Abstract
Fluoroquinolones (FQ) are important second-line drugs to treat tuberculosis; however, FQ resistance is an emerging problem. Resistance has been mainly attributed to mutations in a 21-bp region of the Mycobacterium tuberculosis gyrA gene, often called the quinolone resistance-determining region (QRDR). We have developed a simple, rapid, and specific assay to detect FQ resistance-determining QRDR mutations. The assay amplifies the M. tuberculosis gyrA QRDR in an asymmetrical PCR followed by probing with two sloppy molecular beacons (SMBs) spanning the entire QRDR. Mutations are detected by melting temperature (Tm) shifts that occur when the SMBs bind to mismatched sequences. By testing DNA targets corresponding to all known QRDR mutations, we found that one or both of the SMBs produced a Tm shift of at least 3.6°C for each mutation, making mutation detection very robust. The assay was also able to identify mixtures of wild-type and mutant DNA, with QRDR mutants identified in samples containing as little as 5 to 10% mutant DNA. The assay was blindly validated for its ability to identify the QRDR mutations on DNA extracted from clinical M. tuberculosis strains. Fifty QRDR wild-type samples, 34 QRDR mutant samples, and 8 heteroresistant samples containing mixtures of wild-type and mutant DNA were analyzed. The results showed 100% concordance to conventional DNA sequencing, including a complete identification of all of the mixtures. This SMB Tm shift assay will be a valuable molecular tool to rapidly detect FQ resistance and to detect the emergence of FQ heteroresistance in clinical samples from tuberculosis patients.
INTRODUCTION
The flouroquinolone (FQ) antibiotics have become increasingly important in the treatment of tuberculosis (TB). The FQs are already a critical component of second-line therapy against multidrug-resistant (MDR) TB (3, 14, 31). They are also under study for use in first-line treatment of TB due to their potency and potential sterilizing activity (9, 24). Unfortunately, FQ resistance is being observed with increasing frequency in Mycobacterium tuberculosis isolates that are otherwise fully drug susceptible (15) and in isolates that are already MDR (17, 28, 30). FQ resistance is also an important part of the case definition of extensively drug-resistant (XDR) TB (14–17, 25). These emerging trends have placed a new urgency on the development of rapid methods to detect FQ resistance.
In M. tuberculosis, FQ resistance appears to be principally caused by single-nucleotide polymorphisms in the M. tuberculosis gyrA gene. Between 60 and 90% of FQ-resistant clinical M. tuberculosis isolates have mutations in a short 21-bp “quinolone resistance-determining region” (QRDR) of gyrA, particularly in codons 90, 91, and 94. Mutations in the M. tuberculosis gyrB gene are also associated with FQ resistance, but at a much lower frequency and usually in association with gyrA mutations (9, 12, 18, 21, 23, 27). While most M. tuberculosis strains with gyrA QRDR mutations are FQ resistant, virtually all strains that are wild type in this region are FQ susceptible. The exception to this rule is a known polymorphism at gyrA codon 95. FQ-susceptible M. tuberculosis strains can have either a threonine (T) or a serine (S) allele at this location (95S or 95T, respectively) (14). Consequently, a molecular test that can differentiate between the two wild-type QRDR sequences and any other possible QRDR mutation is able to identify FQ resistance with high sensitivity and specificity. In fact, the gyrA QRDR has emerged as an important biomarker for rapid detection of FQ resistance in M. tuberculosis and is increasingly being used as a target for rapid molecular drug susceptibility tests (6, 13, 20, 21, 26, 29).
A number of molecular methods to detect gyrA QRDR mutations have been described recently, including line probe assays, locked nucleic acid probes, multiplex PCR amplimer conformation analysis (MPAC), denaturing high-performance liquid chromatography (HPLC) heteroduplex analysis, and direct sequencing (6, 11, 13, 20, 21, 26, 29). However, most of these methods are technically challenging and involve handling of PCR amplicons in open tubes, which can lead to amplicon cross-contamination and diminished assay specificity. The extensive washing steps and on-membrane hybridization required by some of the most commonly used techniques also complicate assays and increase assay time (21). No rapid real-time PCR-based method to detect FQ resistance has been yet described. This is likely due to the technical challenge of identifying multiple different mutations in a simple real-time PCR assay.
We have recently described a melting temperature (Tm)-based method that uses asymmetrical PCR in conjunction with sloppy molecular beacons (SMBs) to identify hundreds of different target sequences (4). Here, we investigate whether this approach can be adapted to detect all of the gyrA QRDR mutations associated with FQ-resistant M. tuberculosis in a simple, rapid, and highly specific single-well assay. In addition, we studied the capacity of this approach to detect mixtures of wild-type and mutant DNA, since we had identified a subset of clinical FQ-resistant TB cases that had heteroresistance in their clinical cultures.
MATERIALS AND METHODS
Clinical DNA samples.
DNA samples from FQ-resistant and FQ-susceptible clinical M. tuberculosis isolates were collected from the National Masan Tuberculosis Hospital by the International Tuberculosis Research Center (ITRC), Masan, South Korea; the Public Health Research Institute (PHRI), Newark, NJ; and the WHO TDR Tuberculosis Specimen Bank (5). M. tuberculosis cultures from drug-susceptible and FQ-resistant XDR patients were selected from patients enrolled in a natural history study in Korea. All samples underwent DNA sequencing, and 30 samples representing a wide variety of QRDR mutations, including some wild-type control samples, were selected for testing with the SMB assay. Another set of 41 DNA samples originated from patients with culture-positive tuberculosis from the New York/New Jersey area that had been tested for QRDR mutations by mass spectrometry analysis. This set of DNA samples was also chosen to represent a wide repertoire of the common QRDR mutations, along with a sampling of isolates with wild-type QRDR sequences. Finally a set of 21 DNA samples from MDR TB isolates (with no known XDR cases) from Asia, Africa, and Latin America maintained by the United Nations Children's Fund/UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases were chosen to represent wild-type gyrA QRDR controls to complete our study sample set (Table 1). We confirmed the sequence of the gyrA QRDR in all 92 DNA samples by DNA sequencing. Out of the 21 samples from the WHO TDR collection chosen to represent wild-type gyrA QRDR, two samples turned out to harbor mutant QRDR sequences, one having a D94A T allele mutation and the other being a mixture of wild-type and D89N T allele sequences. Each DNA sample was quantified with Nanodrop microvolume spectrophotometer (Thermo Scientific), and 2 to 4 ng of the DNA sample was used for PCR. All samples were independently coded and randomly redistributed to blind the sample sequence and origin before assay testing.
Table 1.
Two Tm codes for the clinical isolate DNA and their concordance with DNA sequence analysis
| Sample no. |
Tm value (°C) by: |
QRDR type bya: |
||
|---|---|---|---|---|
| QDR1-FAM | QDR2-Cy5 | Tm prediction | Sequencing data | |
| 1 | 64.6 | 63.2 | WT T | WT T |
| 2 | 64.6 | 63.2 | WT T | WT T |
| 3 | 64.6 | 63.3 | WT T | WT T |
| 4 | 64.5 | 63.1 | WT T | WT T |
| 5 | 64.6 | 63.2 | WT T | WT T |
| 6 | 64.5 | 63.5 | WT T | WT T |
| 7 | 64.5 | 63.3 | WT T | WT T |
| 8 | 64.6 | 63.3 | WT T | WT T |
| 9 | 64.5 | 63.1 | WT T | WT T |
| 10 | 64.9 | 63.8 | WT T | WT T |
| 11 | 64.5 | 63.2 | WT T | WT T |
| 12 | 64.5 | 63.2 | WT T | WT T |
| 13 | 64.6 | 63.4 | WT T | WT T |
| 14 | 64.5 | 63.1 | WT T | WT T |
| 15 | 64.4 | 63.2 | WT T | WT T |
| 16 | 64.4 | 63.2 | WT T | WT T |
| 17 | 64.6 | 63.3 | WT T | WT T |
| 18 | 64.5 | 63.2 | WT T | WT T |
| 19 | 64.6 | 63.3 | WT T | WT T |
| 20 | 64.5 | 63.3 | WT T | WT T |
| 21 | 64.4 | 63.0 | WT T | WT T |
| 22 | 64.4 | 63.8 | WT T | WT T |
| 23 | 64.6 | 63.2 | WT T | WT T |
| 24 | 64.4 | 63.2 | WT T | WT T |
| 25 | 64.3 | 63.0 | WT T | WT T |
| 26 | 64.3 | 63.1 | WT T | WT T |
| 27 | 64.4 | 63.1 | WT T | WT T |
| 28 | 64.3 | 63.0 | WT T | WT T |
| 29 | 64.3 | 62.9 | WT T | WT T |
| 30 | 64.4 | 63.2 | WT T | WT T |
| 31 | 64.3 | 63.1 | WT T | WT T |
| 32 | 64.3 | 63.0 | WT T | WT T |
| 33 | 64.3 | 63.1 | WT T | WT T |
| 34 | 64.4 | 63.2 | WT T | WT T |
| 35 | 64.3 | 63.0 | WT T | WT T |
| 36 | 64.6 | 63.2 | WT T | WT T |
| 37 | 64.3 | 62.9 | WT T | WT T |
| 38 | 64.4 | 62.9 | WT T | WT T |
| 39 | 64.4 | 62.9 | WT T | WT T |
| 40 | 64.3 | 62.8 | WT T | WT T |
| 41 | 64.2 | 62.9 | WT T | WT T |
| 42 | 64.3 | 62.8 | WT T | WT T |
| 43 | 64.3 | 62.9 | WT T | WT T |
| 44 | 64.3 | 62.8 | WT T | WT T |
| 45 | 64.3 | 62.8 | WT T | WT T |
| 46 | 64.3 | 62.9 | WT T | WT T |
| 47 | 61.3 | 61.3 | WT S | WT S |
| 48 | 61.4 | 61.3 | WT S | WT S |
| 49 | 61.6 | 61.5 | WT S | WT S |
| 50 | 61.3 | 61.5 | WT S | WT S |
| 51 | 61.6 | 57.3 | G88A T | G88A T |
| 52 | 65.5 | 65.4 | A90V S | A90V S |
| 53 | 65.7 | 65.4 | A90V S | A90V S |
| 54 | 68.3 | 66.9 | A90V T | A90V T |
| 55 | 68.3 | 66.9 | A90V T | A90V T |
| 56 | 68.3 | 66.9 | A90V T | A90V T |
| 57 | 68.3 | 67.3 | A90V T | A90V T |
| 58 | 68.3 | 66.7 | A90V T | A90V T |
| 59 | 68.2 | 66.6 | A90V T | A90V T |
| 60 | 68.3 | 66.7 | A90V T | A90V T |
| 61 | 68.2 | 66.7 | A90V T | A90V T |
| 62 | 68.5 | 66.8 | A90V T | A90V T |
| 63 | 68.3 | 66.7 | A90V T | A90V T |
| 64 | 68.3 | 66.7 | A90V T | A90V T |
| 65 | 58.8 | 58.6 | S91P T | S91P T |
| 66 | 58.6 | 58.3 | S91P T | S91P T |
| 67 | 58.6 | 58.2 | S91P T | S91P T |
| 68 | 58.6 | 58.3 | S91P T | S91P T |
| 69 | 58.5 | 58.3 | S91P T | S91P T |
| 70 | 66.2 | 59.1 | D94G S | D94G S |
| 71 | 68.3 | 60.6 | D94G T | D94G T |
| 72 | 68.4 | 60.5 | D94G T | D94G T |
| 73 | 68.4 | 60.5 | D94G T | D94G T |
| 74 | 68.3 | 60.5 | D94G T | D94G T |
| 75 | 68.3 | 60.5 | D94G T | D94G T |
| 76 | 68.5 | 60.7 | D94G T | D94G T |
| 77 | 68.2 | 60.3 | D94G T | D94G T |
| 78 | 68.3 | 60.4 | D94G T | D94G T |
| 79 | 68.2 | 60.3 | D94G T | D94G T |
| 80 | 62.9 | 57.4 | D94H/N/Y T | D94N T |
| 81 | 64.2 | 58.6 | G88C T or D94A T | D94A T |
| 82 | 60.0 | 54.5 | G88C S or D94H/N/Y S | D94H S |
| 83 | 71.7 | 64.0 | New mutant/double mutant | A90V T D94G T |
| 84 | 69.8 | 63.3 | New mutant/double mutant | A90V T D94G T |
| 85 | 63.6 | 57.7–63.5 | WT T + G88C or D94A T | WT T + D94A T |
| 86 | 63.7–68.0 | 62.5–67.3 | WT T + A90V T | WT T + A90V T |
| 87 | 59.1–64.6 | 58.6–63.3 | WT T + S91P T | WT T + A90V + S91P T |
| 88 | 64.2–68.5 | 63.12–66.9 | WT T + A90V T | WT T + A90V T |
| 89 | 58.5–64.5 | 62.9 | Mixed DNA | WT T + D89N T |
| 90 | 65.4–69.9 | 59.8 | Mixed DNA | D94G T + D94A T |
| 91 | 64.2 | 57.6–63.3 | WT T + D94H/N/Y T | WT T + D94N T |
| 92 | 63.0 | 57.5–63.3 | WT T + D94H/N/Y T | WT T + D94N T |
WT, wild type; T, 95T allele (threonine); S, 95S allele (serine).
DNA samples from nontuberculous mycobacteria (NTM) and Gram-positive and Gram-negative bacteria.
One hundred twenty-one clinical NTM DNA isolates representing 26 species, including M. abcessus, M. aubagnense, M. avium, M. avium subsp. paratuberculosis, M. bollettii, M. celatum, M. chelonae, M. flavescens, M. fortuitum, M. gastri, M. genevense/M. simiae, M. gordonae, M. intermedium, M. intracellulare, M. kansasii, M. malmoense, M. mucogenicum, M. peregrinum, M. porcinum, M. scrofulaceum, M. senegalense, M. shimoidei, M. smegmatis, M. simiae, M. szulgai, M. terrae, and M. xenopi, with several mixed samples containing more than one NTM species, were obtained from ITRC. For evaluation of assay specificity, a panel of 18 species of Gram-negative and Gram-positive bacteria representing the most common bloodstream infection and nosocomial pathogens was also selected: Acinetobacter baumanii, Bacillus cereus, Clostridium difficile, Campylobacter jejuni, Escherichia coli, Haemophilus influenzae, Klebsiella pneumoniae, Listeria grayi, Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus hominis, Streptococcus agalactiae, Streptococcus dysgalactiae, Streptococcus pyogenes, Streptococcus pneumoniae, Vibrio cholerae, and Yersinia enterocolitica. DNA was either isolated from pure clinical strains by boiling a loopful of culture in Instagene matrix (Bio-Rad), or pure genomic DNA from ATCC strains was obtained from BEI Resources (Manassas, VA). The DNA was quantified as described above and used to determine the analytical specificity of the assay, using 5 to 10 ng of the DNA samples for PCR.
SMBs and primers.
A 107-bp fragment (nucleotides 222 to 330, with numbering based on the gene start site) containing the gyrA QRDR was amplified with the target primer gyrA-F (5′-CCGGTCGGTTGCCGAGACC-3′) and the antisense primer gyrA-R (5′-CCAGCGGGTAGCGCAGCGACCAG-3′). These primers were designed to be specific to the M. tuberculosis complex, verified by an alignment of M. tuberculosis and all of the NTM gyrA sequences available in the database (http://www.ncbi.nlm.nih.gov/GenBank/) (7). The two SMB sequences were QDR1 (5′-6-carboxyfluorescein CCGTGCgcgcaccagggtgccctagatcgacacgtcGCACGG-DABCYL-3′) and QDR2 (5′-cyanine 5-CCCGAGGgItgtcgtagattgacacgtcgccgcgcggCCTCGGG-BHQ1-3′), where the underlined uppercase sequences represent the stem portion and the lowercase sequences represent the probe portion of the SMBs, DABCYL represents 4-(4-dimethylamino)phenylazobenzoic acid, BHQ represents Black Hole quencher, and I represents deoxyinosine. Both probes were targeted against the gyrA QRDR region. The SMBs were designed by using the in silico DNA folding program from http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi, and the probe-target hybrid folding program from http://dinamelt.bioinfo.rpi.edu/twostate.php was used to predict the possible probe-target hybrid structures and melting temperatures (Tms). The probes were designed to generate a maximum Tm difference between wild-type and mutant sequences, so as to enable their unambiguous identification. Primers were obtained from Sigma Genosys, and the SMB probes were obtained from Biosearch, CA.
Artificial templates containing gyrA core region mutations.
We designed artificial oligonucleotide templates for all of the documented QRDR mutations. These were then used to test the efficacy of the probes to identify each mutation and the two wild-type polymorphisms. Each oligonucleotide template included the sequences of the gyrA QRDR and the binding sites of the assay's primers. Separate templates were made for each of the mutations G88C, G88A, D89N, A90V, S91P, D94A, D94G, D94N, D94Y, and D94H on both 95S and 95T gyrA allele wild-type sequence backgrounds [designated as gyrA(95S) and gyrA(95T), respectively]. Approximately 105 molecules of each template were used in each PCR assay containing the primers and the SMB probes.
Sloppy molecular beacon melt assay.
A PCR-Tm analysis assay was carried out on artificial templates and clinical sample DNA as follows. All studies of DNA from clinical samples were performed and decoded by an experimenter who was blinded to the nature and identity of the sample. PCR was performed with the Roche Light Cycler 480 II real-time PCR system (Roche Molecular Systems, Inc.), in 20-μl reaction volumes containing 1 μM target primer and 100 nM antisense primer, 0.8 ng/μl of each SMB probe, 4 mM MgCl2, 250 mM deoxynucleoside triphosphates (dNTPs), 1× PCR buffer, 5% glycerol, 0.06 U/μl of AmpliTaq gold Stoffel DNA polymerase (Applied Biosystems), and 2 to 5 ng of sample DNA or an equivalent volume of water. PCR was carried out as follows: activation of the enzyme for 2 min at 95°C, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at 65°C for 30 s, and extension at 72°C for 10 s. Following PCR cycling, post-PCR-Tm analysis was performed by denaturation at 95°C for 5 min, followed by cooling down to 45°C and then gradual heating to 85°C with continuous monitoring of the fluorescence during the process at a rate of 15 data acquisitions per °C. Tm calls were performed at the end of the reaction by using the automated Tm calling software (Light Cycler 480 software), and the resulting Tm values for each probe were determined. Double peaks were automatically identified by the Tm calling software, indicating the presence of mixed DNA.
DNA mixtures containing wild-type and mutant DNA.
DNA mixtures containing wild-type and mutant DNA were prepared to evaluate the performance of the assay to detect presence of heteroresistance. Various amounts of mutant genomic DNA (obtained from the clinical isolates) were added to the wild-type DNA to generate DNA mixtures containing 5, 10, 20, 30, and 40% mutant DNA in a total amount of 1 ng (105 genome equivalents) of DNA. The most common gyrA QRDR mutant DNA types, A90V, S91P, and D94G (11, 21), were individually mixed with wild-type DNA [all having gyrA(95T) alleles], and SMB Tm shift assays were performed to determine the limit of the assay to identify the mutant sequence in the presence of a large wild-type DNA background.
RESULTS
SMB design.
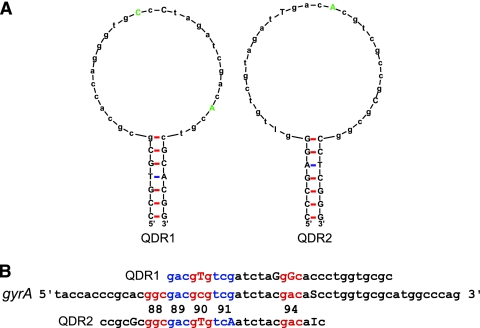
We have previously shown that SMBs can be used to identify NTM species (10) or to distinguish among a wide variety of pathogenic Gram-positive and Gram-negative bacteria (4). SMBs differ from conventional molecular beacons in having unusually long probe sequences, which enable them to query long stretches of DNA and to increase their mismatch tolerance (4). To identify FQ resistance mutations, we designed two SMBs, QDR1 and QDR2, with 30-bp probe sequences and 6- to 7-bp stem sequences (Fig. 1). The probes were designed to generate stable stem-loop structures while avoiding the formation of secondary structures in the loop region (Fig. 1A). The QDR1 probe spanned gyrA codons 89 to 98, while the QDR2 probe spanned gyrA codons 86 to 95 (Fig. 1B). We deliberately introduced mutations into the SMB probe sequences to disrupt secondary structures and palindromes and to generate a stable stem-loop structure that optimized probe-target hybridization kinetics. This is in contrast to conventional molecular beacons, which are designed to be fully complementary to their target sequences. In selecting probe sequence mutations, we attempted to incorporate commonly occurring FQ resistance-associated gyrA QRDR mutations if possible. However, unrelated mutations were also introduced into the probes if naturally occurring mutations did not result in good probe structures. The QDR1 probe contained the A90V and D94G FQ resistance-associated mutations. It also contained an unrelated mutation in codon 93. The QDR2 probe contained the same A90V FQ resistance-associated mutation. It also contained two unrelated mutations in codons 87 and 91 (Fig. 1A and B). As a result, the similarity of probe QRD1 to both A90V and D94G mutations and that of QDR2 to the D94G mutation was higher than their relatedness to the wild-type QRDR sequence. Likewise, the similarity of the SMBs to the S91P mutant and less common FQ-resistant mutants was lower than their similarity to FQ-susceptible wild-type M. tuberculosis. Therefore, each SMB had a gradient of probe-target relatedness, with common mutants being recognized more effectively than the wild type and the wild type more effectively than less common mutations. This approach increased the range of Tms produced by the assay, thereby increasing assay precision (Table 1). It also aided in the identification of mutant DNA in wild-type DNA mixtures (Fig. 2).
Fig. 1.
Assay probes and target. (A) SMB probe structures. The structures for SMB QDR1 and QDR2 are shown. Mutations in the loop region are indicated in uppercase, and those corresponding to known QRDR mutations are shown in green. Uppercase letters in black show the probe stems. (B) Probe target alignments. Among the three sequences shown, “gyrA” indicates the gyrA QRDR, “QDR1” indicates the reverse complement of the QDR1 SMB probe sequence, and “QDR2” indicates the reverse complement of the QDR2 SMB probe sequence. The gyrA codons associated with FQ resistance mutations are marked in red and blue, and their codon numbers are indicated. The mutations introduced into the two SMB sequences are identified by uppercase letters. S denotes either cytosine or guanine, and I denotes deoxyinosine.
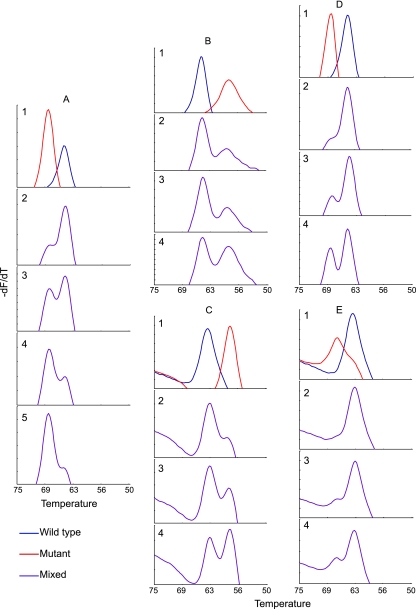
Fig. 2.
Tm profiles of DNA mixtures. The Tm curve for the indicated SMB is shown in an assay containing various mixtures of wild-type and mutant clinical M. tuberculosis DNA. (A) SMB QDR1 tested with wild-type plus D94G mutant DNA. Panel 1, individual wild-type and mutant DNA samples; panel 2, 5% mutant and 95% wild-type DNA; panel 3, 10% mutant and 90% wild-type DNA; panel 4, 20% mutant and 80% wild-type DNA; panel 5, 30% mutant and 70% wild-type DNA. (B) SMB QDR1 tested with wild-type plus S91P mutant DNA; (C) SMB QDR2 tested with wild-type plus S91P mutant DNA; (D) SMB QDR1 tested with wild-type plus A90V mutant DNA; (E) SMB QDR2 tested with wild-type plus A90V mutant DNA. In panels B to E, panel 1 contains individual wild-type and mutant DNA samples, panel 2 contains 10% mutant and 90% wild-type DNA, panel 3 contains 20% mutant and 80% wild-type DNA, and panel 4 contains 30% mutant and 70% wild-type DNA. All of the wild-type and mutant DNA had the gyrA(95T) allele.
Melting temperature profiles.
We tested the ability of the two SMBs to identify QRDR mutations in both the gyrA(95S) and gyrA(95T) allele backgrounds. Artificial QRDR targets were constructed containing all known wild-type and QRDR mutant sequences and tested in the two-SMB assay. Table 2 shows the average Tm values obtained from 10 individual assays for each artificial template. We found that the assay produced distinct and reproducible Tm values that distinguished the two wild-type polymorphisms from all of the QRDR mutations, regardless of the gyrA(95T or 95S) allele background. Notably, the difference between the Tm values (dTm) of the wild-type target and each mutant target was at least 3.6°C for at least one of the two SMBs, indicating that differentiation between wild-type and mutant sequences was likely to be robust (Table 2). We had previously shown that SMB-generated Tms can be used in combination to derive an allele-specific code that identifies bacterial species (4). In this study, we tested whether the two SMBs used in the assay could generate dual Tm codes that specifically identified each mutation. The results show that the two QRDR-specific SMBs generated Tm codes that distinguished between the two wild-type polymorphisms and the most highly prevalent QRDR A90V, S91P, and D94G mutants (11, 21) [for both gyrA(95S) and gyrA(95T) alleles], as well as the G88A [gyrA(95T)] and D89N [gyrA(95S)] mutants. The Tm codes generated in the assay divided the other mutants into four groups containing (i) the G88A [gyrA(95S)] and D89N [gyrA(95T)] mutants, (ii) G88C [gyrA(95S)] and D94H/N/Y [gyrA(95S)] mutants, (iii) G88C [gyrA(95T)] and D94A [gyrA(95T) mutants, and (iv) the D94H/N/Y [gyrA(95T)] mutant. In clustering the mutant and wild-type sequences based on their individual Tm codes, we considered Tm codes to be identical, unless at least one of the Tm values between the Tm codes differed by 2°C and above to compensate for experimental variability. Importantly, all mutations were clearly differentiated from wild-type sequences (Table 2).
Table 2.
Post-PCR Tm values with artificial templates harboring QRDR mutationsa
| QRDR typeb | QDR1 |
QDR2 |
||
|---|---|---|---|---|
| Tm (°C) | dTm (°C)c | Tm (°C) | dTm (°C)c | |
| WT T | 66.2 | 65.4 | ||
| G88A T | 61.7 | 4.4 | 58.1 | 7.4 |
| G88C T | 66.0 | 0.2 | 60.5 | 5.0 |
| D89N T | 60.3 | 5.9 | 58.3 | 7.2 |
| A90V T | 69.8 | −3.6 | 68.0 | −2.6 |
| S91P T | 61.3 | 4.9 | 62.6 | 2.8 |
| D94A T | 66.1 | 0.1 | 60.3 | 5.1 |
| D94G T | 70.3 | −4.2 | 63.5 | 1.9 |
| D94H T | 65.0 | 1.2 | 58.8 | 6.6 |
| D94N T | 64.9 | 1.3 | 58.8 | 6.6 |
| D94Y T | 64.8 | 1.4 | 58.2 | 7.3 |
| WT S | 63.8 | 64.6 | ||
| G88A S | 59.0 | 4.8 | 57.4 | 7.3 |
| G88C S | 63.5 | 0.3 | 57.4 | 7.2 |
| D89N S | 57.2 | 6.6 | 58.4 | 6.2 |
| A90V S | 67.5 | −3.8 | 67.8 | −3.2 |
| S91P S | 58.9 | 4.9 | 59.3 | 5.3 |
| D94A S | 63.9 | −0.1 | 59.3 | 5.4 |
| D94G S | 68.3 | −4.6 | 62.0 | 2.7 |
| D94H S | 62.5 | 1.3 | 56.7 | 8.0 |
| D94N S | 62.5 | 1.3 | 56.6 | 8.0 |
| D94Y S | 62.4 | 1.4 | 56.6 | 8.0 |
Results represent the average of 10 separate reactions. Tm values did not differ by more than 0.5 to 0.8°C.
WT, wild type; T, 95T allele (threonine); S, 95S allele (serine).
dTm = WT Tm − mutant Tm. Negative dTm values imply a higher Tm for the probe with the mutant sequence than that for the wild-type sequence.
Clinical FQ-resistant M. tuberculosis DNA with either the gyrA(95S) or the gyrA(95T) allele harboring all of the known QRDR mutations, except G88C, D89N, and D94Y, were available in our study sample set. We tested the assay with each of these 11 QRDR mutant DNA samples, and the resulting dual Tm codes paralleled the results with the oligonucleotide templates, except each Tm was 1.5 to 2°C lower on average (see Table S1 in the supplemental material). This difference in Tm values is consistent with our previous experience in moving from oligonucleotides to genomic DNA samples (10) and does not affect assay performance, since the change is maintained across both mutant and wild-type DNAs.
Analytical specificity of the assay for M. tuberculosis.
The analytical specificity of the assay was tested against a panel of 139 DNA samples representing 26 different NTM species and 18 Gram-negative and Gram-positive pathogenic bacteria. No signals were obtained with any of bacteria, except for 9/22 M. abcessus samples (data not shown). However, the positive M. abcessus samples caused only one of the two SMBs in the assay (QDR2) to generate a Tm of 55 to 56°C. This resulted in a Tm code that was unique to M. abcessus and that could not be confused with either wild-type or mutant M. tuberculosis.
Detection of DNA mixtures.
We have observed that a number of samples from patients with XDR TB contain mixtures of wild type (FQ-susceptible) and QRDR mutant (FQ-resistant) M. tuberculosis. Mutant detection is challenging in real-time PCR tests in a background of large amounts of wild-type DNA (2). However, we postulated that our probe design criteria, which resulted in large dTms between wild-type and mutant targets, would enable us to detect mutant targets even when they were present as a minor proportion. To test this, we mixed M. tuberculosis genomic DNA from strains that had a wild-type QRDR with strains that had a mutant QRDR to create artificial mixtures with various proportions of mutant target. Three different common QRDR mutants (S91P, A90V, and D94G) were tested in this manner with the SMB assay. Mixtures of wild-type and mutant DNA were detected as double Tm peaks from the QDR1 SMB, the QDR2 SMB, or both (Fig. 2). We observed that mutant detection was somewhat dependent on the actual mutation that was present in the mixture. Thus, the assay could detect 5% D94G DNA mixed with 95% wild-type DNA (Fig. 2A), but required at least 10% S91P and 10% A90V mutant DNA in the mixture for efficient detection (Fig. 2B to E). The QDR1 SMB was better able to detect mixtures than the QDR2 SMB. This is likely due to the larger dTm between wild-type and mutant DNA observed with QDR1 for these three mutations (Fig. 2 and Table 2).
We tested a series of mixtures in a blinded manner to further validate the assay for heteroresistance detection. Mutant and wild-type DNA was mixed in various proportions, and then each sample was split into eight different aliquots. All aliquots were then tested with the assay after blinding. We found that samples containing at least 10% D94G or S91P mutants could be detected in 100% of test replicates. Samples containing at least 20% A90V mutants could be detected in 100% of the test replicates, although samples containing 10% A90V could be detected in only 50% of the test replicates. Thus, our assay showed good consistency in the ability to detect mutant and wild-type DNA mixtures.
Assay performance on blinded clinical DNA samples.
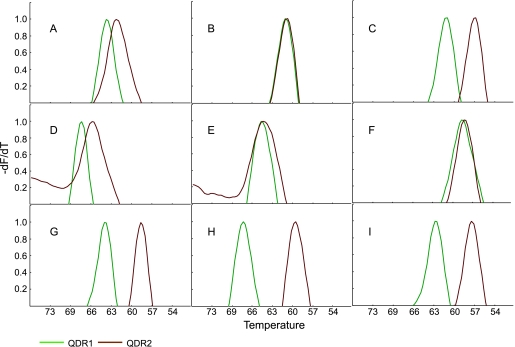
We next tested the performance of the assay on 92 clinical M. tuberculosis isolates, including genomic DNA isolated from FQ-sensitive and FQ-resistant clinical cultures. Each DNA sample was coded, and the SMB assay was performed blindly. The assay showed 100% concordance with sequencing results for wild-type, mutant, double mutant, and mutant–wild-type mixed-DNA samples (Table 1). Each type of individual QRDR mutation and the two wild-type polymorphisms showed distinct peak profiles (Fig. 3). Thus, each mutation could be identified by the distinct dual Tm codes that it generated in the assay (Fig. 4A). The Tm values were highly reproducible. Each SMB generated Tm values that differed by less than 1°C in the presence of different samples with identical QRDR sequences (Fig. 4A). Forty-two out of 46 wild-type QRDR strains had the gyrA(95T) allele, while the other four strains had the gyrA(95S) allele. Among the QRDR mutants, the A90V mutation predominated (13 mutants), followed by D94G (10 mutants) and S91P (5 mutants), consistent with previous studies (11, 21). Only one G88A, D94N, D94A, and D94H mutation each was detected in our study set, and two strains had both A90V and D94G mutations in the QRDR. As with the wild-type strains, the gyrA(95T) allele predominated in the QRDR mutants (Table 1).
Fig. 3.
Dual Tm profiles of wild-type and QRDR mutant DNA. The Tm profiles of SMB QDR1 and QDR2 are shown in the presence of wild-type clinical M. tuberculosis DNA and various clinical QRDR mutants. The dual Tm peaks (°C) of each profile are listed in parentheses. (A) Wild-type gyrA(95T) (64.6 and 63.2); (B) wild-type gyrA(95S) (61.5 and 61.4); (C) G88A gyrA(95T) mutant (60.8 and 57.3); (D) A90V gyrA(95T) mutant (68.3 and 65.4); (E) A90V gyrA(95S) mutant (65.5 and 65.4); (F) S91P gyrA(95T) mutant (58.8 and 58.6); (G) D94A gyrA(95T) mutant (64.2 and 58.6); (H) D94G gyrA(95T) mutant (68.3 and 60.6); and (I) D94N gyrA(95T) mutant (62.9 and 57.4).
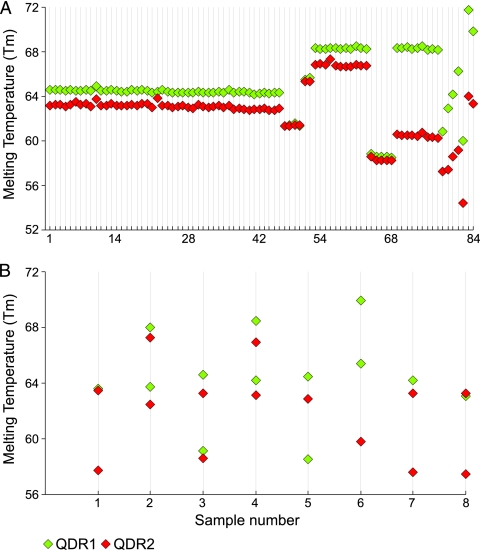
Fig. 4.
Dual Tm codes of the clinical M. tuberculosis DNA tested in this study. The Tm values of each SMB are shown for 92 clinical M. tuberculosis isolates ordered by mutation and gyrA(95S or 95T) allele. Each QRDR sequence type is seen to generate distinct dual Tm profiles. (A) Tm codes of homogeneous DNA samples. Samples 1 to 46, wild-type gyrA(95T); samples 47 to 50, wild-type gyrA(95S); samples 51 and 52, A90V gyrA(95S) mutant; samples 53 to 63, A90V gyrA(95T) mutant; samples 64 to 68, S91P gyrA(95T) mutant; samples 69 to 77, D94G gyrA(95T) mutant; sample 78, G88A gyrA(95T) mutant; sample 79, D94N gyrA(95T) mutant; sample 80, D94A gyrA(95T) mutant; sample 81, D94G gyrA(95S) mutant; sample 82, D94H gyrA(95S) mutant; and samples 83 and 84, A90V D94G gyrA(95T) double mutant. (B) Tm codes of the mixed clinical samples. Either one or both probes can be seen to have generated double peaks with distinct Tm values. Sample 1, wild type and D94A mutant; samples 2 and 4, wild type and A90V mutant; sample 3, wild type and A90V and S91P mutants; sample 5, wild type and D89N mutant; sample 6, D94A and D94G mutants; samples 7 and 8, wild type and D94N mutant. All of the samples contained the gyrA(95T) allele.
Nearly 9% (8/92) of the clinical DNA included in our study showed the presence of heteroresistance, with seven samples containing both wild-type and QRDR mutant sequences and one containing a mixture of D94A and D94G mutants, which was confirmed by DNA sequencing. Our assay identified all of the mixed samples as mixtures by detecting distinct double peaks in either one or both of the SMBs. Deconvolution of the Tm peaks to generate multiple Tm codes allowed us to correctly identify a mixture of mutant and wild-type sequence in five of the eight DNA mixtures (Table 1 and Fig. 4B). One sample was found to contain a mixture of A90V, S91P, and wild-type gyrA(95T) allele DNA on sequencing, but an analysis of the dual Tm codes only revealed a mixture of A90V and wild-type sequences. Two other samples could be identified only as mixed DNA as the double Tm peaks could not be deconvoluted into meaningful Tm codes. DNA sequencing showed the presence of wild-type and D89N gyrA(95T) alleles in one sample and a mixture of D94G and D94A gyrA(95T) allele mutants in the other.
An additional two samples contained two QRDR mutations on the same DNA sequence (samples 83 and 84 in Table 1). Assays performed on these samples resulted in unique single Tm peaks for each SMB, and their Tm codes did not match any known Tm code for the wild-type alleles or the individual QRDR mutations (Fig. 4A, samples 83 and 84). DNA sequencing revealed that these two samples contained A90V and D94G gyrA(95T) allele mutations (Table 1). These two cases demonstrate that combinations of mutations may produce new Tm codes that are specific for that combination and different from the wild type or the single QRDR mutants.
DISCUSSION
We have successfully designed SMBs that identify specific gyrA QRDR mutations and mutant–wild-type mixtures. The dual Tm codes generated in the assay easily differentiated between wild-type DNA derived from FQ-susceptible M. tuberculosis and mutant DNA derived from FQ-resistant M. tuberculosis. The codes also enabled us to identify each mutation in the QRDR and to identify mixed samples containing 5 to 20% mutant DNA. The assay was rapid and robust, and unlike techniques which involve open DNA hybridization, the assay was technically quite simple. The SMB approach allowed the query of the entire gyrA QRDR region with only two probes in an enclosed system. This approach is technically much simpler and robust than existing assays, which use as many as nine probes to identify the gyrA QRDR mutations (21). There was a 100% concordance between our assay results and DNA sequencing.
Our assay was also highly specific for M. tuberculosis. To the best of our knowledge, this is the first study of a QRDR assay which has looked at assay specificity. The M. tuberculosis gyrA QRDR is quite similar to other organisms, and the mutations encountered in FQ-resistant M. tuberculosis clinical isolates have been found in similar regions of the gyrA gene from other FQ-resistant bacteria (27). Assays with more limited specificity might falsely identify NTM or other bacteria as FQ-resistant M. tuberculosis. Although this problem could be partially addressed by including an M. tuberculosis-specific probe in each assay, samples containing mixtures of NTM and M. tuberculosis might still be falsely identified as FQ-resistant TB. This is especially important in settings where NTM infections are highly prevalent. We tested our assay against an extensive panel of NTM and other pathogenic bacteria: only M. abcessus resulted in any Tm signal, and this was only with one of the two probes (QDR2). However this Tm was different from any Tm we have observed in any M. tuberculosis QRDR mutant. Also, none of the M. abcessus strains generated a Tm value in QDR1. Thus, it is highly unlikely that our assay would mistakenly identify an M. abcessus sample as FQ-resistant M. tuberculosis, even when M. tuberculosis was mixed into the sample.
FQ heteroresistance appears to be a relatively common occurrence (1, 9, 11, 21, 22), with some studies reporting that as many as 22 to 31% of patients with FQ-resistant TB were infected with both wild-type and QRDR mutant M. tuberculosis strains (9, 21). We tested our assay's ability to detect the presence of mutant DNA mixed into a large wild-type DNA background and found that the assay detected as little as 5% of the mutant sequence present in a DNA sample. Mutations were consistently detected at 10 to 20% concentrations. These results suggest that the assay will be useful for detecting early development of FQ-resistant TB. Almost 9% of our test panel came from patients who were found to have heteroresistance, and the assay detected the mixed infection in every case.
Our assay has the potential to complement and extend the utility of other real-time PCR tests such as the Xpert MTB/RIF assay, recently released commercially. The Xpert assay rapidly detects the presence of M. tuberculosis with sensitivity close to that of culture and identifies the presence of rifampin resistance (19). Patients identified as rifampin resistant by the Xpert assay are at increased risk for XDR TB. Our assay could be used to screen for FQ resistance in these rifampin-resistant patients, thereby identifying the subset of patients most likely to have XDR. Our assay will have much broader utility if FQs become recommended as part of primary TB therapy. Recent reports suggest that FQ exposure for more than 10 days that predated 60 days before the diagnosis of active tuberculosis was associated with as high as a 20.8% risk of resistance (8). This suggests that FQ-resistant TB will become more common if FQs continue to be used to treat respiratory infections. First-line therapy with an FQ-containing regime could have disastrous consequences in patients with unrecognized primary FQ resistance. We suggest that the incidence of primary FQ resistance is already sufficiently high to justify prescreening all new TB patients with a rapid FQ resistance assay before they are treated with an FQ-containing regime. In summary, we have developed a rapid, simple, and robust method to detect M. tuberculosis gyrA QRDR mutations associated with FQ resistance. This assay will be a useful tool for rapid molecular drug susceptibility testing of FQ resistance and for detecting the early emergence of FQ heteroresistance in TB patients.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI082174 and AI080653 (to D.A., K.T., and S.C.); Northeast Bio-Defense Career training grant 3185-07 (to B.A.); funding from the Intramural Research Program of the NIAID, NIH (to C.E.B.); and the South Korean Ministry of Health, Welfare and Family Affairs (to S.-N.C. and C.E.B.).
D.A. is among a group of inventors who earn royalties for molecular beacon usage.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 29 December 2010.
REFERENCES
- 1. Blaas S. H., et al. 2008. Extensively drug resistant tuberculosis in a high income country: a report of four unrelated cases. BMC Infect. Dis. 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blakemore R., et al. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blumberg H. M., et al. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603–662 [DOI] [PubMed] [Google Scholar]
- 4. Chakravorty S., et al. 2010. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J. Clin. Microbiol. 48:258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chakravorty S., et al. 2008. Rifampin resistance, Beijing-W clade-single nucleotide polymorphism cluster group 2 phylogeny, and the Rv2629 191-C allele in Mycobacterium tuberculosis strains. J. Clin. Microbiol. 46:2555–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng A. F., et al. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dauendorffer J. N., et al. 2003. Identification of mycobacterial species by PCR sequencing of quinolone resistance-determining regions of DNA gyrase genes. J. Clin. Microbiol. 41:1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Devasia R. A., et al. 2009. Fluoroquinolone resistance in Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 180:365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duong D. A., et al. 2009. Beijing genotype of Mycobacterium tuberculosis is significantly associated with high-level fluoroquinolone resistance in Vietnam. Antimicrob. Agents Chemother. 53:4835–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El-Hajj H. H., et al. 2009. Use of sloppy molecular beacon probes for identification of mycobacterial species. J. Clin. Microbiol. 47:1190–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feuerriegel S., et al. 2009. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob. Agents Chemother. 53:3353–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fluit A. C., Visser M. R., Schmitz F. J. 2001. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev. 14:836–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giannoni F., et al. 2005. Evaluation of a new line probe assay for rapid identification of gyrA mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ginsburg A. S., Grosset J. H., Bishai W. R. 2003. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect. Dis. 3:432–442 [DOI] [PubMed] [Google Scholar]
- 15. Ginsburg A. S., et al. 2003. Fluoroquinolone resistance in patients with newly diagnosed tuberculosis. Clin. Infect. Dis. 37:1448–1452 [DOI] [PubMed] [Google Scholar]
- 16. Ginsburg A. S., et al. 2005. Emergence of fluoroquinolone resistance in Mycobacterium tuberculosis during continuously dosed moxifloxacin monotherapy in a mouse model. Antimicrob. Agents Chemother. 49:3977–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grimaldo E. R., et al. 2001. Increased resistance to ciprofloxacin and ofloxacin in multidrug-resistant mycobacterium tuberculosis isolates from patients seen at a tertiary hospital in the Philippines. Int. J. Tuberc. Lung Dis. 5:546–550 [PubMed] [Google Scholar]
- 18. Guillemin I., Jarlier V., Cambau E. 1998. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob. Agents Chemother. 42:2084–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Helb D., et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Higuchi R., Fockler C., Dollinger G., Watson R. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 11:1026–1030 [DOI] [PubMed] [Google Scholar]
- 21. Hillemann D., Rusch-Gerdes S., Richter E. 2009. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 47:1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaplan G., et al. 2003. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect. Immun. 71:7099–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Musser J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Brien R. J. 2003. Development of fluoroquinolones as first-line drugs for tuberculosis—at long last! Am. J. Respir. Crit. Care Med. 168:1266–1268 [DOI] [PubMed] [Google Scholar]
- 25. Reichman L. B. 2008. Tuberculosis drug resistance comes full circle. Lancet 371:1052–1053 [DOI] [PubMed] [Google Scholar]
- 26. Shi R., Otomo K., Yamada H., Tatsumi T., Sugawara I. 2006. Temperature-mediated heteroduplex analysis for the detection of drug-resistant gene mutations in clinical isolates of Mycobacterium tuberculosis by denaturing HPLC, SURVEYOR nuclease. Microbes Infect. 8:128–135 [DOI] [PubMed] [Google Scholar]
- 27. Takiff H. E., et al. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Umubyeyi A. N., et al. 2007. Limited fluoroquinolone resistance among Mycobacterium tuberculosis isolates from Rwanda: results of a national survey. J. Antimicrob. Chemother. 59:1031–1033 [DOI] [PubMed] [Google Scholar]
- 29. van Doorn H. R., et al. 2008. Fluoroquinolone resistance detection in Mycobacterium tuberculosis with locked nucleic acid probe real-time PCR. Int. J. Tuberc. Lung Dis. 12:736–742 [PubMed] [Google Scholar]
- 30. Xu P., et al. 2009. Prevalence of fluoroquinolone resistance among tuberculosis patients in Shanghai, China. Antimicrob. Agents Chemother. 53:3170–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yew W. W., et al. 2000. Outcomes of patients with multidrug-resistant pulmonary tuberculosis treated with ofloxacin/levofloxacin-containing regimens. Chest 117:744–751 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.