France in 1999 (23). However, the prevalence reported by Pinel et al. was probably greatly underestimated, since a prevalence of 14.5% was obtained with our qPCR assay.
qPCR can detect all forms of Blastocystis parasites, including cysts, and allows quantification of the parasite. We confirmed that the poor sensitivity of DLM could be explained by the lack of detection of cysts, as was the case with patient 34 (see Table 1), who was found to harbor >107 Blastocystis/g of stool sample by qPCR whereas the corresponding DLM result was negative. XIVC was also able to detect cysts; however, compared to our qPCR, its sensitivity was only 52% and it failed to give a positive result for 3 samples containing more than 106 Blastocystis/g of stool. Stensvold et al. (30) compared their PCR assay to XIVC and concluded that the sensitivity of XIVC was 89%. They suggested that some fecal samples did not contain viable Blastocystis or that cultured organisms were so inconspicuous that the XIVC analysis missed them (30). We also demonstrated that our qPCR assay was more sensitive than XIVC: among the 13 qPCR-positive/XIVC-negative samples, 8 contained fewer than 103 Blastocystis/g of stool sample, which was near the lower limit of detection of our qPCR assay (102 Blastocystis/g). The overall prevalence in our study was 14.5%, but only one stool sample was tested per patient. The diagnostic procedure could probably be made more sensitive by sequential analysis of 2 to 3 stool samples, as is routinely done by laboratories running standard parasitological assays. Multiple assays could also be valuable for optimizing the quantification of parasite burden for each patient. To improve the sensitivity of qPCR, it is also necessary to control the absence of PCR inhibitors. Indeed, inhibitors were detected in 16 samples, and 1 of them gave a positive result after 10-fold dilution of the corresponding DNA extract. One study developed a qPCR assay in which the limit of detection was down to 760 Blastocystis per 100 mg of stool sample, detecting a 152-bp fragment from an unknown region (10). That assay, which was tested with ATCC strains (including ST1, ST3, and ST4 strains) and applied to stool samples from 3 patients, highlighted the difficulty of developing a qPCR assay that allows detection of all Blastocystis subtypes due to the broad genetic variability involved. Our qPCR assay was designed to allow detection of the 10 subtypes of Blastocystis, and we checked its ability to detect Blastocystis from DNA extracts of typed isolates of the 9 subtypes (ST1 to ST9; see Fig. 2A) already found in human stools (38). As stated above, the specificity of our SYBR green-based qPCR assay was 95%. Specificity would be enhanced by using probe-based qPCR assays. However, the only sequence available for all the subtypes was the SSU rRNA sequence, and the extensive genetic variability made it impossible for us to design probe-based qPCR assays. We chose to use this high variability to set up primers amplifying a variable region of the SSU rRNA gene among the 10 subtypes, thus allowing subtyping by direct sequencing of qPCR products.
Interestingly, sequencing of Blastocystis-positive samples revealed that ST4 was the most prevalent subtype (63.0%), followed by ST3 (14.8%) and ST7 (11.1%) and then ST1, ST2, and ST6 (3.7% each). ST3 is usually the major subtype found in humans, whereas ST4 is a minor subtype found in rodents (38). Only one study in Spain reported a very high prevalence of ST4 associated with symptomatic patients (4). Although the lack of a significant association of subtype with disease may have been due to the small number of Blastocystis-positive patients in our study, we observed that patients carrying ST4 were no more subject to symptomatic infections than those carrying another ST. The significance of the high prevalence of ST4 remains unclear, and there is a need for molecular studies to be performed on potential reservoirs in the same area. We identified 3 patients carrying ST7 and one carrying ST6, which was an unexpected finding, given that these avian STs are rarely found in Western countries (38). Our results differ significantly from recently published French data reporting ST3 (53.5%) as the most prevalent ST, followed by ST1 (25.6%), ST4 and ST2 (9.3%), and ST7 (2.3%) (28). These differences could be due to the methodology used to isolate Blastocystis, as shown in Fig. 3. Indeed, the distribution of subtypes was dependent on the diagnostic method used, as DLM and XVIC failed to detect a large fraction of ST4 carriers that were detected by qPCR. Our report is the first to clearly demonstrate the impact of detection methods on relative distributions of subtypes. It can also be hypothesized that ST distribution differs between areas depending on reservoirs and modes of contamination, and even in the same country, as shown on a large scale in China (16). Data published by Souppart et al. (28) came from patients hospitalized in Lille, a big city in northern France, whereas animal reservoirs and food habits may differ significantly between the two areas studied, with Clermont-Ferrand hospitals probably receiving more “rural area” patients than Lille hospitals. Finally, subtyping by direct sequencing of qPCR products did not allow detection of mixed infections by several STs known to occur only in 2.6% to 14.6% of positive samples (17, 43). The results of recent work in mouse studies suggest that Blastocystis species are able to infiltrate the digestive epithelium, invading the muscle layer (5). Nevertheless, the pathogenic potential of Blastocystis remains unclear, and relationships between symptoms and parasite density and/or subtype have never been unambiguously demonstrated. Stenzel and Boreham correlated symptomatic patients with a parasite count higher than 5 per ×400 field by DLM, whereas Leder et al. did not find any correlation (13, 34). We did not find any correlation between parasite load determined by qPCR (ranging from <102 to >107 Blastocystis parasites/g of stools from symptomatic patients) and symptoms. This suggests that parasite shedding in stools does not reflect interactions with the host in the digestive tract. Also, qPCR was able to quantify both cyst and vacuolar forms, and it is possible that the vacuolar form gives a better reflection of pathogenicity, as suggested by Elwakil and Hewedi (5).
In our study, Giardia intestinalis was the only other pathogen found to be associated with Blastocystis parasites. We found no patients positive for Dientamoeba fragilis, which is frequently associated with Blastocystis infections (32). This may be due to poor sensitivity of standard parasitological assays (31). Eight symptomatic patients—4 from the control group (nonimmunocompromised) and 4 from the immunocompromised group presenting hematological malignancies (HM) group–were shown to be infected by only Blastocystis parasites. Among those 8 symptomatic patients, 5 were subtyped as harboring ST4, 2 as harboring ST3, and 1 as harboring ST7. Our study was initially designed to integrate host factors by comparing HM patients to controls. The few clinical data available on Blastocystis infections in immunocompromised patients essentially concern HIV patients. An Ethiopian study found a higher prevalence of Blastocystis parasites in a HIV cohort (9), while Kurniawan et al. (12) studied 318 HIV patients before antiretroviral therapy was administered and found that Blastocystis species were more prevalent when the lymphocyte CD4 count was below 200 CD4/mm3, and Cirioni et al. associated the presence of Blastocystis with symptoms at a CD4 count of ≤200 CD4/mm3 (3). Other categories of immunocompromised patients, such as renal transplant recipients, can develop blastocystosis (24). HM patients are at risk for opportunistic infections, especially before and in the weeks following stem cell transplantation. Gosh et al. reported a case of exacerbated intestinal symptoms in a patient with digestive graft versus host disease (6). Taşova et al. studied the prevalence of Blastocystis in HM patients with gastrointestinal complaints (GC) compared to the prevalence in a group of nonimmunocompromised patients with GC (39). In contrast with our data, Blastocystis species were more frequently involved in GC in patients from the HM group than in the nonimmunocompromised symptomatic patients. In our study, we also looked at the white cell blood count in the HM patients with positive qPCR results for Blastocystis parasites. Three of the 4 symptomatic HM patients had lymphopenia, and two had neutropenia. However, other patients from the HM group with positive test results had the same hematological abnormalities without gastrointestinal symptoms (see Table 2).
In summary, we developed a highly sensitive qPCR assay enabling the detection and quantification of Blastocystis isolates directly from human stool samples. Using this qPCR method, we detected 2-fold-more positive patients than with the culture approach that to date has been accepted as the gold standard method. The qPCR assay can be easily applied to large epidemiological studies, with rapid results compared to xenic cultures. The molecular method was validated through a prospective epidemiological study of 2 cohorts—one group consisting of patients presenting hematological malignancies and one control group of nonimmunocompromised patients. Host factors and intrinsic parasite factors such as parasite density and/or subtype were not correlated with pathogenicity. Further studies are now required on larger patient populations. All Blastocystis-positive samples were easily subtyped by direct sequencing of qPCR products, revealing an unexpected subtype distribution with a high prevalence of subtype 4. This study demonstrated the impact of detection methods on relative subtype distributions, thus underlining the need to harmonize the diagnostic tools used for epidemiological studies.
ACKNOWLEDGMENTS
We are grateful to Eric Viscogliosi and Rune Stensvold for providing strains and DNA samples. We also thank David Biron and ATT-Auvergne Traduction Technique for their useful criticism on the manuscript.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1. Abe N. 2004. Molecular and phylogenetic analysis of Blastocystis isolates from various hosts. Vet. Parasitol. 120:235–242 [DOI] [PubMed] [Google Scholar]
- 2. Amin O. M. 2002. Seasonal prevalence of intestinal parasites in the United States during 2000. Am. J. Trop. Med. Hyg. 66:799–803 [DOI] [PubMed] [Google Scholar]
- 3. Cirioni O., Giacometti A., Drenaggi D., Ancarani F., Scalise G. 1999. Prevalence and clinical relevance of Blastocystis hominis in diverse patient cohorts. Eur. J. Epidemiol. 15:389–393 [DOI] [PubMed] [Google Scholar]
- 4. Domínguez-Márquez M. V., Guna R., Munoz C., Gomez-Munoz M. T., Borras R. 2009. High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain). Parasitol. Res. 105:949–955 [DOI] [PubMed] [Google Scholar]
- 5. Elwakil H. S., Hewedi I. H. 2010. Pathogenic potential of Blastocystis hominis in laboratory mice. Parasitol. Res. 107:685–689 [DOI] [PubMed] [Google Scholar]
- 6. Ghosh K., Ayyaril M., Nirmala V. 1998. Acute GVHD involving the gastrointestinal tract and infestation with Blastocystis hominis in a patient with chronic myeloid leukaemia following allogeneic bone marrow transplantation. Bone Marrow Transplant. 22:1115–1117 [DOI] [PubMed] [Google Scholar]
- 7. Giacometti A., Cirioni O., Fiorentini A., Fortuna M., Scalise G. 1999. Irritable bowel syndrome in patients with Blastocystis hominis infection. Eur. J. Clin. Microbiol. Infect. Dis. 18:436–439 [DOI] [PubMed] [Google Scholar]
- 8. Gupta R., Parsi K. 2006. Chronic urticaria due to Blastocystis hominis. Australas. J. Dermatol. 47:117–119 [DOI] [PubMed] [Google Scholar]
- 9. Hailemariam G., et al. 2004. Intestinal parasitic infections in HIV/AIDS and HIV seronegative individuals in a teaching hospital, Ethiopia. Jpn. J. Infect. Dis. 57:41–43 [PubMed] [Google Scholar]
- 10. Jones M. S., Jr., et al. 2008. Detection of Blastocystis from stool samples using real-time PCR. Parasitol. Res. 103:551–557 [DOI] [PubMed] [Google Scholar]
- 11. Jones W. R. 1946. The experimental infection of rats with Entamoeba histolytica with a method for evaluating the anti-amoebic properties of new compounds. Ann. Trop. Med. Parasitol. 40:130–140 [DOI] [PubMed] [Google Scholar]
- 12. Kurniawan A., et al. 2009. Intestinal parasitic infections in HIV/AIDS patients presenting with diarrhoea in Jakarta, Indonesia. Trans. R. Soc. Trop. Med. Hyg. 103:892–898 [DOI] [PubMed] [Google Scholar]
- 13. Leder K., Hellard M. E., Sinclair M. I., Fairley C. K., Wolfe R. 2005. No correlation between clinical symptoms and Blastocystis hominis in immunocompetent individuals. J. Gastroenterol. Hepatol. 20:1390–1394 [DOI] [PubMed] [Google Scholar]
- 14. Leelayoova S., et al. 2002. In-vitro cultivation: a sensitive method for detecting Blastocystis hominis. Ann. Trop. Med. Parasitol. 96:803–807 [DOI] [PubMed] [Google Scholar]
- 15. Levy Y., George J., Shoenfeld Y. 1996. Severe Blastocystis hominis in an elderly man. J. Infect. 33:57–59 [DOI] [PubMed] [Google Scholar]
- 16. Li L. H., et al. 2007. Cross-sectional surveys and subtype classification of human Blastocystis isolates from four epidemiological settings in China. Parasitol. Res. 102:83–90 [DOI] [PubMed] [Google Scholar]
- 17. Li L. H., et al. 2007. Molecular epidemiology of human Blastocystis in a village in Yunnan province, China. Parasitol. Int. 56:281–286 [DOI] [PubMed] [Google Scholar]
- 18. Moe K. T., et al. 1997. Experimental Blastocystis hominis infection in laboratory mice. Parasitol. Res. 83:319–325 [DOI] [PubMed] [Google Scholar]
- 19. Nassir E., et al. 2004. Blastocystis hominis as a cause of hypoalbuminemia and anasarca. Eur. J. Clin. Microbiol. Infect. Dis. 23:399–402 [DOI] [PubMed] [Google Scholar]
- 20. Noël C., et al. 2005. Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species, and zoonosis. J. Clin. Microbiol. 43:348–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parkar U., et al. 2010. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet. Parasitol. 169:8–17 [DOI] [PubMed] [Google Scholar]
- 22. Pegelow K., et al. 1997. Parasitological and nutritional situation of school children in the Sukaraja district, West Java, Indonesia. Southeast Asian J. Trop. Med. Public Health 28:173–190 [PubMed] [Google Scholar]
- 23. Pinel C., et al. 1999. Blastocystis hominis: epidemiological and clinical remarks from more than 3,500 stool examinations. Ann. Biol. Clin. (Paris) 57:601–604 [PubMed] [Google Scholar]
- 24. Rao K., Sekar U., Iraivan K. T., Abraham G., Soundararajan P. 2003. Blastocystis hominis—an emerging cause of diarrhoea in renal transplant recipients. J. Assoc. Physicians India 51:719–721 [PubMed] [Google Scholar]
- 25. Rene B. A., Stensvold C. R., Badsberg J. H., Nielsen H. V. 2009. Subtype analysis of Blastocystis isolates from Blastocystis cyst excreting patients. Am. J. Trop. Med. Hyg. 80:588–592 [PubMed] [Google Scholar]
- 26. Rossignol J. F., Kabil S. M., Said M., Samir H., Younis A. M. 2005. Effect of nitazoxanide in persistent diarrhea and enteritis associated with Blastocystis hominis. Clin. Gastroenterol. Hepatol. 3:987–991 [DOI] [PubMed] [Google Scholar]
- 27. Scicluna S. M., Tawari B., Clark C. G. 2006. DNA barcoding of Blastocystis. Protist 157:77–85 [DOI] [PubMed] [Google Scholar]
- 28. Souppart L., et al. 2009. Molecular epidemiology of human Blastocystis isolates in France. Parasitol. Res. 105:413–421 [DOI] [PubMed] [Google Scholar]
- 29. Stensvold C. R., et al. 2009. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 39:473–479 [DOI] [PubMed] [Google Scholar]
- 30. Stensvold C. R., Arendrup M. C., Jespersgaard C., Molbak K., Nielsen H. V. 2007. Detecting Blastocystis using parasitologic and DNA-based methods: a comparative study. Diagn. Microbiol. Infect. Dis. 59:303–307 [DOI] [PubMed] [Google Scholar]
- 31. Stensvold C. R., Arendrup M. C., Molbak K., Nielsen H. V. 2007. The prevalence of Dientamoeba fragilis in patients with suspected enteroparasitic disease in a metropolitan area in Denmark. Clin. Microbiol. Infect. 13:839–842 [DOI] [PubMed] [Google Scholar]
- 32. Stensvold C. R., Nielsen H. V., Molbak K., Smith H. V. 2009. Pursuing the clinical significance of Blastocystis—diagnostic limitations. Trends Parasitol. 25:23–29 [DOI] [PubMed] [Google Scholar]
- 33. Stensvold C. R., et al. 2007. Terminology for Blastocystis subtypes—a consensus. Trends Parasitol. 23:93–96 [DOI] [PubMed] [Google Scholar]
- 34. Stenzel D. J., Boreham P. F. 1996. Blastocystis hominis revisited. Clin. Microbiol. Rev. 9:563–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suresh K., et al. 1993. In vitro encystment and experimental infections of Blastocystis hominis. Parasitol. Res. 79:456–460 [DOI] [PubMed] [Google Scholar]
- 36. Suresh K., Smith H. 2004. Comparison of methods for detecting Blastocystis hominis. Eur. J. Clin. Microbiol. Infect. Dis. 23:509–511 [DOI] [PubMed] [Google Scholar]
- 37. Suresh K., Smith H. V., Tan T. C. 2005. Viable Blastocystis cysts in Scottish and Malaysian sewage samples. Appl. Environ. Microbiol. 71:5619–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tan K. S. 2008. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 21:639–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taşova Y., Sahin B., Koltas S., Paydas S. 2000. Clinical significance and frequency of Blastocystis hominis in Turkish patients with hematological malignancy. Acta Med. Okayama 54:133–136 [DOI] [PubMed] [Google Scholar]
- 40. Termmathurapoj S., et al. 2004. The usefulness of short-term in vitro cultivation for the detection and molecular study of Blastocystis hominis in stool specimens. Parasitol. Res. 93:445–447 [DOI] [PubMed] [Google Scholar]
- 41. van Saanen-Ciurea M., El Achcachi H. 1985. Blastocystis hominis: culture and morphological study. Experientia 41:546. [PubMed] [Google Scholar]
- 42. Yakoob J., et al. 2004. Irritable bowel syndrome: in search of an etiology: role of Blastocystis hominis. Am. J. Trop. Med. Hyg. 70:383–385 [PubMed] [Google Scholar]
- 43. Yan Y., et al. 2006. Genetic variability of Blastocystis hominis isolates in China. Parasitol. Res. 99:597–601 [DOI] [PubMed] [Google Scholar]
- 44. Zierdt C. H., Tan H. 1976. Endosymbiosis in Blastocystis hominis. Exp. Parasitol. 39:422–430 [DOI] [PubMed] [Google Scholar]
- 45. Zman V., Khan K. Z. 1994. A comparison of direct microscopy with culture for the diagnosis of Blastocystis hominis. Southeast Asian J. Trop. Med. Public Health 25:792–793 [PubMed] [Google Scholar]