Abstract
Here, we report a laboratory-developed colorimetric-plate method for rapid disk diffusion susceptibility testing of Escherichia coli. One hundred isolates were evaluated. Categorical agreement between the colorimetric plate and the standard disk diffusion method was 99%. Mean time to results was 7.07 h (95% confidence interval, 5.96 to 8.19).
Escherichia coli is the most frequent cause of urinary tract infections and among the leading pathogens causing bloodstream infections (10, 31). High levels of antibiotic resistance are observed in E. coli (6, 13, 29, 31). Severe adverse outcomes such as increased length of hospital stay and delay of appropriate therapy are shown to be associated with multidrug-resistant Enterobacteriaceae (26, 30). The overall cost of antibiotic-resistant urinary tract infections to the health care system is high (2, 23). Appropriate antibiotic use is a priority (24, 25, 28, 32). Inappropriate initial antimicrobial therapy is independently associated with adverse outcomes, especially when not adjusted quickly with rapid susceptibility test results (17, 18, 20). Thus, a rapid, simple, and inexpensive alternative to commercial and higher-technology platforms for the susceptibility testing of E. coli isolates from urinary specimens is needed (25, 28, 32). Typically, standard antimicrobial susceptibility test methods require 16 to 18 h of incubation to produce interpretable results (7, 9). For rapid susceptibility testing in daily laboratory work, there are several automated systems, providing results within 8 to 12 h (27). Noncommercial rapid colorimetric assays employing oxidation-reduction indicators for antimicrobial susceptibility testing with MICs are described for various bacteria (1). A commercial rapid susceptibility testing medium for disk diffusion susceptibility testing is also reported (19). So far, all of the studies have reported either MIC-adapted methods or commercial disk diffusion methods. However, a simpler or cheaper alternative to commercial and higher-technology platforms does not exist. Such an alternative is much needed, especially for the susceptibility testing of E. coli isolates from the urinary specimens of outpatients, because the isolates constitute most of the daily laboratory workload, and conventional abbreviated identification of E. coli is easy, cheap, and sufficient (8). In this study, we have developed a rapid colorimetric medium for disk diffusion susceptibility testing of E. coli.
The study was conducted on nonrepetitive clinical isolates (n = 100) of E. coli. The study collection was characterized with conventional tests and the Vitek 2 (bioMérieux, France) automated microbiology system. Sixty-eight strains were isolated from urine samples, and 28 were extended-spectrum beta-lactamase (ESBL) producers. For the preparation of colorimetric plates, resazurin additive reagent was freshly prepared by dissolving 2.5 mg of resazurin sodium salt (Sigma-Aldrich, Germany) in 10 ml distilled water and sterilized using a syringe with an 0.2-μm filter (Schleicher & Schuell, Germany). Modified Mueller-Hinton agar medium was prepared by adding 10 ml of resazurin reagent to 990 ml of freshly prepared and cooled (45 to 50°C) Mueller-Hinton agar (Merck, Germany) medium (pH 7.0). Colorimetric plates were prepared by pouring ca. 70 ml of the modified Mueller-Hinton agar medium into each of the 150-mm by 15-mm bacteriological petri dishes to obtain a 4-mm depth. The plates were protected from light. When not used the same day, plates were stored at 4°C for up to 7 days. The procedure described by the Clinical and Laboratory Standards Institute (CLSI) standard was employed for performing the disk diffusion susceptibility tests by the colorimetric-plate method (9). All isolates were also tested with the standard method (9). Performance standards defined in the CLSI standard were used in the interpretation of test results (7). The color change of the medium from blue (resazurin) to pink (resorufin) was observed with the unaided eye. From the start of the incubation period, the observations were done at hourly intervals up to 5 h and then at 10-min intervals. When a visible color change was observed, the completed period was recorded as time to results (i.e., hours). The results of the experiments were valid if the results obtained with the quality control strain were within the acceptable limits given in the CLSI standard and there were no growth and color change in the negative-control plate after the incubation period (7). For reproducibility testing, 10 organisms were tested with the new method, at three distinct laboratory sites (by different personnel on each site) on three separate days in triplicate with a different inoculum prepared for each test (16). Reproducibility results were acceptable if the results of the overall reproducibility study from all test sites for any antimicrobial agent showed ≥95% category agreement compared to the test mode (i.e., the most frequent test result for the isolate) as a reference (16). Acceptable performance criteria were as follows: category agreement, ≥90%; very major discrepancy, ≤1.5%; major discrepancy, ≤3%; minor discrepancy, ≤10% (16). Performance metrics of the new method in terms of time to results were reported as the mean value with 95% confidence limits. The kappa statistic was calculated to measure agreement between the new method and the reference method. The differences between mean times to results of fresh and stock isolates were compared by the Student t test. The significance level for statistical tests was accepted as P < 0.05.
Results of the reproducibility tests were evaluated in comparison to the test mode as a reference (Table 1). Overall reproducibility was ca. 99.5%. There was no very major or major discrepancy between the results of the colorimetric-plate method and the standard method (Table 2). Overall category agreement of the colorimetric-plate method was ca. 99%. There was very good agreement between the standard and the new method as shown by the kappa statistic (κ = 0.983; P < 0.001). Times to results were determined by the observation of a visible color change (Fig. 1). Overall mean time to results was 7.07 h (95% confidence interval, 5.96 to 8.19). Mean times to results were 6.25 h (95% confidence interval, 5.23 to 7.27) for fresh clinical isolates and 8.17 h (95% confidence interval, 6.28 to 10.06) (P = 0.015) for clinical stock isolates.
Table 1.
Reproducibility results of the colorimetric-plate method in antimicrobial susceptibility testing of Escherichia coli
| Antimicrobiala | No. of isolatesb | No. of testsc | No. of test results in interpretive categorye: |
Reproducibility result, % (no.)d |
|||||
|---|---|---|---|---|---|---|---|---|---|
| S | I | R | CA | VMD | MD | mD | |||
| Ampicillin | 10 | 270 | 108 | 0 | 162 | 100 | 0 | 0 | 0 |
| Ampicillin-sulbactam | 10 | 270 | 169 | 47 | 54 | 97.4 | 0 | 0 | 2.6 (7) |
| Cefazolin | 10 | 270 | 162 | 0 | 108 | 100 | 0 | 0 | 0 |
| Cefuroxime | 10 | 270 | 162 | 0 | 108 | 100 | 0 | 0 | 0 |
| Ceftriaxone | 10 | 270 | 163 | 27 | 80 | 99.6 | ∼1.3 (1) | 0 | 0 |
| Cefepime | 10 | 270 | 270 | 0 | 0 | 100 | 0 | 0 | 0 |
| Imipenem | 10 | 270 | 270 | 0 | 0 | 100 | 0 | 0 | 0 |
| Amikacin | 10 | 270 | 216 | 2 | 52 | 99.3 | 0 | 0 | 0.7 (2) |
| Gentamicin | 10 | 270 | 192 | 50 | 28 | 98.5 | 0 | 0 | 1.5 (4) |
| Ciprofloxacin | 10 | 270 | 189 | 0 | 81 | 100 | 0 | 0 | 0 |
| Co-trimoxazole | 10 | 270 | 162 | 0 | 108 | 100 | 0 | 0 | 0 |
| Nitrofurantoin | 10 | 189 | 189 | 0 | 0 | 100 | 0 | 0 | 0 |
| All antimicrobials | 10 | 3,159 | 2,252 | 126 | 781 | ∼99.5 | 0.1 (1) | 0 | 0.4 (13) |
Nitrofurantoin was not tested in isolates of nonurine origin.
E. coli ATCC 25922 (of nonurine origin) and nine clinical E. coli isolates, of which two were of nonurine origin.
Isolates tested in three replicate tests at three distinct sites in three different days.
Reproducibility was acceptable if the results showed ≥95% category agreement compared to the test mode (i.e., the most frequent test result for the isolate) as a reference. CA, categorical agreement; VMD, very major discrepancy; MD, major discrepancy; mD, minor discrepancy.
S, susceptible; I, intermediate; R, resistant.
Table 2.
Comparative performance metrics of colorimetric-plate method in susceptibility testing of Escherichia colid
| Antimicrobiala | Test method | No. of isolatesb | No. of testsa,b | No. of test results in interpretive category: |
Performance analysis result, % (no.)c |
||||
|---|---|---|---|---|---|---|---|---|---|
| S | I | R | CA | VMD, MD | mD | ||||
| Ampicillin | CLSI | 100 | 100 | 5 | 1 | 94 | |||
| CP | 100 | 100 | 5 | 0 | 95 | 99 | 0 | 1 (1) | |
| Ampicillin-sulbactam | CLSI | 100 | 100 | 38 | 15 | 47 | |||
| CP | 100 | 100 | 40 | 12 | 48 | 97 | 0 | 3 (3) | |
| Cefazolin | CLSI | 100 | 100 | 50 | 1 | 49 | |||
| CP | 100 | 100 | 50 | 4 | 46 | 97 | 0 | 3 (3) | |
| Cefuroxime | CLSI | 100 | 100 | 54 | 0 | 46 | |||
| CP | 100 | 100 | 54 | 0 | 46 | 100 | 0 | 0 | |
| Ceftriaxone | CLSI | 100 | 100 | 53 | 4 | 43 | |||
| CP | 100 | 100 | 53 | 5 | 42 | 99 | 0 | 1 (1) | |
| Cefepime | CLSI | 100 | 100 | 89 | 8 | 3 | |||
| CP | 100 | 100 | 90 | 7 | 3 | 99 | 0 | 1 (1) | |
| Imipenem | CLSI | 100 | 100 | 100 | 0 | 0 | |||
| CP | 100 | 100 | 100 | 0 | 0 | 100 | 0 | 0 | |
| Amikacin | CLSI | 100 | 100 | 97 | 0 | 3 | |||
| CP | 100 | 100 | 96 | 1 | 3 | 99 | 0 | 1 (1) | |
| Gentamicin | CLSI | 100 | 100 | 65 | 0 | 35 | |||
| CP | 100 | 100 | 65 | 0 | 35 | 100 | 0 | 0 | |
| Ciprofloxacin | CLSI | 100 | 100 | 43 | 1 | 56 | |||
| CP | 100 | 100 | 43 | 1 | 56 | 100 | 0 | 0 | |
| Co-trimoxazole | CLSI | 100 | 100 | 41 | 0 | 59 | |||
| CP | 100 | 100 | 41 | 0 | 59 | 100 | 0 | 0 | |
| Nitrofurantoin | CLSI | 68 | 68 | 61 | 5 | 2 | |||
| CP | 68 | 68 | 61 | 5 | 2 | 100 | 0 | 0 | |
| All antimicrobials | CLSI | 100a | 1,168a,b | 696 | 35 | 437 | |||
| CP | 100a | 1,168a,b | 698 | 35 | 435 | ∼99.1 | 0 | ∼0.9 (10) | |
| FDA criteriac | ≥90 | ≤1.5, ≤3 | ≤5.5–10c | ||||||
Nitrofurantoin was not tested in isolates of nonurine origin.
Not including the quality control strain E. coli ATCC 25922.
FDA definitions, formulae, and acceptable performance criteria were used in performance analysis. Acceptable performance criteria for mD rate as an acceptable criterion or improved criterion were defined for the purposes of this study by the authors.
Abbreviations: CLSI, standard disk diffusion method of the Clinical and Laboratory Standards Institute; CP, colorimetric-plate method; S, susceptible; I, intermediate; R, resistant; CA, categorical agreement; VMD, very major discrepancy; MD, major discrepancy; mD, minor discrepancy; FDA, Food and Drug Administration.
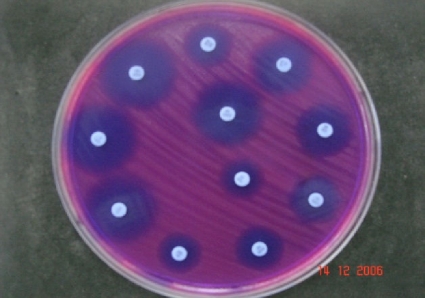
Fig. 1.
View of the in-house colorimetric plate for susceptibility testing of E. coli after an incubation period of 7 h. Due to the redox indicator resazurin, a background color change from blue (resazurin) to pink (resorufin) is observed in the zones of the plate where E. coli colonies grow.
For rapid antimicrobial susceptibility testing in daily laboratory work, there are various methods already available. One approach is using automated antimicrobial susceptibility test systems, providing results within 8 to 12 h, with proven performance and the added value of integrated data analysis and expert system utilities (3, 4, 11, 12, 27). As a noncommercial alternative, the in-house colorimetric-plate method produced results in 7 h with a minor extra cost to Mueller-Hinton agar medium. Additionally, the colorimetric-plate method has the advantage of potentially being used in gradient-based MIC detection methods on agar media, such as gradient plate, gradient strip, or agar dilution methods. Unlike automated antimicrobial susceptibility test systems, the colorimetric-plate method should also be usable in rapid synergy testing by the gradient strip method on agar media. However, such promising uses of the colorimetric-plate method require further research and evidence. There are also various automated zone reader platforms, at least one of which can be used with a commercially developed rapid disk diffusion susceptibility testing medium providing first results within 4 to 7 h (22). Recently, a rapid susceptibility testing medium for disk diffusion susceptibility testing was reported (19). This system is only commercially available. Previously, various other noncommercial rapid colorimetric assays employing oxidation-reduction indicators for antimicrobial susceptibility testing were described for various bacteria (1). However, MIC-based methods are technically demanding to prepare and perform and are not suitable for daily use. There are also chromogenic media commercially developed for the rapid detection of ESBL-producing organisms (14, 15).
In conclusion, the in-house colorimetric-plate method produced disk diffusion susceptibility test results rapidly and accurately in clinical E. coli isolates. However, the interlaboratory reproducibility of the colorimetric-plate method should be evaluated in the future. Further research should focus on analyzing the performance of the colorimetric-plate method in rapid ESBL detection and in rapid susceptibility testing of other important Gram-negative pathogens with an abbreviated identification procedure, potential use in rapid synergy testing, and clinical and economic impact in patient care (5, 11, 21).
Acknowledgments
There are no conflicts of interest to declare.
Footnotes
Published ahead of print on 19 January 2011.
REFERENCES
- 1. Acuner I. C., Eroglu C. 2006. Unacceptable performance and the lack of reproducibility results in the report of colorimetric methods for early detection of vancomycin and oxacillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 44:2318–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alam M. F., et al. 2009. The additional costs of antibiotics and re-consultations for antibiotic-resistant Escherichia coli urinary tract infections managed in general practice. Int. J. Antimicrob. Agents 33:255–257 [DOI] [PubMed] [Google Scholar]
- 3. Barenfanger J., Drake C., Kacich G. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 37:1415–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bayramoglu G., Acuner I. C., Sinirtas M., Gedikoglu S., Durupinar B. 2006. Performance evaluation of the BD Phoenix automated microbiology system in meropenem susceptibility testing of clinical Pseudomonas aeruginosa isolates. Saudi Med. J. 27:1921–1923 [PubMed] [Google Scholar]
- 5. Bruns D. E. 2001. Laboratory-related outcomes in healthcare. Clin. Chem. 47:1547–1552 [PubMed] [Google Scholar]
- 6. Cantón R., et al. 2008. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14(Suppl. 1):144–153 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI document M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2008. Abbreviated identification of bacteria and yeast; approved guideline, 2nd ed. CLSI document M35-A2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial disk susceptibility tests; approved standard, 10th ed. CLSI document M02-A10 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Denton M. 2007. Enterobacteriaceae. Int. J. Antimicrob. Agents 29(Suppl. 3):S9–S22 [DOI] [PubMed] [Google Scholar]
- 11. Doern G. V., Vautour R., Gaudet M., Levy B. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eigner U., Schmid A., Wild U., Bertsch D., Fahr A. M. 2005. Analysis of the comparative workflow and performance characteristics of the VITEK 2 and Phoenix systems. J. Clin. Microbiol. 43:3829–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erb A., Stürmer T., Marre R., Brenner H. 2007. Prevalence of antibiotic resistance in Escherichia coli: overview of geographical, temporal, and methodological variations. Eur. J. Clin. Microbiol. Infect. Dis. 26:83–90 [DOI] [PubMed] [Google Scholar]
- 14. Ercis S., et al. 2007. Rapid 4 to 6 hour detection of extended-spectrum beta-lactamases in a routine laboratory. Scand. J. Infect. Dis. 39:781–785 [DOI] [PubMed] [Google Scholar]
- 15. Färber J., et al. 2008. Extended-spectrum beta-lactamase detection with different panels for automated susceptibility testing and with a chromogenic medium. J. Clin. Microbiol. 46:3721–3727 (Erratum, 47:285, 2009.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Food and Drug Administration 2009. Guidance for industry and FDA. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems. Center for Devices and Radiological Health, Food and Drug Administration, U.S. Department of Health and Human Services, Silver Spring, MD [Google Scholar]
- 17. Kang C. I., et al. 2005. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 49:760–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kerremans J. J., et al. 2008. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J. Antimicrob. Chemother. 61:428–435 [DOI] [PubMed] [Google Scholar]
- 19. Kocagoz T., et al. 2007. Quicolor: a novel system for rapid antimicrobial susceptibility testing. Ann. Microbiol. 57:131–135 [Google Scholar]
- 20. Kollef M. H. 2000. Inadequate antimicrobial treatment: an important determinant of outcome for hospitalized patients. Clin. Infect. Dis. 31(Suppl. 4):S131–S138 [DOI] [PubMed] [Google Scholar]
- 21. Niederman M. S. 2001. Impact of antibiotic resistance on clinical outcomes and the cost of care. Crit. Care Med. 29(Suppl. 4):N114–N120 [DOI] [PubMed] [Google Scholar]
- 22. Nijs A., et al. 2003. Comparison and evaluation of Osiris and Sirscan 2000 antimicrobial susceptibility systems in the clinical microbiology laboratory. J. Clin. Microbiol. 41:3627–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peterson L. R. 2008. Antibiotic policy and prescribing strategies for therapy of extended-spectrum beta-lactamase-producing Enterobacteriaceae: the role of piperacillin-tazobactam. Clin. Microbiol. Infect. 14(Suppl. 1):181–184 [DOI] [PubMed] [Google Scholar]
- 24. Pfaller M. A., Segreti J. 2006. Overview of the epidemiological profile and laboratory detection of extended-spectrum beta-lactamases. Clin. Infect. Dis. 42(Suppl. 4):S153–S163 [DOI] [PubMed] [Google Scholar]
- 25. Pitout J. D., Laupland K. B. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8:159–166 [DOI] [PubMed] [Google Scholar]
- 26. Ramphal R., Ambrose P. G. 2006. Extended-spectrum beta-lactamases and clinical outcomes: current data. Clin. Infect. Dis. 42(Suppl. 4):S164–S172 [DOI] [PubMed] [Google Scholar]
- 27. Richter S. S., Ferraro M. J. 2007. Susceptibility testing instrumentation and computerized expert systems for data analysis and interpretation, p. 245–256 In Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. (ed.), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC [Google Scholar]
- 28. Rodríguez-Baño J., et al. 2006. Clinical and molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli as a cause of nosocomial infection or colonization: implications for control. Clin. Infect. Dis. 42:37–45 [DOI] [PubMed] [Google Scholar]
- 29. Rossolini G. M., Mantengoli E. 2008. Antimicrobial resistance in Europe and its potential impact on empirical therapy. Clin. Microbiol. Infect. 14(Suppl. 6):2–8 [DOI] [PubMed] [Google Scholar]
- 30. Schwaber M. J., et al. 2006. Clinical and economic impact of bacteremia with extended-spectrum-beta-lactamase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 50:1257–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. von Baum H., Marre R. 2005. Antimicrobial resistance of Escherichia coli and therapeutic implications. Int. J. Med. Microbiol. 295:503–511 [DOI] [PubMed] [Google Scholar]
- 32. Warren R. E., Harvey G., Carr R., Ward D., Doroshenko A. 2008. Control of infections due to extended-spectrum beta-lactamase-producing organisms in hospitals and the community. Clin. Microbiol. Infect. 14(Suppl. 1):124–133 [DOI] [PubMed] [Google Scholar]