Abstract
Previous studies have demonstrated that presecretory proteins such as maltose binding protein (MBP) and outer membrane protein A (OmpA) are targeted to the Escherichia coli inner membrane by the molecular chaperone SecB, but that integral membrane proteins are targeted by the signal recognition particle (SRP). In vitro studies have suggested that trigger factor binds to a sequence near the N terminus of the mature region of OmpA and shunts the protein into the SecB pathway by blocking an interaction between SRP and the signal peptide. By contrast, we have found that the targeting pathway of a protein under physiological conditions is dictated by the composition of its targeting signal. Replacement of the MBP or OmpA signal peptide with the first transmembrane segment of AcrB abolished the dependence on SecB for transport and rerouted both proteins into the SRP targeting pathway. More modest alterations of the MBP signal peptide that simply increase its hydrophobicity also promoted SRP binding. Furthermore, we obtained evidence that SRP has a low affinity for typical signal peptides in vivo. These results imply that different classes of E. coli proteins are targeted by distinct pathways because bacterial SRP binds to a more restricted range of targeting signals than its eukaryotic counterpart.
In mammalian cells, both presecretory and integral membrane proteins are targeted to the endoplasmic reticulum by a single mechanism. The central player in the targeting reaction is the signal recognition particle (SRP), a ribonucleoprotein complex consisting of six polypeptides and a 300 nucleotide RNA (reviewed in 1). In an initial step, the 54 kDa subunit of SRP (SRP54) recognizes specific hydrophobic targeting signals as they emerge from translating ribosomes (2, 3). The targeting signals are either N-terminal signal sequences or, in the case of many membrane proteins that lack discrete signal peptides, the first transmembrane segment (TMS). Once SRP has engaged a targeting signal, the ribosome-nascent chain complex migrates to the endoplasmic reticulum where an interaction between SRP and a heterodimeric SRP receptor (SR) catalyzes release of the nascent chain and its insertion into a translocation channel (4, 5).
By contrast, presecretory proteins and inner membrane proteins (IMPs) are targeted to the inner membrane (IM) by distinct mechanisms in Escherichia coli. As in higher eukaryotic cells, polytopic IMPs that lack cleaved signal peptides are targeted to the IM by SRP (6–9). The SRP found in many bacteria is a simplified version of the eukaryotic particle that consists of an SRP54 homolog (Ffh) and a truncated ≈100-nt RNA (4.5S RNA) (10, 11), and the SR consists of only an α-subunit homolog (FtsY) (12, 13). Nevertheless, a variety of evidence indicates that the basic biochemical properties of the SRP pathway are highly conserved (8, 9, 14–17). Like its eukaryotic counterpart, E. coli SRP appears to recognize TMSs as specific targeting signals cotranslationally (8, 18). By contrast, many presecretory proteins are targeted to the IM by molecular chaperones, such as SecB and DnaK (19), that bind promiscuously to a wide variety of polypeptide sequences. Chaperones bind to the mature region of presecretory proteins late in translation or posttranslationally and keep them in a loosely folded conformation that is required for their translocation across the membrane (20). By binding to presecretory proteins, the chaperones also keep signal peptides available for interactions with the translocation machinery. Although there appears to be significant redundancy among chaperone-based targeting pathways, the export of a subset of presecretory proteins including the periplasmic maltose binding protein (MBP) and many outer membrane proteins (OMPs) is particularly SecB-dependent (19).
Because all hydrophobic targeting signals are recognized as synonymous by mammalian SRP, the mechanism by which presecretory proteins are routed into SRP-independent targeting pathways in bacteria is not immediately apparent. Although E. coli SRP has been shown to interact with signal peptides in vitro (15–17, 21), it is conceivable that, under physiological conditions, the particle binds efficiently only to TMSs, which contain a longer stretch of consecutive hydrophobic amino acids than signal peptides. If so, presecretory proteins would be routed into chaperone-based targeting pathways by default. This hypothesis is supported by a study on the biogenesis of the M13 procoat protein, an unusual small IMP that contains a signal peptide. Although integration of the wild-type protein does not require SRP, a mutation that greatly increases the hydrophobicity of the signal peptide reroutes the protein into the SRP pathway (22). Moreover, the notion that SRPs can distinguish between targeting signals based on hydrophobicity is supported by the finding that yeast SRP selectively targets presecretory proteins that contain especially hydrophobic signal peptides to the endoplasmic reticulum (23). An alternative hypothesis, however, is that E. coli presecretory proteins are recognized by another factor that occludes the binding of SRP. This view is supported by in vitro cross-linking experiments in which the binding of trigger factor to a sequence within the first 125 amino acids of pro-OmpA (but beyond the signal peptide) was shown to prevent the association of SRP (18). These data led to the proposal that presecretory proteins are routed into the SecB pathway by virtue of their preferential recognition by trigger factor.
In this study, we have analyzed the basis for the differential targeting of E. coli presecretory and membrane proteins under physiological conditions. We found that we could switch the targeting pathway of presecretory proteins by increasing the hydrophobicity of their targeting signals. Although both MBP and OmpA were targeted by the SecB pathway, derivatives that contain more hydrophobic signal peptides or a TMS in place of the signal peptide were targeted by the SRP pathway. Overproduction of SRP did not alter the targeting pathway of native MBP, suggesting that SRP effectively has a much higher affinity for TMSs than for typical signal peptides in vivo. Thus, the data imply that the nature of the targeting signal plays a dominant role in determining the targeting pathway for a given protein.
Materials and Methods
Reagents, Media, and Bacterial Strains.
Polyclonal antiserum against MBP, chloramphenicol acetyl transferase (CAT), and influenza hemagglutinin epitope HA.11 (HA) were obtained from New England Biolabs, 5 Prime → 3 Prime, and Covance, respectively. Affinity-purified antibodies against Ffh and FtsY have been described (7). Media preparation and basic bacterial manipulations were performed by standard methods (24). Selective media contained ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml) as required. All bacterial cultures were grown at 37°C. The bacterial strains used in this study were MC4100 (F− araD139 Δ(argF-lac)U169 rpsL150 relA1 thi fib5301 deoC1 ptsF25 rbsR), TST1 (MC4100 malE52∷Tn10), CK1953 (MC4100 secB∷Tn5; ref. 19), WAM113 [MC4100 ara+ ffh∷kan-1 λ(Para-ffh Apr); ref. 25], HDB50 (TST1 secB∷Tn5), HDB51 (WAM113 secB+ zic-4901∷Tn10), and HDB52 (WAM113 secB∷Tn5 zic-4901∷Tn10). MC4100 and TST1 were obtained from the American Type Culture Collection and the E. coli Genetic Stock Center, respectively.
Plasmid Construction.
To construct pJH28 and pJH29, pMAL-p2X and pMAL-c2x (New England Biolabs), respectively, were digested with BspHI and SacI to excise bla and lacZα. A DNA fragment containing the CAT gene from pACYC184 was then generated by PCR, digested with BspHI and SacI, and ligated to the prepared malE expression vectors. A DNA fragment corresponding to the first TMS of AcrB was generated by PCR using the oligonucleotides 5′-AGGAGCCGTTAACATATGCCTAATTTCTTTATC-3′ (TMS1) and 5′-GTAGGATATTGCGCCACCGGCATATGGAGGATCGC-3′, digested with NdeI, and cloned into the NdeI site of the modified pMAL-c2X plasmid. To construct plasmids encoding MBP signal peptide mutants, point mutations were introduced into pJH28 by using the QuikChange mutagenesis kit (Stratagene). OmpA was amplified by PCR using E. coli genomic DNA as a template and cloned behind the lac promoter in plasmid RB11 (26). An HA tag (YPYDVPDYASL) was then attached to the C terminus of OmpA by PCR to generate RB11-OmpA-HA. A DNA fragment containing the CAT gene from pACYC184 that was generated by PCR was then cloned into the ScaI site of RB11-OmpA-HA. NdeI and EagI sites were introduced at the start of ompA and at the end of the signal peptide, respectively, by site-directed mutagenesis as described above, to create plasmid pHL11. A DNA fragment corresponding to the first TMS of AcrB was generated by PCR using the oligonucleotides TMS1 and 5′-GTAGGATATTCGGCCGCCGGCAGTTTGAGGATCGC-3′, digested with NdeI and EagI, and cloned into pHL11 to produce pHL12. Plasmids pHL13 and pHL14 were constructed by inserting oligonucleotides 5′-GGCCGCTGGTAAACCAGGTGTACAGGGCGATGG-3′ and 5′-GGCCGCTGGTAAACCAGGTGTACAGGGCGATGGTCGTTATGGTGGCTC-3′ and their complements, respectively, into the EagI site of pHL11.
A SalI–BamHI fragment containing ffh and its promoter was excised from pHDB1 (7) and cloned into pACYC184 to produce pHDB7. To construct pHQ3, a DNA fragment containing the 4.5S RNA gene (ffs) and its promoter that was generated by PCR was cleaved with XbaI and ClaI and cloned into pHDB7. To construct pHQ4, a PCR fragment containing ftsY and its promoter was digested with BamHI and HindIII and cloned into pHQ3. The fidelity of all PCRs was verified by testing amplified genes in complementation assays.
Protein Export/IMP Insertion Assays.
For most experiments, cells were grown in M9 containing 0.2% glucose. Overnight cultures were washed and diluted into fresh medium at an OD550 of 0.025. For Ffh depletion studies, cells were grown overnight in M9 containing 0.2% fructose and 0.2% arabinose, washed in medium lacking arabinose, and then added at an OD550 of 0.005 to medium containing fructose and either arabinose or glucose (0.2%). When cultures reached an OD550 of 0.2–0.3 (after 4–5 h or 6–7 h, in the Ffh depletion studies), synthesis of plasmid-borne MBP and OmpA (and their derivatives) was then induced by the addition of 50 μM and 1 mM isopropyl β-d-thiogalactoside, respectively, and synthesis of endogenous MBP was induced by the addition of 0.2% maltose. To analyze the fate of MBP, OmpA, and their derivatives, aliquots were removed from each culture 30 min after the addition of isopropyl β-d-thiogalactoside or maltose. Cells were then pulse-labeled by the addition of Tran35S-label (Amersham; 30 μCi/ml; 1 Ci = 37 GBq) for 30 s. In some experiments, proteins were immediately collected by precipitation with 10% trichloroacetic acid (TCA). Otherwise, spheroplasts were generated, and half of each sample was treated with proteinase K as described (9) before TCA precipitation. Immunoprecipitations were performed essentially as described (26). Unless otherwise noted, proteins were resolved by SDS/PAGE on 8%–16% minigels (NOVEX, San Diego).
Western Blotting.
TCA-precipitated proteins were solubilized, resolved by SDS/PAGE, and transferred to nitrocellulose as described (7). After incubating filters with a primary antibody, antibody–antigen complexes were detected with 35S-labeled protein A (Amersham). The level of radioactivity in individual bands was quantitated by using a Fuji BAS-2500 phosphorimager.
Results
Replacement of the Signal Sequence of a Presecretory Protein with a TMS Alters Its Targeting Pathway.
To examine the role of the targeting signal in specifying the targeting pathway for a given protein, we replaced the signal peptide of two SecB-dependent presecretory proteins, MBP and OmpA, with the first TMS of the polytopic IMP AcrB to create TM-MBP and TM-OmpA (Fig. 1). Previous studies have shown that a truncated version of AcrB that contains only the first TMS is targeted to the IM by the SRP pathway (27). Thus, if the entry into a targeting pathway is dictated by the targeting signal rather than sequences elsewhere in a protein, then the presence of the AcrB TMS should reroute MBP and OmpA into the SRP pathway. Because the cleavage of the MBP and OmpA signal peptides requires the presence of a leader peptidase recognition site (alanines at positions −1 and −3), we expected that the introduction of the AcrB TMS would convert the presecretory proteins into IMPs. This prediction was confirmed in experiments in which we analyzed the intracellular localization of TM-MBP and TM-OmpA by cell fractionation and Western blotting. As expected, both proteins localized to the membrane fraction (data not shown) and had a slightly slower mobility on SDS/PAGE than the mature periplasmic forms of MBP and OmpA (see below).
Figure 1.
PreMBP and ProOmpA variants used in this study. The sequence of the N terminus of preMBP and proOmpA and the leader peptidase cleavage site (arrow) is shown. The point mutations, substitutions, and insertions in each variant are underlined. TM-MBP and TM-OmpA do not have a cleaved signal peptide. The proOmpA analyzed herein contains a proline in place of the naturally occurring glutamine at position −2 (italics). This change was introduced to create a restriction site that could be used to make the proOmpA variants.
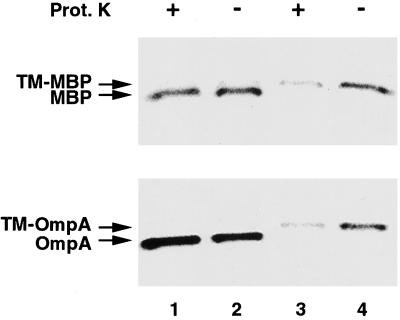
To determine the targeting pathway of the chimeric proteins, we developed a simple protease protection assay to measure the efficiency of their insertion into the IM. TST1 (MC4100 malE−) was first transformed with a plasmid encoding either a native presecretory protein (pJH28 or pHL11) or the corresponding membrane-anchored version (pJH29 or pHL12) and then grown in M9 medium. After pulse-labeling, cells were converted to spheroplasts and half of each sample was treated with proteinase K. Total protein was recovered by TCA precipitation, and MBP- or OmpA-containing polypeptides were isolated by immunoprecipitation. Cleavage of the signal peptide showed that the wild-type presecretory proteins were efficiently secreted (Fig. 2, lanes 1 and 2). The exported proteins were resistant to protease digestion presumably because they rapidly acquired a tightly folded conformation after translocation and, in the case of OmpA, assembly into the outer membrane. By contrast, TM-MBP and TM-OmpA were susceptible to protease digestion (Fig. 2, lanes 3 and 4). These results imply that the mature region of each protein is translocated into the periplasm during the normal insertion process but does not fold completely. Indeed, it has been shown that OmpA cannot fold into its native state unless it reaches the outer membrane (28). Presumably, the attachment of MBP to the IM also hinders attainment of a protease-resistant conformation. As a corollary, the results predict that TM-MBP and TM-OmpA should be protected from protease digestion under conditions that promote retention in the cytoplasm.
Figure 2.
Protease-sensitivity of TM-MBP and TM-OmpA. TST1 transformed with pJH28, pJH29, pHL11, or pHL12 were pulse-labeled, converted to spheroplasts, and treated with proteinase K. MBP- and OmpA-containing polypeptides were immunoprecipitated with anti-MBP and anti-HA antibodies, respectively. Lanes: 1 and 2, cells producing MBP or OmpA; 3 and 4, cells producing TM-MBP or TM-OmpA. Proteinase K was added to the samples in lanes 1 and 3.
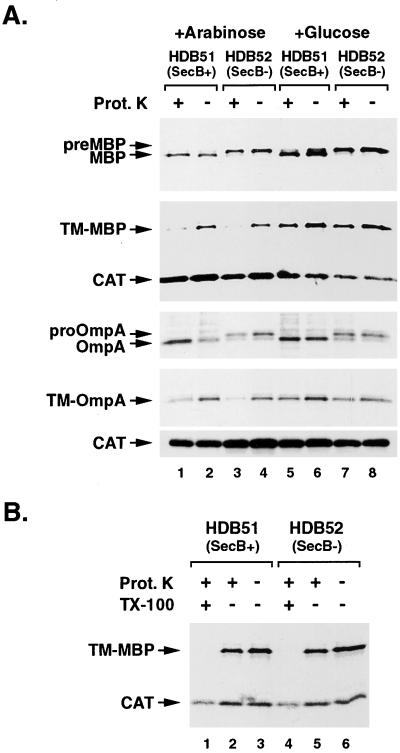
With this method, we obtained strong evidence that the presence of a TMS diverts MBP and OmpA into the SRP pathway. To manipulate the SecB and SRP pathways simultaneously, we constructed isogenic secB+ and secB− strains (HDB51 and HDB52, respectively) in which ffh expression is under the control of an arabinose-inducible promoter. Cells were transformed with a plasmid encoding MBP, OmpA, TM-MBP, or TM-OmpA and grown in M9 containing arabinose. Ffh was then depleted from half of the cells by replacing the arabinose with glucose. Because ffh is essential for viability (25), cells were labeled before growth defects were observed to minimize secondary effects of Ffh depletion. Protein transport across the IM was assayed as described above, except that the cytoplasmic protein CAT was also immunoprecipitated to verify that the spheroplasts were intact. As expected, the export of pre-MBP and pro-OmpA was SecB-dependent. Although both proteins were efficiently translocated in HDB51 (Fig. 3A, lanes 1 and 2), the lack of signal peptide processing indicated that translocation was blocked in HDB52 (Fig. 3A, lanes 3 and 4). Consistent with previous results (8, 9, 12, 13), we found that SRP depletion had little to no effect on the export of either protein (Fig. 3A, lanes 5–8). By contrast, the insertion of TM-MBP and TM-OmpA was SRP-dependent. Proteinase K degraded the membrane-anchored proteins in spheroplasts derived from both HDB51 and HDB52, indicating they were properly inserted into the IM, even in the absence of SecB (Fig. 3A, lanes 1–4). Both TM-MBP and TM-OmpA were largely resistant to protease digestion, however, when SRP was depleted from either strain (Fig. 3A, lanes 5–8). The observation that TM-MBP was degraded by proteinase K when Triton X-100 was added to disrupt the IM (Fig. 3B, lanes 1 and 4) confirmed that the protease-resistance was caused by retention of the protein in the cytoplasm in the absence of SRP.
Figure 3.
Replacement of the MBP and OmpA signal peptides with a TMS reroutes the proteins into the SRP targeting pathway. (A) HDB51 (PBAD-ffh secB+) and HDB52 (PBAD-ffh secB−) transformed with pJH28, pJH29, pHL11, or pHL12 were grown in M9 medium containing arabinose (lanes 1–4) or glucose (lanes 5–8), radiolabeled, converted to spheroplasts, and treated with proteinase K. MBP- and OmpA-containing polypeptides and CAT were immunoprecipitated. Lanes: 1, 2, 5, and 6, HDB51 cells; 3, 4, 7, and 8, HDB52 cells. Proteinase K was added to the samples shown in lanes 1, 3, 5, and 7. (B) HDB50 and HDB51 transformed with pJH29 were grown in M9 medium containing glucose. The experiment shown in A was repeated except that 1% Triton X-100 was added to the samples in lanes 1 and 4 immediately before proteinase K treatment. Lanes: 1–3, HDB51 cells; 4–6, HDB52 cells. Proteinase K was added to the samples in lanes 1, 2, 4, and 5.
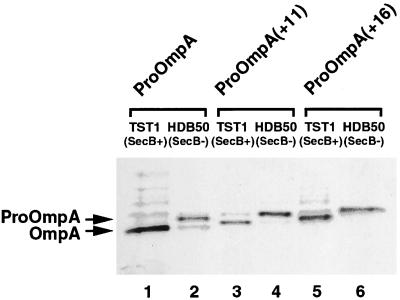
Although these results suggested that the targeting pathway of an E. coli protein is influenced by the sequence of its targeting signal, it was also possible that TM-MBP and TM-OmpA were routed into the SRP pathway simply because they had a longer targeting signal than the native presecretory proteins. If the entry of a presecretory protein into a chaperone-based targeting pathway requires that trigger factor bind to a specific site in the mature region before SRP can recognize the signal peptide, then increasing the distance between the hydrophobic core of the targeting signal and the downstream site might favor SRP binding. To test this hypothesis, we inserted random 11- and 16-amino acid peptides between the OmpA signal sequence and the mature region of the protein to create proOmpA(+11) and proOmpA(+16) (Fig. 1). We then transformed TST1 and HDB50 (TST1 secB−) with a plasmid encoding either proOmpA or one of the two variants and determined whether SecB was required for export. The precursor form of all three proteins was rapidly processed to the mature form in TST1 (Fig. 4, lanes 1, 3, and 5) but accumulated in the cytoplasm of HDB50 (Fig. 4, lanes 2, 4, and 6). These results demonstrate that proOmpA(+11) and proOmpA(+16) are targeted by SecB and strongly suggest that the distance between the signal sequence and a hypothetical trigger factor binding site in the mature region is not a critical determinant for entry into the SecB pathway.
Figure 4.
Increasing the distance between the signal peptide and the mature region of proOmpA does not change its targeting pathway. TST1 and HDB50 (secB−) transformed with pHL11, pHL13, or pHL14 were radiolabeled, and OmpA-containing polypeptides were immunoprecipitated with anti-HA antibodies. The precursor encoded on each plasmid is indicated at the top. Lanes: 1, 3, and 5, TST1 cells; 2, 4, and 6, HDB50 cells.
Increasing the Hydrophobicity of the MBP Signal Peptide Routes the Protein into the SRP Targeting Pathway.
We next analyzed the basis by which proteins that contain one or more TMSs are selected for entry into the SRP targeting pathways in more detail. One possibility is that E. coli SRP binds preferentially to targeting signals that surpass a threshold level of hydrophobicity. Alternatively, SRP may recognize subtle structural features of TMSs that are absent from signal peptides. To distinguish between these two possibilities, we constructed five MBP mutants (MBP*1–5) in which polar amino acids in the middle of the signal peptide were replaced with large hydrophobic residues (see Fig. 1). MBP*1 is a triple-point mutant that contains 12 consecutive hydrophobic amino acids in its signal peptide, and MBP*2–5 are single- and double-point mutants that contain shorter hydrophobic stretches. None of the mutations affect signal peptide cleavage or the rate of translation (Fig. 5 and data not shown). HDB51 and HDB52 were transformed with a plasmid encoding a preMBP* mutant and grown in M9 containing arabinose. Ffh was depleted in half of the cells by switching the carbon source as described above. All of the MBP* proteins were quantitatively exported in HDB52 when the SRP concentration was high (Fig. 5, lane 4), indicating that the mutations abolished SecB dependence. By contrast, a significant amount of preMBP*1 was detected in HDB51 after Ffh depletion (Fig. 5 Top, lane 5). This result implies that a large increase in the hydrophobicity of the MBP signal peptide reroutes the protein into the SRP pathway. Interestingly, more preMBP*1 was detected in HDB52 after Ffh depletion than in HDB51, and the export of MBP*2–5 was significantly impaired only when both SRP and SecB were absent (Fig. 5, lane 6). Thus, the results strongly suggest that although slight increases in signal peptide hydrophobicity are sufficient to reroute MBP into the SRP pathway, the targeting of MBP variants that contain modified signal peptides can be partially or completely rescued by SecB when the SRP pathway is compromised.
Figure 5.
Increasing the hydrophobicity of the MBP signal peptide reroutes the protein into the SRP pathway. HDB51 and HDB52 transformed with a plasmid encoding the indicated MBP* mutant were grown in M9 medium containing arabinose (lanes 3 and 4) or glucose (lanes 5 and 6) and radiolabeled. TST1 and HDB50 transformed with pJH28 were grown in M9 medium containing glucose and radiolabeled to provide molecular weight markers (lanes 1 and 2). MBP-containing polypeptides were immunoprecipitated and resolved on a 10% NuPage gel with a standard Mops buffer (NOVEX). Lanes: 3 and 5, HDB51 cells; 4 and 6, HDB52 cells.
E. coli SRP Has a Much Lower Affinity for Signal Peptides than for TMSs in Vivo.
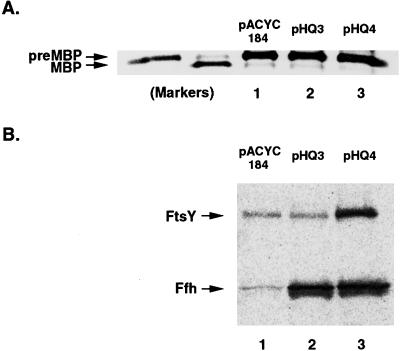
In light of the observation that E. coli SRP binds effectively to a variety of signal peptides in vitro and to a slightly modified version of the MBP signal peptide in vivo, we considered the possibility that a minor alteration of cell physiology might reroute naturally occurring presecretory proteins into the SRP pathway. Even if SRP has only a moderately higher affinity for TMSs than for signal peptides, it is conceivable that cells exploit this affinity difference and set the SRP concentration so that the particle bypasses most signal peptides. In biochemical assays, increasing the SRP concentration promotes recognition of relatively weak substrates (29). Thus, increasing the SRP level in vivo might drive presecretory proteins into the SRP pathway and abolish SecB dependence. To test this hypothesis, we analyzed the effect of overproducing SRP and FtsY on MBP export in CK1953 (secB−) cells. Cells were transformed with pHQ3 (ffh+ ffs+) or pHQ4 (ffh+ ffs+ ftsY+), which contain SRP genes expressed under the control of their natural promoters, or pACYC184. The presence of these plasmids does not have a detectable effect on either IMP biogenesis or cell growth (data not shown). MBP export was analyzed as described above. Virtually no mature MBP was detected in any of the cells (Fig. 6A), indicating that export was SecB-dependent. Western blot analysis showed that cells harboring pHQ3 and pHQ4 contained approximately 20- and 7-fold more Ffh and FtsY, respectively, than control cells (Fig. 6B). Because free Ffh is highly unstable (30), most of the excess protein was likely complexed with 4.5S RNA. By showing that the MBP targeting pathway is not influenced by SRP concentration, these results imply that the E. coli translation and targeting systems have evolved so that SRP can discriminate very precisely between targeting signals that differ only slightly in composition.
Figure 6.
MBP remains SecB-dependent in cells that overproduce SRP and FtsY. (A) CK1953 (secB−) transformed with the indicated plasmid were pulse-labeled and endogenous MBP-containing polypeptides were immunoprecipitated. PreMBP and MBP markers were generated as described in Fig. 5. (B) cK1953 transformed with the indicated plasmid were grown as in A but not radiolabeled. Ffh and FtsY were detected by Western blot.
Discussion
In this study we have obtained strong evidence that the targeting signal dictates the primary targeting mechanism of an E. coli presecretory protein or IMP. By replacing the signal peptide of two presecretory proteins with a TMS, we completely abolished their dependence on SecB for proper localization. These results suggested that TM-MBP and TM-OmpA are targeted by another pathway even in secB+ cells. Consistent with this view, we found that the transport of the chimeric proteins required SRP. On one level, these data provide proof that the SecB and SRP targeting pathways are completely independent. More significantly, the data show that alteration of the targeting signal is sufficient to reroute a protein from one targeting pathway to another. Because increasing the hydrophobicity of the MBP signal peptide and the attachment of a TMS produced similar effects on targeting, it is likely that the composition of the targeting signal, rather than its identity as a TMS or a signal peptide, determines the targeting pathway. Thus, the data strongly suggest that the binding of trigger factor to the mature region of presecretory proteins does not exclude proteins from the SRP pathway in vivo. However, they do not exclude the possibility that a chaperone routes presecretory proteins into the SecB pathway by binding to signal peptides with higher affinity than SRP. This scenario is very unlikely, although, because trigger factor does not have a marked affinity for hydrophobic sequences (21) and DnaK, the only other chaperone that is known to bind to short nascent polypeptide chains, is not required for the entry of MBP or OmpA into the SecB pathway (31).
Our results now provide an explanation for several observations that were puzzling when they were reported in 1989. With a genetic screen, Collier and Bassford (32) isolated MBP signal peptide mutants that improved export in a secB− strain. All of the mutations increased the net hydrophobicity of the signal peptide by effectively lengthening the hydrophobic core. MBP is normally exported in a posttranslational fashion but, remarkably, at least some of the mutants exhibited a significant degree of cotranslational translocation. In light of our observations on the MBP* mutants, it is likely that these investigators unknowingly isolated mutants that reroute MBP into the SRP pathway.
Perhaps the most noteworthy implication of our results is that the entry of proteins into the SRP pathway is far more sensitive to the composition of targeting signals in E. coli than in mammalian cells. This conclusion was not anticipated from in vitro studies on E. coli SRP. It is particularly intriguing to note that, unlike the minimally hydrophobic OmpA signal peptide, the MBP signal peptide is sufficiently hydrophobic [1.60 on the Goldman–Engelman–Steitz (GES) scale] to be scored as a TMS by the TopPred II algorithm (33). The observation that native MBP is targeted by SecB but that the MBP* mutants are targeted by SRP suggests that the hydrophobicity threshold for entry into the SRP pathway is quite high. Thus, our results raise the possibility that IMPs that contain a moderately hydrophobic first TMS are targeted by an SRP-independent mechanism or are targeted by SRP only after a highly hydrophobic TMS is synthesized. Alternatively, moderately hydrophobic TMSs may contain as yet unrecognized structural features that promote SRP binding. These possibilities remain to be explored because the first TMS of most of the IMPs that have been shown to use the SRP pathway so far including AcrB, MalF, MtlA, Lep, and FtsQ (6, 7, 9, 34) are extremely hydrophobic (>2.0 on the GES scale).
The ability of E. coli SRP to discriminate precisely between closely related targeting signals may reflect the extremely rapid rate of translation under physiological conditions. Although E. coli Ffh has a smaller substrate binding domain than mammalian SRP54 that might account for an altered substrate specificity, the observation that the bacterial protein interacts with typical signal peptides in vitro suggests that the fundamental binding properties are evolutionarily conserved. Ffh may bind preferentially to long hydrophobic sequences in vivo, however, partly because the time window for interaction is greater. In biochemical assays, SRP is generally added to relatively slow cell-free translation systems or to static ribosome–nascent-chain complexes. Under these conditions, Ffh may bind to signal peptides because they are accessible for a prolonged period. Regardless of the exact mechanism by which presecretory proteins are excluded from the SRP pathway in vivo, the finding that MBP targeting is immune to changes in SRP concentration suggests that the system is calibrated to effectively produce large differences in the affinity of SRP for TMSs and signal peptides.
Multiple targeting pathways may have evolved in bacteria (and other unicellular organisms) because rapid growth requires the targeting of different classes of proteins to the secretory pathway via distinct mechanisms. Because ribosomes are in vast excess over translocation sites, a targeting mechanism in which translation and translocation are uncoupled probably provides the most efficient means of exporting proteins rapidly (35). Posttranslational modes of translocation appear to work well for relatively hydrophilic molecules that can exist in a metastable conformation during their transit through the cytoplasm. By contrast, an SRP-based cotranslational targeting mechanism that promotes rapid association of ribosome–nascent-chain complexes with transport channels appears to be required for effective biogenesis of extremely hydrophobic IMPs that would tend to aggregate if exposed to the cytoplasmic environment. Moreover, recent results suggest that a highly efficient targeting system for IMPs is required partly to protect cells from the toxic effects of mislocalizing hydrophobic proteins in the cytoplasm (36). Nevertheless, a significant fraction of many SRP substrates can be transported across the IM by secondary pathways (6, 7, 22, 27). In the experiments shown herein, about half of the TM-OmpA and TM-MBP is successfully transported after SRP depletion, and the less hydrophobic MBP* mutants seem to represent an intermediate class of proteins that can use SRP and SecB pathways with similar efficiency.
Finally, it should be noted that our results may also have significant practical implications. Despite the presence of a typical signal peptide, many heterologous presecretory proteins are not exported efficiently in E. coli. Some of these proteins may not be secreted efficiently by chaperone-based posttranslational mechanisms because they readily fold into a translocation-incompetent conformation. It may be possible to target such proteins to the IM at an early stage of their synthesis via the SRP pathway and thereby overcome the problem of misfolding in the cytoplasm by simply increasing the hydrophobicity of their signal peptides.
Acknowledgments
We thank Janine Hyndman for excellent technical assistance, Hai-Yan Qi for constructing plasmids pHQ3 and pHQ4, and Rob Boulianne and Brenda Peculis for critical reading of the manuscript.
Abbreviations
- CAT
chloramphenicol acetyltransferase
- HA
hemagglutinin
- IM
inner membrane
- IMP
inner membrane protein
- MBP
maltose binding protein
- OMP
outer membrane protein
- SRP
signal recognition particle
- TMS
transmembrane segment
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Walter P, Johnson A E. Ann Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 2.Krieg U C, Walter P, Johnson A E. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurzchalia T V, Wiedmann M, Girshovich A S, Bochkareva E S, Bielka H, Rapoport T A. Nature (London) 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 4.Gilmore R, Walter P, Blobel G. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer D I, Krause E, Dobberstein B. Nature (London) 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- 6.deGier J-WL, Mansournia P, Valent QA, Phillips G J, Luirink J, von Heijne G. FEBS Letters. 1996;399:307–309. doi: 10.1016/s0014-5793(96)01354-3. [DOI] [PubMed] [Google Scholar]
- 7.Ulbrandt N D, Newitt J A, Bernstein H D. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 8.Valent Q A, Scotti P A, High S, deGier J-WL, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch H G, Hengelage T, Neumann-Haefelin C, MacFarlane J, Hoffschulte H K, Schimz KL, Mechler B, Müller M. Mol Biol Cell. 1999;10:2163–2173. doi: 10.1091/mbc.10.7.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poritz M A, Bernstein H D, Strub K, Zopf D, Wilhelm H, Walter P. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 11.Ribes V, Römisch K, Giner A, Dobberstein B, Tollervey D. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- 12.Römisch K, Webb J, Herz S, Prehn S, Frank R, Vingron M, Dobberstein B. Nature (London) 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein H D, Poritz M A, Strub K, Hoben P, Brenner S, Walter P. Nature (London) 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 14.Miller J D, Wilhelm H, Gierasch L, Gilmore R, Walter P. Nature (London) 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein H D, Zopf D, Freymann D M, Walter P. Proc Natl Acad Sci USA. 1993;90:5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luirink J, High S, Wood H, Giner A, Tollervey D, Dobberstein B. Nature (London) 1992;359:741–743. doi: 10.1038/359741a0. [DOI] [PubMed] [Google Scholar]
- 17.Powers T, Walter P. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck K, Wu L-F, Brunner J, Muller M. EMBO J. 2000;19:134–143. doi: 10.1093/emboj/19.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumamoto C A, Beckwith J. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collier D N, Bankaitis V A, Weiss J B, Bassford P J., Jr Cell. 1988;53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 21.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.deGier J-W, Scotti P A, Sääf A, Valent Q A, Kuhn A, Luirink J, von Heijne G. Proc Natl Acad Sci USA. 1998;95:14646–14651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng D T, Brown J D, Walter P. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 25.Phillips G J, Silhavy T J. Nature (London) 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 26.Newitt J A, Bernstein H D. J Biol Chem. 1998;273:12451–12456. doi: 10.1074/jbc.273.20.12451. [DOI] [PubMed] [Google Scholar]
- 27.Newitt J A, Ulbrandt N D, Bernstein H D. J Bacteriol. 1999;181:4561–4567. doi: 10.1128/jb.181.15.4561-4567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freundl R, Schwarz H, Stierhof Y-D, Gamon K, Hindennach I, Henning U. J Biol Chem. 1986;261:11355–11361. [PubMed] [Google Scholar]
- 29.Walter P, Ibrahimi I, Blobel G. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen C G, Pedersen S. J Bacteriol. 1994;176:7148–7154. doi: 10.1128/jb.176.23.7148-7154.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wild J, Altman A, Yura T, Gross C A. Genes Dev. 1992;6:1165–1175. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 32.Collier D N, Bassford P J., Jr J Bacteriol. 1989;171:4640–4647. doi: 10.1128/jb.171.9.4640-4647.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 34.Tian H, Boyd D, Beckwith J. Proc Natl Acad Sci USA. 2000;97:4730–4735. doi: 10.1073/pnas.090087297. . (First Published April 18, 2000; 10.1073/pnas.090087297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnsson N, Varshavsky A. EMBO J. 1994;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein, H. D. & Hyndman, J. B. (2001) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]