Abstract
A culture confirmation test for the detection of Mycobacterium tuberculosis complex strains that uses a lateral-flow immunochromatographic assay to detect the MPB64 antigen, the MGIT TBc identification (TBc ID) test, has been developed. We evaluated the performance of the TBc ID test in the detection of the M. tuberculosis complex in 222 primary-positive liquid cultures. We compared these results to those of nucleic acid-based identification and conventional biochemical tests. The validity of the TBc ID test was determined, and all of the nontuberculous mycobacteria (NTM) and Nocardia species tested were found to be negative. The detection limit of the TBc ID test was 5 × 105 CFU/ml, and for IS6110 real-time PCR it was 5 CFU/ml. All of the M. tuberculosis and M. africanum cultures were found to be positive, while M. bovis and M. bovis BCG cultures were negative. With the exception of 1 contaminated culture, the 221 culture-positive isolates contained 171 (77.5%) M. tuberculosis isolates, 39 (17.6%) NTM species, and 11 (5.0%) unidentified species. Two culture-positive isolates harbored a 63-bp deletion at position 196 of the mpb64 gene. The sensitivity, specificity, positive predictive values, and negative predictive values of the TBc ID test were 98.8, 100, 100, and 95.1%, respectively. Furthermore, the approximate turnaround time for real-time PCR was 4 h (including buffer and sample preparation), while for the TBc ID test it was less than 1 h. We suggest an algorithm for the primary identification of M. tuberculosis in liquid culture using the TBc ID test as an alternative to conventional subculture followed by identification using biochemical methods.
INTRODUCTION
In 2007, the World Health Organization (WHO) adopted a policy that recommended the use of liquid culture methods for culture and drug susceptibility tests as a standard for tuberculosis (TB) diagnosis and case management (28). The Taiwan Centers for Disease Control (CDC) recommended that liquid and solid media be used simultaneously for mycobacterial culture (7), and approximately 90% of clinical mycobacteriology laboratories in Taiwan use a liquid culture system for the isolation of the Mycobacterium tuberculosis complex from clinical specimens. Acid-fast bacillin (AFB) smear tests then are performed on positive cultures to dismiss contamination (4, 20). The turnaround time (TAT) for the recovery of the M. tuberculosis complex thus is reduced to 10 to 14 days (8, 18). Although the recovery of mycobacteria can be accelerated by using liquid culture systems, this practice provides only partial benefits if it is not accompanied by a rapid species identification test (16). Differentiating M. tuberculosis from nontuberculous mycobacteria (NTM) as soon as possible is important, particularly in situations in which NTM strains represent a considerable share of the clinical isolates.
The identification of M. tuberculosis is time-consuming using conventional biochemical methods. The subculturing of isolated mycobacteria from liquid cultures onto solid media and their subsequent identification using conventional biochemical methods requires an additional 3 to 5 weeks (25). In addition, ambiguous biochemical reactions can confuse the test results. Thus, the identification of M. tuberculosis using biochemical methods is a complex, labor-intensive, and time-consuming process. Nucleic acid amplification (NAA) methods, such as real-time PCR, are both rapid and specific but are technically challenging, and they require the use of sophisticated instruments. For this reason, the WHO also recommends the use of rapid and affordable methods for the identification to the species level of the M. tuberculosis complex and NTM organisms (http://www.who.int/tb/dots/laboratory/policy/en/index.html).
The Taiwan CDC conducted a TB laboratory diagnosis survey in 2009. Their results showed that there are 36 clinical mycobacteriology laboratories in the country that perform four conventional bacteriological tests (smear, culture, identification, and drug susceptibility testing) for TB diagnosis. Of these 36 laboratories, 19 (52.8%) use biochemical tests, 9 (25%) use NAA, and 8 (22.2%) use both methods for the culture identification of M. tuberculosis complex strains. Among these same laboratories, 13 use commercial molecular diagnosis assays, and 10 perform in-house PCR using IS6110 (11) or other probes. Therefore, to strengthen TB laboratory services and facilitate the timely management of TB cases, the implementation of a simple and reliable test is needed in Taiwan.
In August 2009, BD Diagnostics (a division of Becton, Dickson, and Company) launched the BD MGIT TBc identification (TBc ID) test for the identification of the M. tuberculosis complex from liquid culture in Africa and the European Union. The TBc ID test is a lateral-flow immunochromatographic assay based on the detection of MPB64 in liquid cultures using an MPT64-specific monoclonal antibody. MPB64 is a mycobacterial protein that is secreted by M. tuberculosis and certain strains of M. bovis (1, 22, 29). However, some substrains of M. bovis BCG in the M. tuberculosis complex produce no MPT64 antigen (19). This qualitative test is rapid (readable in 15 min), easy to use, and requires no processing or additional instrumentation. No clinical trial of the TBc ID test was ever reported. In this study, we evaluated the performance of the TBc ID, NAAs (such as IS6110 real-time PCR), and biochemical tests for the identification of culture-positive mycobacteria in liquid media.
MATERIALS AND METHODS
Reference strains, clinical specimens, and mycobacterial culture.
The validation of the TBc ID test was conducted using 24 NTM strains, 18 mixtures of M. tuberculosis and NTM strains, 2 M. bovis strains, 1 M. africanum strain, and 1 Nocardia strain. For prospective analysis, clinical respiratory specimens were digested and decontaminated using N-acetyl-l-cysteine (Sigma Chemical Company, St. Louis, MO) and 2% sodium hydroxide (NaLC-NaOH). The detection limits of both the TBc ID test and real-time PCR were evaluated using serial dilutions of an M. tuberculosis stock (5 × 107 CFU/ml). We performed 10-fold serial dilutions from stocked H37Rv DNA (5 × 107 CFU/ml) to 0.05 CFU/ml as the lowest concentration, and we separately evaluated the limit of each assay. The procedures were repeated twice. The processed specimens were concentrated using centrifugation, inoculated in Bactec MGIT 960 culture tubes, and incubated in the BD Bactec MGIT system (Becton Dickinson Microbiology Systems, Cockeysville, MD) (10). Once a positive signal was detected using the system, a smear microscopy test was performed to screen the deposit in the culture tubes for AFB using an auramine fluorescent stain. The results were confirmed after staining using the Ziehl-Neelsen method, and samples were classified as either scanty, 1+, 2+, 3+, or 4+ (3). From January to February 2010, consecutive culture-positive MGIT samples recovered from 3,214 clinical specimens were included in this study.
Assays for culture-positive MGIT media.
Both TBc ID and conventional biochemical tests were performed in the TB Laboratory at the Taipei Medical University-Wan Fang Hospital, one of the contracted mycobacteriology laboratories of the Taiwan CDC. NAA tests and sequencing were performed in the Reference Laboratory of Mycobacteriology at the Taiwan CDC.
(i) Conventional biochemical tests.
Bactec cultures that were AFB positive were subcultured onto solid Löwenstein-Jensen (L-J) slants and incubated at 37°C to obtain colonies for identification. Conventional biochemical tests, including niacin production, nitrate reduction, 3-day acrylsulfatase, 3-day Tween 80 hydrolysis, urease, semiquantitative catalase, and tolerance to 5% NaCl, were performed to identify the M. tuberculosis complex (6).
(ii) NAA tests.
For the identification of M. tuberculosis complex strains in AFB-positive Bactec cultures (2 to 3 days after a positive signal was detected), IS6110 real-time PCR was performed according to the method of Cleary et al. (9). The primers used for this assay were IS6 (5′-GGCTGTGGGTAGCAGACC-3′) and IS7 (5′-CGGGTCCAGATGGCTTGC-3′), as well as an internal oligonucleotide probe (5′-[6-carboxyfluorescein]-TGTCGACCTGGGCAGGGTTCG-[6-carboxytetramethylrhodamine]-3′).
(iii) TBc ID.
TBc ID devices were inoculated with 0.1 ml of Bactec cultures within 3 days of the detection of AFB-positive growth using an MGIT 960 instrument. Cultures were used directly for the TBc ID assay according to the manufacturer's recommendations (5). All inoculated devices were incubated for 15 min at room temperature before the results were visually assessed for positive detection (i.e., a visible test line) and reagent function (i.e., a visible control line).
(iv) PCR-RFLP.
To identify mycobacteria at the species level, we performed PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of a 65-kDa protein according to the method described by Telenti et al. (26). We compared the obtained patterns to those in the database at http://app.chuv.ch/prasite/index.html (Prasite; Centre Hospitalier Universitaire Vaudois Lausanne).
Analysis of discrepant results.
Conventional confirmation tests were considered the gold standard for performance calculations. When discrepant results between the NAA, TBc ID assays, and conventional biochemical confirmation tests were observed, the results from mpb64 gene sequencing were used to resolve the discrepancies. Mutations in the mpb64 gene were analyzed using PCR amplification, and the resulting product was sequenced using the following oligonucleotide primers: AMS-50-F (5′-TCGATCTGCTAGCTTGAGTCTGGT-3′) and AMS-51-R (5′-ACCACCGCACCAAGGCTGCTGTCTA-3′) (23). The PCR products were sequenced using an ABI 3730 automated sequencer (Applied Biosystems, Life Technologies Corporation, CA) under standardized conditions. The data were analyzed using Sequencing Analysis 5.2.0 software (Applied Biosystems, Life Technologies Corporation, CA).
Performance analysis.
Using the results of the conventional biochemical identification methods as the gold standard, the performance of the TBc ID and IS6110 real-time PCR tests was analyzed. To assess the performance of these identification tests, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated after discrepant analysis (2). We also evaluated the TAT of the tests based on the protocols suggested by the manufacturers and the Taiwan CDC.
RESULTS
Validity of the TBc ID test.
The detection limits of the TBc ID and IS6110 real-time PCR tests were 5 × 105 and 5 CFU/ml, respectively. The results of the identification of reference mycobacterial strains are summarized in Table 1. The analytical specificity of the TBc ID test was assessed using reference strains. Using the TBc ID test, 3 M. tuberculosis strains, 1 M. africanum strain, and 18 mixtures of M. tuberculosis and NTM strains were identified as positive, while 18 NTM strains, 1 Nocardia strain, 1 Gordonia sp., 1 M. bovis strain, and 1 M. bovis BCG strain were identified as negative.
Table 1.
Validation of the BD MGIT TBc identification test using reference mycobacterial strains
| Mycobacterium sp. (no.) | TBc ID | Real-time PCR |
|---|---|---|
| M. tuberculosis (3) | + | + |
| M. africanum TMC 5122 [Rist 3414] | + | + |
| M. bovis TMC1011 [BCG Pasteur] | − | + |
| M. bovis [BCG Pasteur] | − | + |
| M. kansasii TMC 1201 | − | − |
| M. intracellulare TMC 1411 [P-54; Wilson] | − | − |
| M. avium 1982 [McKee 1] | − | − |
| M. simiae 3055 [N14] | − | − |
| M. haemophilum 1 [TMC 804] | − | − |
| M. gordonae TMC 1325 [Kowal] | − | − |
| M. fortuitum [TMC 1529] | − | − |
| M. malmoense Mo 816 [CIP 105775; TMC 802] | − | − |
| M. marinum Aronson [TMC 1218] | − | − |
| M. chelonae TMC 1544 [Friedmann] | − | − |
| M. abscessus CDC T-2366-6 [PS 308] | − | − |
| M. flavescens D-25 [TMC 1541] | − | − |
| M. triviale [V subgroup of Runyon group III] | − | − |
| M. xenopi TMC 1482 | − | − |
| M. tuberculosis and NTM (18) | + | + |
| Nocardia sp. | − | − |
| Gordonia sp. | − | − |
Mycobacterial species identification.
We obtained 221 (6.9%) AFB smear-positive Bactec cultures out of 3,214 different clinical specimens. However, 11 cultures (5.0%) yielded no colonies after being subcultured onto L-J slants; thus, no identification results were obtained for these cultures. We identified 210 L-J culture-positive samples using standard biochemical tests: 171 (81.4%) were culture positive for M. tuberculosis, and 39 (18.6%) were culture positive for NTM. The most prominent NTM species identified using PCR-RFLP were M. abscessus (48.7%), M. kansasii (15.4%), M. gordonae (10.3%), and M. intracellulare (7.7%) (Table 2). None of the 210 culture-positive samples collected from patients were identified as containing mixed cultures of M. tuberculosis and NTM.
Table 2.
Comparison of the IS6110 real-time PCR and BD MGIT TBc identification tests to culture confirmation tests
| Mycobacterial species identified using biochemical methods and hsp65 PCR-RFLPa (no. of specimens) | IS6110 real-time PCR (no. of specimens = 210) |
BD MGIT TBc identification test (no. of specimens = 210) |
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| M. tuberculosis (171) | 171 | 0 | 169 | 2 |
| NTM (39) | 0 | 39 | 0 | 39 |
| M. abscessus (19) | 19 | 19 | ||
| M. kansasii (6) | 6 | 6 | ||
| M. gordonae (4) | 4 | 4 | ||
| M. intracellulare (3) | 3 | 3 | ||
| M. arupense (1) | 1 | 1 | ||
| M. avium (1) | 1 | 1 | ||
| M. chelonae (1) | 1 | 1 | ||
| M. fallax (1) | 1 | 1 | ||
| M. fortuitum (1) | 1 | 1 | ||
| M. holsaticum (1) | 1 | 1 | ||
| M. nonchromogenicum (1) | 1 | 1 | ||
A total of 222 specimens were primary positive in liquid culture, 11 were culture negative on L-J medium, and 1 was contaminated.
TBc ID and NAA tests.
The results of the NAA and TBc ID tests were compared to the culture results, and the results are summarized in Table 2. The NAA test performed as well as gold-standard biochemical methods. In contrast, the TBc ID test successfully yielded 39 true-negative results for 39 NTM cultures and 2 false-negative results for M. tuberculosis cultures. The gene sequencing of the two false-negative samples revealed a 63-bp deletion at position 196 in the mpb64 gene in both. In addition, of the 11 AFB smear-positive Bactec cultures with no L-J growth, the results of all of the TBc ID tests were negative, while one true-positive NAA test result was observed. This false-negative TBc ID result also was seen in tests of two other specimens from the same patient that yielded culture-positive M. tuberculosis. Of the 39 samples that were identified as negative by the TBc ID and NAA tests, each tested positive for NTM (Table 2).
Correlation between tests.
Compared to conventional biochemical methods, the sensitivity of the TBc ID test was 98.8%, the specificity was 100%, the positive predictive value was 100%, and the negative predictive value was 95.1% (Table 3). In contrast, the same four performance indexes all were 100% for the NAA method. For the primary identification of M. tuberculosis from liquid culture, the TAT was 1 h using the TBc ID test and 1 to 3 days using the NAA test; in contrast, the average TAT for conventional biochemical tests was 24 days (range, 7 to 49 days).
Table 3.
Correlation between culture confirmation assays and culture results
| Assay (no. of samples) | No. of cultured specimens with indicated result |
Sensitivity (%) | Specificity (%) | Predictive value (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
M. tuberculosis |
NTM |
|||||||||
| All | Assay (+) | Assay (−) | All | Assay (+) | Assay (−) | Positive | Negative | |||
| IS6110 real-time PCR (210) | 171 | 171 | 0 | 39 | 0 | 39 | 100 | 100 | 100 | 100 |
| BD MGIT TBc identification test (210) | 171 | 169 | 2 | 39 | 0 | 39 | 98.8 | 100 | 100 | 95.1 |
DISCUSSION
In mycobacterial laboratories in Taiwan, the average recovery rate of the M. tuberculosis complex in all clinical specimens has decreased from approximately 70% in 2000 to 50% in 2009 (unpublished data). Therefore, a fast and cost-effective method that allows for the qualitative detection of the M. tuberculosis complex from acid-fast bacillus (AFB)-positive cultures is necessary. We conducted and reported the first clinical trial of the TBc ID test. We evaluated the use of a simple MPB64-based lateral-flow immunochromatographic assay, the TBc ID test, to enhance TAT and facilitate the rapid identification of the M. tuberculosis complex directly from liquid culture. The lateral-flow immunochromatographic assay is useful because of its specificity and ease of use. Thus, several such tests are commercially available, including the Capilia TB assay (Tauns Laboratories, Inc., Numazu, Japan), the Tibilia rapid test (Hangzhou, China), the SD Bioline TB Ag MPT64 rapid test (Standard Diagnostics, South Korea), and the MGIT TBc ID test. Previous studies have demonstrated that these assays all had outstanding performance, exhibiting 92.4 to 100% sensitivity and 94 to 100% specificity in identifying the M. tuberculosis complex (Table 4) (13, 14, 21, 23, 24, 27, and M. Warns et al., presented at the 30th Annual Congress of the European Society of Mycobacteriology, Porto, Portugal, July 2009). In this study, we found that the TBc ID test exhibited 98.8% sensitivity and 100% specificity compared to biochemical tests. However, to improve the accuracy of the identifications and confirm negative TBc ID test results, a secondary NAA test must be employed after reculturing MGIT-positive cultures onto solid medium.
Table 4.
Summary of the performance of MPB64-based lateral-flow immunochromatographic assays in the detection of Mycobacterium tuberculosis in different studies
| Category | Study method | No. of NTM with positive results | Species (no.) | No. of MTB with negative results | Mutation(s) in mpb64 gene (no. of isolates) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Reference | Country(ies) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Capilia TB assay | 0 | 3 | 63-bp deletion at 196 (3), 1-bp deletion at 266 (1), G→A at 402 (2), IS6110 insertion at 501 (1), 176-bp deletion at 512 (5) | 99.2 | 100 | 14 | Japan | |||
| Accuprobe | 0 | 0 | 100 | 100 | |||||||
| 2 | Capilia TB assay | 0 | 3 | C insertion at 287 (1), A→T at 388 (1), IS6110 insertion at 177 (1) | 92.4 | 100 | 13 | Germany | |||
| 3 | Capilia TB assay | 2 | MAC (1), M. chelonae (1) | 2 | 98.6 | 97.9 | 98.6 | 97.9 | 27 | Taiwan | |
| BD ProbeTec ET system | 2 | M. fortuitum (1), M. triviale (1) | 3 | 97.3 | 97.1 | 98.2 | 95.8 | ||||
| 4 | Capilia TB assay | 1 | MTBC and NTM mix | 6 | 63-bp deletion at 196 (5), 2-bp insertion at 436 (1) | 97 | 100 | 23 | Thailand | ||
| 5 | Capilia TB assay | 1 | M. intracellulare (1) | 5 | 96.9 | 98.6 | 99.4 | 93.5 | 24 | Taiwan | |
| BD ProbeTec ET system | 2 | M. intracellulare (1), M. terrae (1) | 1 | 99.4 | 97.3 | 98.8 | 98.6 | ||||
| 6 | MGIT TBc ID Test | 0 | 0 | 100 | 100 | M. Warns et al., presented at the 30th Annual Congress of the European Society of Mycobacteriology, Porto, Portugal, July 2009 | |||||
| Capilia TB assay | 1 | M. marinum (1) | 0 | 100 | 94 | ||||||
| SD bioline TB Ag MPT64 Rapid Test | 1 | M. gastri (1) | 0 | 100 | 94 | ||||||
| 7 | Capilia TB assay | 2 | M. gordonae (1), unidentified NTM (1) | 1 | GC insertion (1) | 99.6 | 99.5 | 7 | Zambia, South Africa | ||
| 8 | MGIT TBc ID Test | 0 | 2 (from 1 patient) | 63-bp deletion at 196, downstream 50 A→G (2) | 98.8 | 100 | 100 | 95.1 | This study | Taiwan | |
| IS6110 real-time PCR | 0 | 0 | 100 | 100 | 100 | 100 |
The specificity of the TBc ID test was shown to be 100% in two studies. However, cross-reactivity with M. avium complex (MAC), M. chelonae, M. intracellulare, M. marinum, and M. gordonae was observed with the Capilia test, and cross-reactivity was observed with M. gastri with the Bioline device (Table 4). The sensitivity of the TBc ID test ranged from 98.8 to 100%, while the sensitivity of the Capilia test ranged from 92.4 to 99.6% in similar studies. In our study, the TBc ID test yielded two false-negative results for two M. tuberculosis complex cultures from the same patient, using biochemical tests as a gold standard. These two isolates were confirmed as true positives using IS6110 real-time PCR and were found to contain a 63-bp deletion at position 196 of the mpb64 gene. Mutations (13, 14), deletions (14, 23), and IS6110 or CG insertions (13, 14, 21, 23) in the coding region (22, 29) of the mpb64 gene that result in false-negative test results have been reported previously.
A recent clinical evaluation performed in South Africa revealed that the TBc ID test exhibited excellent performance compared to that of the niacin test (21) (Table 4). Similar results were obtained using the BD ProbeTec ET system (BD Diagnostic Systems, Sparks, MD) in two studies performed in Taiwan (24, 27). However, both the Gen Probe AccuProbe and IS6110 real-time PCR tests outperformed the TBc ID, which occasionally was cross-reactive with NTM strains or yielded false negatives for M. tuberculosis complex species with mutations in the mpb64 gene (15). Furthermore, the immunochromatographic assay was less labor-intensive than other identification methods. In this study, the TAT for identification was 24 days in the clinical setting using standard biochemical tests, and it was 1 day for the TBc ID test. Thus, to avoid false-negative results and shorten the TAT of routine clinical practices, we suggest a new algorithm for the primary identification of M. tuberculosis strains from liquid culture that uses the TBc ID test as an alternative to the currently available biochemical methods (Fig. 1).
Fig. 1.
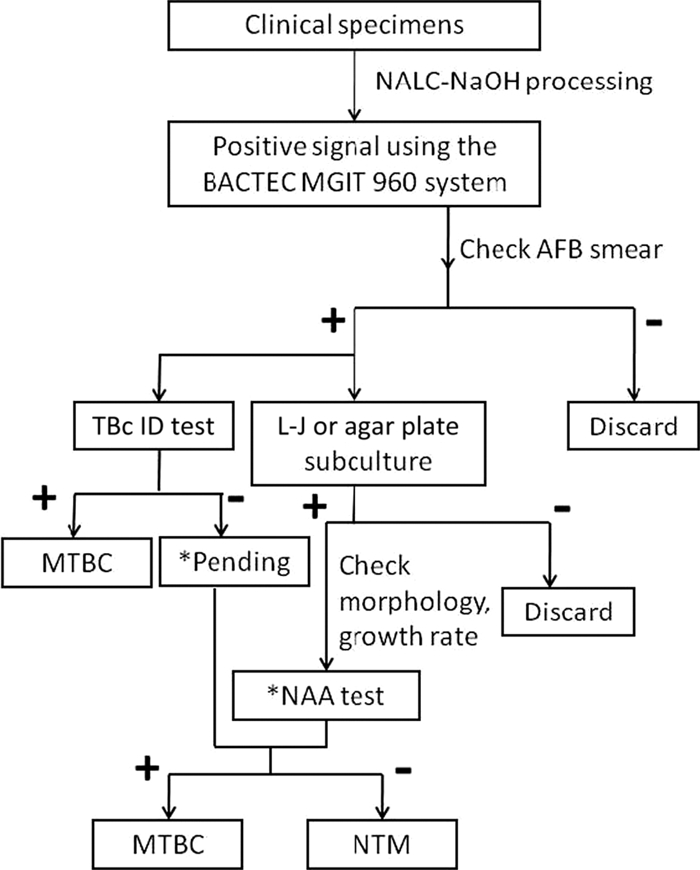
Suggested algorithm for the culture and identification of the Mycobacterium tuberculosis complex. The asterisk indicates that for pending samples, a nucleic acid amplification (NAA) test should be performed when the morphology and growth rate suggest the presence of the M. tuberculosis complex.
Taiwan is considered a country with moderate TB prevalence, with an incidence rate of 62 per 100,000 in 2008 (http://www.cdc.gov.tw/public/Data/9123117221971.pdf). However, an increased incidence of NTM infections, from 32.3% in 2000 to 49.8% in 2008, was reported in a recent hospital-based survey; this rise in NTM infections was due primarily to M. avium and M. abscessus infections (17). Therefore, to avoid inappropriate antituberculous treatment decisions, the accurate differential diagnosis of M. tuberculosis is imperative. The increase in the number of NTM isolations has been attributed to the implementation of liquid culture systems (12) and the use of improved identification/differentiation techniques. In this study, we proved that the TBc ID test could unambiguously differentiate M. tuberculosis from NTM strains and mixtures of both in a clinical setting.
In conclusion, the use of liquid culture systems together with a rapid, simple identification test has the potential to increase the speed of diagnosis and, most importantly, aid in the identification of drug-resistant TB cases. Our findings suggest that lateral-flow immunochromatographic assays for the detection of MPB64, such as the TBc ID test, are crucial for the rapid and accurate diagnosis of TB to facilitate early treatment and reduce the spread of infections.
ACKNOWLEDGMENTS
We thank colleagues at the Laboratory of Mycobacteriology of the Taipei Medical University-Wan Fang Hospital for their technical assistance and BD Taiwan for providing the 250 MGIT TBc identification test kits to the hospital.
Footnotes
Published ahead of print on 29 December 2010.
REFERENCES
- 1. Abe C., Hirano K., Tomiyama T. 1999. Simple and rapid identification of the Mycobacterium tuberculosis complex by immunochromatographic assay using anti-MPB64 monoclonal antibodies. J. Clin. Microbiol. 37:3693–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altman D. G., Bland J. M. 1994. Statistics notes: diagnostic tests 2: predictive values. BMJ 309:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. American Thoracic Society 1981. Diagnostic standards and classification of tuberculosis and other mycobacterial disease. Am. Rev. Respir. Dis. 123:343–358 [DOI] [PubMed] [Google Scholar]
- 4. Attorri S., Dunbar S., Clarridge J. E. I., Jr 2000. Assessment of morphology for rapid presumptive identification of Mycobacterium tuberculosis and Mycobacterium kansasii. J. Clin. Microbiol. 38:1426–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. BD Diagnostic Systems 2009. BD MGIT™ TBc ID identification test package insert, BD document L8085917(01). BD Diagnostic Systems, Sparks, MD [Google Scholar]
- 6. Centers for Disease Control 1981. Public health mycobacteriology. A guide for the level II laboratory. Centers for Disease Control, Atlanta, GA [Google Scholar]
- 7. Centers for Disease Control, Taipei, Taiwan 2004. Laboratory manual of mycobacteria. Centers for Disease Control, Taipei, Taiwan [Google Scholar]
- 8. Chien H. P., Yu M. C., Wu M. H., Lin T. P., Luh K. T. 2000. Comparison of the Bactec MGIT 960 with Lowenstein-Jensen medium for recovery of mycobacteria from clinical specimens. Int. J. Tuberc. Lung Dis. 4:866–870 [PubMed] [Google Scholar]
- 9. Cleary T., Roudel G., Casillas O., Miller N. 2003. Rapid and specific detection of Mycobacterium tuberculosis by using the smart cycle instrument and a specific fluorogenic probe. J. Clin. Microbiol. 41:4783–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanna B. A., et al. 1999. Multicenter evaluation of the Bactec MGIT 960 system for recovery of mycobacteria. J. Clin. Microbiol. 37:748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hermans P. W., et al. 1990. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol. 28:2051–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hernández-Garduño E., Rodrigues M., Elwood R. K. 2009. The incidence of pulmonary non-tuberculous mycobacteria in British Columbia, Canada. Int. J. Tuberc. Lung Dis. 13:1086–1093 [PubMed] [Google Scholar]
- 13. Hillemann D., Rüsch-Gerdes S., Richter E. 2005. Application of the Capilia TB assay for culture confirmation of Mycobacterium tuberculosis complex isolates. Int. J. Tuberc. Lung Dis. 9:1409–1411 [PubMed] [Google Scholar]
- 14. Hirano K., Aono A., Takahashi M., Abe C. 2004. Mutations including IS6110 insertion in the gene encoding the MPB64 protein of Capilia TB-negative Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 42:390–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ismail N. A., Baba K., Pombo D., Hoosen A. A. 2009. Use of an immunochromatographic kit for the rapid detection of Mycobacterium tuberculosis from broth cultures. Int. J. Tuberc. Lung Dis. 13:1045–1047 [PubMed] [Google Scholar]
- 16. Katila M. L., Katila P., Erkinjuntti-Pekkanen R. 2000. Accelerated detection and identification of mycobacteria with MGIT 960 and COBAS AMPLICOR Systems. J. Clin. Microbiol. 38:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lai C. C., et al. 2010. Increasing incidence of nontuberculous mycobacteria, Taiwan, 2000–2008. Emerg. Infect. Dis. 16:294–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J. J., et al. 2003. Comparative evaluation of the Bactec MGIT 960 system with solid medium for isolation of mycobacteria. Int. J. Tuberc. Lung Dis. 7:569–574 [PubMed] [Google Scholar]
- 19. Li H., et al. 1993. Evidence for absence of the MPB64 gene in some substrains of Mycobacterium bovis BCG. Infect. Immun. 61:1730–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarter Y. S., Ratkiewicz I. N., Robinson A. 1998. Cord formation in Bactec medium is a reliable, rapid method for presumptive identification of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 36:2769–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muyoyeta M., et al. 2010. Evaluation of the Capilia TB assay for culture confirmation of Mycobacterium tuberculosis in Zambia and South Africa. J. Clin. Microbiol. 48:3773–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagai S., Wiker H. G., Harboe M., Kinomoto M. 1991. Isolation and partial characterization of major protein antigens in the culture fluid of Mycobacterium tuberculosis. Infect. Immun. 59:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ngamlert K., et al. 2009. Diagnostic performance and costs of Capilia TB for Mycobacterium tuberculosis complex identification from broth-based culture in Bangkok, Thailand. Trop. Med. Int. Health 14:748–753 [DOI] [PubMed] [Google Scholar]
- 24. Shen G. H., et al. 2009. Combining the Capilia TB assay with smear morphology for the identification of Mycobacterium tuberculosis complex. Int. J. Tuberc. Lung Dis. 13:371–376 [PubMed] [Google Scholar]
- 25. Sun J. R., Lee S. Y., Perng C. L., Lu J. J. 2009. Detecting Mycobacterium tuberculosis in Bactec MGIT 960 cultures by inhouse IS6110-based PCR assay in routine clinical practice. J. Formos. Med. Assoc. 108:119–125 [DOI] [PubMed] [Google Scholar]
- 26. Telenti A., et al. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J. Y., et al. 2007. Performance assessment of the Capilia TB assay and the BD ProbeTec ET system for rapid culture confirmation of Mycobacterium tuberculosis. Diagn. Microbiol. Infect. Dis. 59:395–399 [DOI] [PubMed] [Google Scholar]
- 28. World Health Organization 2007. Use of liquid TB culture and drug susceptibility testing (DST) in low and medium income settings. Summary report of the expert group meeting on the use of liquid culture media. WHO, Geneva, Switzerland [Google Scholar]
- 29. Yamaguchi R., et al. 1989. Cloning and characterization of the gene for immunogenic protein MPB64 of Mycobacterium bovis BCG. Infect. Immun. 57:283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]


