Abstract
Pseudomonas aeruginosa is a common opportunistic bacterial pathogen that causes a variety of infections in humans. Populations of P. aeruginosa are dominated by common clones that can be isolated from diverse clinical and environmental sources. To determine whether specific clones are associated with corneal infection, we used a portable genotyping microarray system to analyze a set of 63 P. aeruginosa isolates from patients with corneal ulcers (keratitis). We then used population analysis to compare the keratitis isolates to a wider collection of P. aeruginosa from various nonocular sources. We identified various markers in a subpopulation of P. aeruginosa associated with keratitis that were in strong disequilibrium with the wider P. aeruginosa population, including oriC, exoU, katN, unmodified flagellin, and the carriage of common genomic islands. The genome sequencing of a keratitis isolate (39016; representing the dominant serotype O11), which was associated with a prolonged clinical healing time, revealed several genomic islands and prophages within the accessory genome. The PCR amplification screening of all 63 keratitis isolates, however, provided little evidence for the shared carriage of specific prophages or genomic islands between serotypes. P. aeruginosa twitching motility, due to type IV pili, is implicated in corneal virulence. We demonstrated that 46% of the O11 keratitis isolates, including 39016, carry a distinctive pilA, encoding the pilin of type IV pili. Thus, the keratitis isolates were associated with specific characteristics, indicating that a subpopulation of P. aeruginosa is adapted to cause corneal infection.
INTRODUCTION
Pseudomonas aeruginosa is a common infective cause of corneal ulceration (bacterial keratitis) (14, 32, 54). P. aeruginosa keratitis is frequently associated with contact lens wear (54), because hypoxia and trauma from the contact lens allows bacterial adhesion, and as the bacterium is a common contaminant of moist areas such as a contact lens case, the exposure of the ocular surface to the bacterium can easily occur. Subsequently, a combination of P. aeruginosa virulence factors and damage to host ocular defenses allows infection to develop. The host immune and inflammatory responses then contribute both to the elimination of the bacteria and to associated damage to tissues (14). P. aeruginosa is an opportunistic pathogen that is capable of causing a range of infections (16, 38), and it carries an impressive array of virulence factors. The virulence factors implicated in keratitis include twitching motility associated with type IV pili (63), flagella (13), a type III secretion system (22), and quorum-sensing regulated exoproducts (58).
The genome of P. aeruginosa consists of core genes (approximately 90%) and accessory genes (approximately 10%) (40). The core genome, carried by all strains of P. aeruginosa, includes many of the recognized virulence genes (61). However, some strain-variable virulence-associated genes also occur, often clustered within genomic islands (3, 33, 47, 50, 60, 61). Such pathogenicity islands are key elements in the development of variable pathogenic behaviors within single bacterial species (15). It has been demonstrated that P. aeruginosa virulence can be combinatorial (34), with different combinations of accessory genome modules contributing to the virulence of different strains. In isolates of P. aeruginosa from cases of keratitis, there is some evidence for preferential association with particular accessory genome modules. For example, we (59) and others (37) have observed an association with the carriage of genes required for the secretion of the cytolytic exotoxin U rather than the alternative exotoxin S. It also has been reported that exoU-positive strains of P. aeruginosa cause more severe corneal disease (35, 53).
P. aeruginosa populations undergo frequent recombination events contributing to the evolution of successful epidemic clones (42, 43). Moreover, individual isolates of clones tend to share specific repertoires of accessory genomic segments (57). In a previous study, we screened a different set of 63 P. aeruginosa isolates from cases of ulcerative keratitis collected from six United Kingdom centers for a limited number of variable virulence-related phenotypes and genotypes (59). Using random amplified polymorphic DNA typing, we found no evidence for a common clone, although 61% of isolates were either serotype O1 or O11.
In this study, we have used a portable bacterial genotyping system to analyze a collection of United Kingdom isolates of P. aeruginosa from cases of keratitis for both the population structure based on the core genome and the carriage of variable genomic islands. Furthermore, we report an analysis of the genome sequence of a serotype O11 isolate that was associated with severe disease and determine the distribution of its genomic islands and variable genes among the wider collection. We found evidence for a subpopulation of P. aeruginosa associated with keratitis infection, and we report an association between a subset of O11 isolates of P. aeruginosa that caused a severe keratitis and the carriage of a novel variant of type IV pilus-associated pilA.
MATERIALS AND METHODS
Bacterial isolates.
Isolates of P. aeruginosa were collected as part of a collaborative venture involving the United Kingdom Microbiology Ophthalmic Group, comprising microbiologists and ophthalmologists from six United Kingdom centers in London (Moorfields Eye Hospital), Birmingham, Newcastle, Bristol, Manchester, and Liverpool. Between April 2003 and March 2004, isolates from the corneas of patients with ulcerative keratitis were sent to the reference laboratory in Liverpool for storage. This study involves 63 isolates confirmed as P. aeruginosa by the PCR amplification of the oprL gene (11) (Table 1). The distribution of serotypes among the 63 isolates has been reported previously (59).
Table 1.
Strains used in this study
| Strain | Serotype | Sourcea | Hexa.c | DB cloned | Novel pilAe |
|---|---|---|---|---|---|
| 39390 | O2 | LON | 0812 | V | − |
| 39291 | O2 | LON | 0812 | V | − |
| 39129 | O2 | LON | 0812 | V | − |
| 39091 | O4 | LON | 7421 | None | − |
| 48017 | O11 | NPM | 9429 | Eb5 | − |
| 39008 | O11 | LIV | 9429 | Eb5 | + |
| 39033 | O6 | BRI | 0C1A | B10 | − |
| 39284 | NTb | LON | 239A | L | − |
| 39421 | O4 | LON | 2C1A | F | − |
| 39285 | NT | LON | 2F92 | B19 | − |
| 39376 | O6 | BRI | 3C2A | U | − |
| 39228 | O11 | LON | 4B9A | None | + |
| 48020 | O4 | LIV | 4C82 | None | − |
| 39130 | O11 | LON | 6B9A | None | + |
| 39255 | O3 | MAN | 744A | None | − |
| 39053 | O6 | BRI | 7C2E | A5 | − |
| 39221 | O6 | LON | 7C2E | A5 | − |
| 39005 | O1 | LIV | 840A | B22 | − |
| 48002 | O1 | BRI | 840A | B22 | − |
| 39050 | O11 | BRI | A429 | B25 | − |
| 39340 | O11 | BIR | A429 | B25 | − |
| 39098 | O1 | LON | A429 | B25 | − |
| 39386 | O1 | BRI | A429 | B25 | + |
| 39274 | O11 | MAN | A429 | B25 | − |
| 48011 | O6 | MAN | AC2A | B27 | − |
| 39047 | O1 | BRI | B429 | B31 | − |
| 39092 | O11 | LON | B429 | B31 | − |
| 39320 | O11 | LON | B429 | B31 | − |
| 39299 | O11 | LON | B469 | B32 | + |
| 39384 | O1 | BRI | C40A | C | − |
| 39201 | O1 | MAN | C40A | C | − |
| 39266 | O1 | NEW | C40A | C | − |
| 39135 | O1 | NEW | C40A | C | − |
| 48005 | O11 | LON | C429 | None | − |
| 39115 | O10 | LON | D421 | A | − |
| 48023 | O10 | NEW | D421 | A | − |
| 48008 | O10 | LON | D421 | A | − |
| 39191 | O10 | LON | D421 | A | − |
| 39004 | O1 | LIV | E40A | C7 | − |
| 39015 | O11 | MAN | E429 | B | − |
| 39158 | O11 | LON | E429 | B | − |
| 39352 | O11 | BIR | E469 | B34 | − |
| 39324 | O10 | LON | E661 | B35 | − |
| 39177 | O1 | MAN | EA0A | A3 | − |
| 39145 | O1 | MAN | EA0A | A3 | − |
| 39055 | O1 | NEW | F419 | V2 | − |
| 39413 | O11 | MAN | F419 | V2 | + |
| 39402 | O11 | MAN | F419 | V2 | + |
| 39302 | O8 | LON | F421 | A2 | − |
| 39103 | O8 | LON | F421 | A2 | − |
| 39061 | O1 | LIV | F429 | I | − |
| 39172 | O11 | LON | F429 | I | − |
| 39096 | O11 | LON | F429 | I | − |
| 39293 | NT | LON | F461 | B37 | − |
| 39181 | NT | LON | F461 | B37 | − |
| 39016 | O11 | MAN | F469 | D | + |
| 39304 | O11 | LON | F469 | D | + |
| 39375 | O11 | BRI | F469 | D | + |
| 39394 | O11 | BRI | F469 | D | + |
| 39212 | O11 | LON | F469 | D | + |
| 39267 | O8 | NEW | F469 | D | − |
| 39087 | O11 | LON | F469 | D | − |
| 39131 | O1 | LON | F469 | D | − |
LIV, Liverpool; LON, Moorfield Eye Hospital, London; NEW, Royal Victoria Infirmary, Newcastle-upon-Tyne; MAN, Manchester Royal Eye Hospital; BRI, Bristol Royal Infirmary; BEH, Bristol Eye Hospital; BIR, Birmingham and Midlands Eye Centre; NPM, Northwick Park & St. Mark's Hospitals, London.
NT, not serotypeable.
Hexa., hexadecimal code.
DB clone, corresponding database clone.
A plus indicates that the sample was PCR positive for the novel pilA gene.
Antimicrobial susceptibilities and clinical outcome data.
The 63 P. aeruginosa isolates in this study are from a larger collection of bacterial isolates that has been used to assess antimicrobial susceptibilities (51) and clinical outcome (24). The MIC was determined using Etest strips (AB bioMérieux, Solna, Sweden) according to the manufacturer's instructions. The MIC for each antimicrobial was recorded as the intersection of the zone of inhibition with the scale on the Etest strip as described previously.
To determine clinical outcome, the following data were collected at the date of presentation and again at the date of healing: the size (orthogonal diameters of the major and minor axes) of the corneal ulcer (41) and scar, minimum distance from the limbus, duration of treatment, corneal surgery (e.g., application of corneal glue or tectonic or penetrating corneal graft), and the loss of the eye (enucleation or evisceration) (24). Healing time was defined as the interval to the closure of the epithelial defect (the absence of an epithelial defect using fluorescein staining on slit-lamp biomicroscopy) and treatment time as the interval during which antimicrobials were prescribed. Because ulcer size is an important determinant of healing, the primary clinical outcome measure used was the ratio of the healing time to ulcer size as previously defined (24).
For both antimicrobial susceptibility and clinical outcome, values obtained for isolate 39016 were compared to those for a larger collection (n = 91) of P. aeruginosa isolates from keratitis patients reported previously (24, 51).
Genotyping using Clondiag AT.
The Clondiag (Jena, Germany) ArrayTubes (AT) genotyping system (57) was used according to the protocol provided by the manufacturer. The AT microarray chip detects two different types of sequences: 13 single-nucleotide polymorphisms (SNPs) for the analysis of the conserved genome and 38 variable genetic markers for the analysis of the accessory genome. The latter include markers for virulence factors and previously reported genomic islands. The data from the 13 SNPs, flagellin type (a or b), and presence of the mutually exclusive type III secretion exotoxins (S or U) can be converted into a hexadecimal code represented by four digits and allowing the published database to be searched (56). For SNP analysis, the PAO1 type allele is designated 0 and the alternative allele is designated 1 (for example, oriC0 or oriC1).
Analyses of P. aeruginosa populations.
An eBurst (version 3.0) analysis was performed with all 63 P. aeruginosa keratitis isolates and a database of strains from three previous studies (39, 46, 56). All strains were analyzed for 14 binary markers (13 SNPs and fliC type a or b). exoS and exoU were not included in the eBurst analyses.
In addition, with the same data a distance plot was carried out using the program TIGR-Mev. A distance matrix based on SNP patterns was constructed, resulting in a distance plot where the major components allow the localization of the positions of all strains in the multidimensional space. The distances of a specific strain from all others represent its position in a 136-dimensional space. To visualize this distribution, the normalized positions were projected onto a two-dimensional surface using the TIGR-Mev software package.
DNA isolation and PCR amplification assays.
Genomic DNA was isolated from P. aeruginosa 39016 using the Wizard Genomic DNA purification kit (Promega). DNA for PCR amplification assays was obtained by resuspending one to three colonies of bacteria in 200 μl of a suspension of 5% (wt/vol) Chelex-100 (Bio-Rad). After vigorous mixing, the suspension was boiled for 5 min and then centrifuged for 2 min at full speed in a benchtop microcentrifuge. Supernatant (150 μl) was removed and stored at −20°C for use directly in PCR assays. Oligonucleotide primers and annealing temperatures used in PCR amplification assays are shown in Table 2.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence | Amplicon size (bp) | Target gene or RODa | Annealing temp (°C) | Reference/comment |
|---|---|---|---|---|---|
| PAL1 | ATGGAAATGCTGAAATTCGGC | 504 | oprL | 57 | 11 |
| PAL2 | CTTCTTCAGCTCGA | ||||
| ORF265F | GTGGGTTTGCAAAAGCGTAT | 234 | ROD1 | 58 | Putative ATPase |
| ORF265R | CACCTCTTCAGGTGTGCTGA | ||||
| ORF624F | GAGCCAGCCTCTTGAGTCAC | 203 | ROD2 | 58 | Hypothetical protein |
| ORF624R | TGCGGGTATAGGTTTTCTGG | ||||
| ORF2240F | GCCAGTCACCAAGATGGAGT | 167 | ROD6 | 58 | Putative cI-like repressor (D3-like phage) |
| ORF2240R | ATCTCAACATGCGGGTTCTC | ||||
| ORF2549F | AGGCAACGAAGCCAAGATAA | 302 | ROD7 | 58 | Putative outer membrane efflux protein |
| ORF2549R | TGCTGAAGCTGACACCGTAG | ||||
| ORF3034F | GCTACACCCTCGTGGAGATG | 306 | ROD11 | 58 | Putative secreted protein |
| ORF3034R | GCCGCTCGAAGCAGATTT | ||||
| ORF4339F | AACTCGCAATCCACCGTATC | 150 | ROD15 | 58 | Putative HlyD-like secretion protein |
| ORF4339R | GATCCGTCCTCCTGTTTCAA | ||||
| ORF5228F | GTCATGCCCACAAACTGATG | 325 | ROD16 | 58 | Conserved hypothetical protein in B3-like prophage |
| ORF5228R | ACCTTGGTGGACCGCTTAC | ||||
| ORF5375F | GTAGCGGCTCCAAACTGAAG | 207 | ROD17 | 58 | Novel hypothetical protein in B3-like prophage |
| ORF5375R | TAAGCTCGGTGGCGATGTAG | ||||
| ORF5388F | TGTTCATGGACATGGAGGAA | 326 | ROD17 | 58 | Putative portal protein in MP38-like prophage |
| ORF5388R | CAGCTCGTTCTGGTCTTCG | ||||
| ORF6116F | TCGAATGTGAAGTGCCTCAG | 218 | ROD18 | 58 | Putative acetyltransferase |
| ORF6116R | GTAACGGATTTCGGTGTTGC | ||||
| novPilAF | CGGGTTCCAGTTTGTTGACT | 184 | pilA | 58 | Targets C-terminal half of novel pilA |
| novPilAR | CAGCCACCATTAACATCACG |
Target ROD indicates that the primers target a gene in a region of difference, as defined in footnote b in Table 5. With the exception of the primers for the oprL assay, all primers were designed during this study.
Genome sequence generation.
Strain 39016 was sequenced using the Roche 454 genome sequencer FLX (GS-FLX) by following the manufacturer's instructions (Roche Life Science, Branford, CT). In brief, (i) a fragment library and (ii) a 3-kb paired-end library were prepared using the standard FLX chemistry for the 454 genome sequencer. The fragment library was prepared by fragmentation, the attachment of adapter sequences, the refinement of the ends, and the selection of adapted molecules. The paired-end library was produced by hydroshear shearing, circularization, the addition of adapters, and selection as described for the fragment library. Both libraries were amplified by emulsion PCR (emPCR), and fragment-containing beads were recovered and enriched. Sequencing primers were added and each library was deposited onto half a PicoTiterPlate plate and sequenced.
Genome assembly and annotation.
Reads were assembled de novo into contigs, which in turn were incorporated into scaffolds using the Roche 454 Newbler assembler (version 2.3) with default settings. The resulting scaffolds were ordered and orientated with respect to Pseudomonas aeruginosa PAO1 (AE004091) using an in-house Perl script alongside the alignment program NUCmer (part of the MUMmer 3.20 software package; http://mummer.sourceforge.net/). A single scaffold then was generated from this ordering with 100-base stretches of Ns separating each scaffold.
For each scaffold-contig in turn, putative open reading frames (ORFs) were called using Glimmer version 3.02 (http://www.cbcb.umd.edu/software/glimmer/). A further in-house Perl script then was run to identify and correct those ORFs likely to have been split due to sequencing errors when handling homopolymer repeats. This involved the BLASTP alignment of ORF protein translations against a database of translations generated from previously annotated P. aeruginosa genomes, the identifications of likely indels within homopolymer regions, and the modification of coding sequence feature positions to correct errors and the merging of relevant ORFs. Such modifications were recorded as metadata (in the form of the eventual GenBank feature note field). Such ORFs also were marked with the exception flag set to “low-quality sequence region” for the final GenBank submission to signify poor-quality sequencing.
Putative function then was assigned to each gene by BLAST (NCBI BLAST 2.2.17) comparison to a database of sequences generated from previously annotated P. aeruginosa genomes (P. aeruginosa PAO1, AE004091; P. aeruginosa UCBPP-PA14, CP000438; P. aeruginosa PA7, CP000744; and P. aeruginosa LESB58, FM209186). Putative tRNA genes were detected using tRNAscan-SE 1.23 (ftp://selab.janelia.org/pub/software/tRNAscan-SE/).
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession number AEEX00000000. The version described in this paper is the first version, AEEX01000000. For convenience, an additional pseudochromosome (a concatenation of contigs guided by the scaffold with 100 Ns separating adjoining contigs) also was prepared from the same data for use with the sequence comparison tool ACT (part of Artemis; http://www.sanger.ac.uk/resources/software/artemis/).
A dendrogram of PilA amino acid sequences was constructed using sequences retrieved from GenBank and aligned using CLUSTAL X (55). A phylogenetic tree was generated from the alignment using the genetic distance-based unweighted-pair group method using average linkage algorithms of the Data Analysis in Molecular Biology software (DAMBE; http://dambe.bio.uottowa.ca/software.asp).
Nucleotide sequence accession numbers.
The sequences determined in the course of this work have been deposited at DDBJ/EMBL/GenBank under the accession numbers AEEX00000000 and AEEX01000000.
RESULTS
Distribution of clones among the keratitis isolates.
We analyzed 63 isolates using the AT genotyping system (56) The core genome data are summarized in Table 1. We assigned each isolate a hexadecimal code for comparison to the database of previously genotyped P. aeruginosa isolates. In a previous study, 16 clones made up more than half of a panel of 240 strains from diverse habitats and geographical origins (clones A to P) (56). Twenty-three (37%) of the 63 keratitis isolates matched one of these major clones. Six of the isolates (10%) did not match any clone in the database. The largest number of isolates sharing the same clone designation was eight representatives of clone D, a clone that was isolated from four different centers (Table 1).
Population structure analysis of P. aeruginosa keratitis isolates.
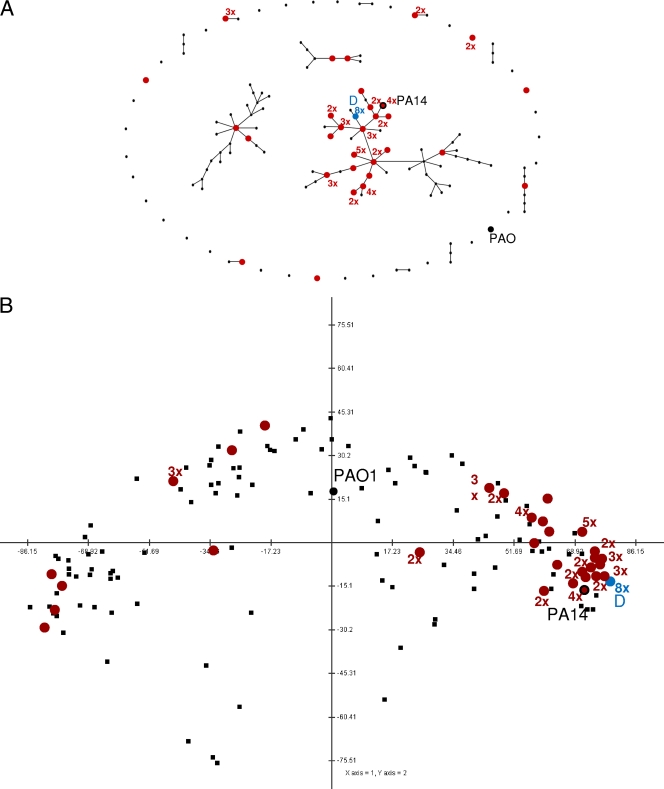
We used two complementary approaches to examine the position of the keratitis P. aeruginosa clones among the wider population structure of P. aeruginosa isolates (Fig. 1). First, eBurst analysis revealed that 47 of the 63 isolates (75%) were located within a major cluster that includes 7 of the 16 most common P. aeruginosa clones (Fig. 1A). All 47 isolates in this cluster are very closely related. The relative positions within the cluster, however, as predicted by the eBurst algorithm, are not unequivocal, and many of the isolates at one branch of the tree are only single-locus variants (SLVs) of isolates from another branch. However, although the order of the branches in this cluster of strains as depicted in Fig. 1A represents only one option, the 47 keratitis isolates were clustered together in all possible variants.
Fig. 1.
Positions of keratitis isolates in the P. aeruginosa population. (A) Position of all analyzed keratitis isolates of this study in an eBurst analysis containing all published, SNP-typed P. aeruginosa strains. Each dot represents a clone based on the AT profile. Two dots connected by a single line differ in only one locus (single-locus variants [SLV]). Most keratitis isolates represent a closely related subgroup. (B) Two-dimensional projection of the distance matrix of SNP-typed P. aeruginosa strains. The plot was performed with the correlations between values in the distance matrix. Hence, this is a plot of the correlations of the genetic distances of the strains to each other, making it easy to see that most keratitis isolates are very closely related. Distance values were normalized and mean centered. The first two components, represented by the axes in the figure, contributed 90% of the variance of the SNP patterns. As for the eBurst analysis, most keratitis isolates fall within a closely related subgroup. Completely unrelated mathematical models were used for A and B, but both plots indicated that the keratitis isolates represent a specific, closely related subpopulation within the global P. aeruginosa population structure and may have arisen from a common ancestor. The locations of keratitis isolates are indicated in red, with the number of keratitis isolates at each location (if more than one) also shown. For example, 2× indicates two isolates at one location. The positions of clone D (blue circle) and the reference strains PAO1 (large black circle) and PA14 (red circle with black circumference, indicating that this clone type also was present among the keratitis isolates) are shown. The same keratitis strains clustered using both methods.
In a complementary and independent approach, the population structure was analyzed by the calculation of differences/distances of the SNP patterns of all 136 different strains. More than 90% of the total variance of the SNP distances could be described in this two-dimensional projection. As in the eBurst analysis, the majority of P. aeruginosa strains were separated into a few major clusters of closely related strains (Fig. 1B). Most keratitis isolates (51 of 63 isolates) belong to the same cluster, underlining the close relationship of these strains.
The analysis of the keratitis isolates confirmed a previously observed association between the oriC0 allele and a lack of exoU (55). The keratitis isolates that carried the oriC1 allele, however, differed from strains in the previous study, which included isolates from diverse sources. Whereas in the collection analyzed by Wiehlmann et al. (56) exoS was present in about 50% of the oriC1 strains, only 10 of 48 (21%) keratitis isolates with oriC1 were exoS positive, while the other 38 carried the exoU gene instead. From a total survey of 322 isolates representing 128 clones, P. aeruginosa carrying oriC0 numbered 179 isolates (56%) in 76 strains (59%), whereas P. aeruginosa isolates carrying oriC1 numbered 143 isolates in 52 strains (39, 46, 56). There was a significantly higher proportion of P. aeruginosa carrying the oriC1 allele (48 of 63 strains [76%]; P < 0.001) in the keratitis isolates. This suggests that P. aeruginosa strains that cause corneal infections predominantly belong to a subpopulation of P. aeruginosa, characterized by the oriC1 allele and the presence of exoU.
Distribution of genomic islands among the keratitis isolates according to AT analysis.
All but three of the keratitis isolates were positive for the common genomic island PAGI-1 (36). PAGI-2 and PAGI-3 are representatives of a family of genomic islands that integrate at the tRNA(Gly) locus (26, 33). The carriage of PAGI-2/PAGI-3-like islands was not demonstrated in more than half of the keratitis isolates. In the other half, only six isolates were positive for more than 3 of the 10 possible hybridization signals on the chip. Six of the AT markers target genes that belong to the conserved part of this kind of island, whereas only four are specific for PAGI-2 or PAGI-3. Hence, we conclude that a genomic island related to the class PAGI-2/3 family was absent from 37 of 63 keratitis isolates (58%). This compares with 159 (49%) from 322 isolates from a larger P. aeruginosa collection from diverse sources (39, 46, 56).
PAPI-1, PAPI-2, and pKLC102 belong to a family of genomic isolates that integrate at tRNA(Lys) loci and often carry virulence factors for attachment or transport (18, 26). The AT markers pKL-1 and pKL-3 target the conserved part of this family of genomic islands, whereas the other AT markers target genes representing specific family members. Among the keratitis isolates, seven carried pKLC102 and three carried PAPI-1. The AT data showed that 38 of the 63 keratitis isolates do not contain any genomic island from this family, compared to 94 of 322 reference isolates (P < 0.001).
The flagellin glycosylation island is specific to P. aeruginosa with type a flagellins, but some strains carry an abbreviated version of the island (2). Twelve (19%) of the keratitis isolates carried the complete island, whereas 91 (28%) isolates from the larger reference collection did so. The incomplete glycosylation island was carried by 35 (56%) keratitis isolates and 112 (35%) isolates from the wider reference collection. Hence, strains that lack the whole island and are unable to modify type a flagellins were overrepresented among the keratitis isolates, but the difference was not statistically significant.
Distribution of other variable genes among the keratitis isolates.
Compared to carriage by the wider strain collection, there were no significant differences in the carriage of the variable ORFs PA0636, PA0722, PA0728, PA2221, and PA3835 in the keratitis isolates. In contrast, 18 of 63 (29%) keratitis isolates were positive for ORF PA2185, whereas 188 of 322 (58%) were positive in the wider collection. Thus, keratitis isolates were significantly less likely (P < 0.0001) to carry PA2185, which encodes the nonheme catalase KatN. The distribution of pyoverdine receptor types closely resembled that of non-cystic fibrosis isolates from the larger reference collection (57).
Genome sequence of isolate 39016.
We selected isolate 39016 for further genome sequencing. This was because it represents the most abundant clone in this study (clone D) and the most common serotype (O11), it occupies a central location within the major cluster of isolates (Fig. 1), and it was associated with a severe keratitis (large corneal ulcer, prolonged healing time, and relatively high resistance to antimicrobial agents [Table 3]). In addition, AT data suggest that it lacks many of the accessory genome genes represented on the microarray. Hence, we hypothesized that its accessory genome carries novel genomic islands shared with other keratitis isolates.
Table 3.
MICs of antimicrobials against isolates of P. aeruginosa, including 39016a
| Antimicrobial | MIC for isolate 39016 | Value for all isolates, including 39016 |
P. aeruginosa systemic breakpoint | |||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Min | Max | |||
| Penicillin | 256 | 512 | 512 | 256 | 512 | NA |
| Cefuroxime | 256 | 300 | 512 | 256 | 512 | NA |
| Ceftazidime | 1.5 | 1 | 1.50 | 0.25 | 3 | 8 |
| Chloramphenicol | 24 | 512 | 512 | 0.09 | 512 | NA |
| Gentamicin | 1.5 | 1.50 | 2 | 0.19 | 12 | 4 |
| Amikacin | 2 | 2 | 3 | 1 | 512 | 8 |
| Vancomycin | 256 | 512 | 512 | 256 | 512 | NA |
| Teicoplanin | 256 | 512 | 512 | 256 | 512 | NA |
| Ciprofloxacin | 0.94 | 0.09 | 0.25 | 0.03 | 1 | 0.5 |
| Ofloxacin | 0.75 | 0.50 | 1.50 | 0.03 | 6 | 2 |
All values are in mg/liter. The table shows the minimum (Min) and maximum (Max) MICs and the MIC at which 50% (MIC50) and 90% (MIC90) of isolates were inhibited for a collection of keratitis isolates reported previously (24, 51). The systemic breakpoint concentrations for which P. aeruginosa is regarded as susceptible or resistant to that antimicrobial according to the British Society of Antimicrobial Chemotherapy (1) are included. NA, not applicable.
Details of the genome sequence data obtained are summarized in Table 4. The 13 scaffolds obtained were ordered with reference to the genome of strain PAO1 to produce a single pseudochromosome. The ACT analysis of the pseudogenome compared to that of the genome of strain PAO1 showed the presence of a number of large regions of additional DNA in the isolate 39016 genome (regions of difference [ROD]) (Table 5). The location of several with reference to the genome of PAO1 corresponded with regions of genomic plasticity (RGP) (40) reported previously (Table 5). In contrast, there were relatively few strain PAO1 genes absent from the genome of isolate 39016 (Table 6), reflecting the larger genome size of the keratitis isolate.
Table 4.
Summary of genome sequence data
| Data source | Total | No. of bases | Avg length (mean, bp) | Avg length (median, bp) | N50 length (bp) | Minimum length (bp) | Maximum length (bp) |
|---|---|---|---|---|---|---|---|
| Fragment library | 310,982 | 73,369,878 | 235.93 | 247 | 28 | 329 | |
| Paired end library | 477,473 | 59,667,177 | 124.96 | 116 | 20 | 481 | |
| Contigs | 486 | 6,760,206 | 13,909.89 | 5,948 | 35,020 | 108 | 214,365 |
| Scaffolds | 13 | 6,864,765 | 528,058.85 | 270,819 | 810,981 | 2,219 | 2,193,829 |
Table 5.
Summary of regions of difference (>5 kb) in the genome of P. aeruginosa strain 39016 compared to the genome of PAO1
| RODa | Size (kb) | PAO1b | PA14c | tRNAd | Comment(s) |
|---|---|---|---|---|---|
| ROD1 (RGP2) | 34.7 | 0263–0264e | Part | Yes | Partial match to strains PA14 and PACS171b; has non-Pseudomonas matches in central region, including restriction-modification proteins |
| ROD2 | 8.3 | 0574–0575 | – | Yes | Non-Pseudomonas matches, includes phage integrase |
| ROD3 (RGP6) | 41.3 | 0819–0827e | – | Prophage; partial match with phage F10 along the whole length, but very divergent from F10; generally low-identity protein matches | |
| ROD4 (RGP7) | 79.0 | 0977–0980 | Part | Yes | High identity shared with strain 6077 ExoU island A |
| ROD5 (RGP9) | 16.5 | 1086–1095 | – | Flagellar and flagellar glycosylation locus | |
| ROD6 (RGP19) | 56.3 | 1964–1971 | – | Prophage; partial match to Pseudomonas phage D3 (<30%) | |
| ROD7 | 23.6 | 2211–2212 | – | Integrated mobile element; same 5-kb match with strain PA7 and plasmid pRA2 (including transposase), some non-P. aeruginosa matches (including two-component regulatory system; 68-81% identity with P. putida and transport proteins) | |
| ROD8 (RGP23) | >50 | 2218–2229e | – | Incomplete because of gap but includes extensive match with PAGI-1 | |
| ROD9 (RGP56) | 29.9 | 2396–2403 | Part | Type II pyoverdine biosynthesis locus | |
| ROD10 (RGP27) | >110 | 2583–2584e | – | Yes | Genomic island; extensive high-identity matches with the genomes of other Gram-negatives, such as Bordetella petrii, Comamonas testosterone, Acidovorax sp. strain JS42, Delftia acidovorans, and P. aeruginosa 2192; includes heavy-metal resistance genes and some non-Pseudomonas matches |
| ROD11 | 9.9 | 2683–2684e | – | 99% Identity with PACS10223 (incomplete sequence; contains a gap); includes putative ABC transporter protein, proline racemase family protein, dihydrodipicolinate synthetase family protein, transporter protein | |
| ROD12 (RGP28) | >26.4 | 2729–2737 | – | Yes | Mostly non-Pseudomonas matches, including restriction-modification proteins |
| ROD13 | >54 | 2748–2749e | – | Putative integrated mobile element; at either end it matches strain PACS171b; the central region contains plasmid and transposase matches and putative sulfate transport proteins (crosses two scaffolds and contains gaps) | |
| ROD14 (RGP31) | 11.6 | 3141–3161 | – | Yes | O11 O-antigen biosynthesis genes |
| ROD15 | >45 | 3621–3622e | Part | Two regions sharing similarity with PA14 either side of a region (∼15 kb) containing non-Pseudomonas matches (including regulatory and transport-related proteins and a putative acriflavin resistance protein); contains some gaps | |
| ROD16 | 39.7 | 4436–4437 | – | Prophage; shares extensive similarity with strain PACS458 and phage B3, but with divergent regions | |
| ROD17 | 37.2 | 4512–4513 | – | Prophage; shares extensive similarity with phages DMS3, MP22, MP28, and D3112 | |
| ROD18 | >13.5 | 5149–5150 | – | Yes | Includes putative integrase and acetyltransferase |
| ROD19 | 6.1 | 5290–5291 | – | Mostly low-identity non-Pseudomonas matches, includes putative resolvase/recombinase protein; very low GC content | |
| ROD20 | >7.8 | 5548–5549 | – | Mostly non-P. aeruginosa matches, includes putative TnsA endonuclease, Tn7-like transposition protein B, helicase (contains gap) |
Table 6.
Regions deleted from the genome of P. aeruginosa isolate 39016 compared to the genome of PAO1
| PAO1 genomic region | Size (kb) | Comment(s) on ORFs in the deleted region |
|---|---|---|
| PA0202-0207 | 6.5 | Putative amidase; transport proteins and a regulatory protein |
| PA0715-0729 | 12.3 | Prophage Pf1 |
| PA0632-0648 | 13.9 | Deletion of phage-tail pyocin F2 genes |
| PA1381-1393 | 18.2 | Glycosyltransferases, PulD and transport protein, possible polysaccharide synthesis locus |
| PA2100-2106 | 7.0 | Putative transcriptional regulator and amino acid biosynthesis and metabolism proteins |
| PA2182-2188 | 5.9 | Hypothetical proteins and nonheme catalase KatN (PA2187 is not deleted) |
| PA2333-2336 | 6.5 | Putative sulfatase; regulatory protein and tonB-dependent protein |
| PA3065-3067 | 2.4 | Hypothetical proteins and putative regulatory protein |
| PA3497-3514 | 16.5 | Putative enzymes; transport, regulatory, and hypothetical proteins |
| PA4101-4107 | 6.6 | Putative two-component regulatory system and hypothetical proteins |
| PA4191-4195 | 4.2 | Putative transport proteins |
Virulence genes.
The vast majority of the 265 P. aeruginosa virulence factor coding sequences described previously for strain PAO1 (61) are present in the genome of strain 39106. The exceptions are ORFs for the probable type II secretion protein (XqhB; PA1382), a hypothetical protein (PA1867), and exotoxin S-related ORFs (PA1710 and PA3841). Some putative virulence genes, such as PA0041, encoding a putative filamentous hemagglutinin (FHA)-like protein, are present but divergent. Such divergence for P. aeruginosa FHA-like proteins has been reported previously (49). The flagellin (FliC; PA1092), a flagellar hook-associated protein (FlgL; PA1087), a capping protein (FliD; PA1094), and PA1095 also are present but divergent compared to PAO1. However, given the difference in the flagellin type of the two strains, this is not surprising. Likewise, the pyoverdine biosynthesis gene pvdD (PA2399) is divergent, reflecting a difference in pyoverdine type.
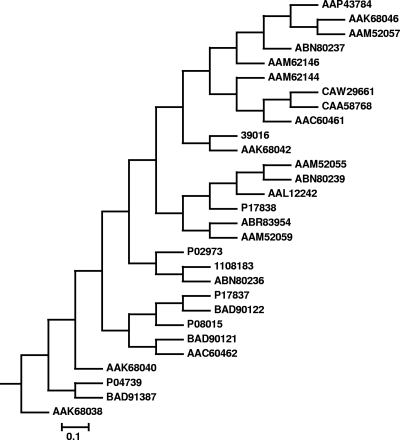
There also is divergence within the type IV pili gene cluster (PA4525 to PA4528), which is associated with adhesion and twitching motility. P. aeruginosa pil gene clusters can be subdivided on the basis of gene organization into five groups (I to V) (29). The genome of P. aeruginosa 39016 contains a group II arrangement of the pilABC genes (29). The predicted type IV pilin (encoded by pilA) of the keratitis isolate 39016 shares 99 to 100% amino acid identity with 3 of the 44 PilA sequences in the database. Two are represented by accession numbers AAK68044 and AK68042 and are from strains 82932 and 82935, which are clinical isolates of unknown source (20). There was also a third match to strain PA5235 (rectal swab; AAM44066) (29). The next-best P. aeruginosa match shared only 54% amino acid sequence identity.
Antimicrobial susceptibility and clinical outcome data.
The MIC data for the P. aeruginosa isolates used in this study are included in Table 3 with the systemic breakpoint data for comparison. Since the MICs are above the systemic breakpoints, isolate 39016 would be regarded as resistant to ciprofloxacin and ofloxacin, fluoroquinolones that are commonly used to treat keratitis due to P. aeruginosa. It should be noted, however, that there currently are no breakpoints for topical antimicrobials used to treat ophthalmic infections (24). The patient from whom 39016 was isolated had an ulcer size of 5 by 3 mm and a healing time of 60 days, with a ratio of healing time to ulcer size of 4 days/mm2. The mean healing time for the group of patients (n = 91) with P. aeruginosa keratitis is 15.23 (standard deviations [SD], 16.82) days, mean ulcer size is 2.81 (SD, 1.91) mm by 2.33 (SD, 1.74) mm, and the mean ratio of healing time to ulcer size is 3.75 (SD, 3.43) days/mm2 (24).
There was no evidence of mutations in gyrA, gyrB, or parC between the genome of isolate 39016 and previously published P. aeruginosa genomes. A single-amino-acid change in parE was found compared to the genome of PAO1, but this mutation has not been associated with resistance. It has been reported that mutations in the PAO1 genes PA1259 and PA2110 can lead to increased resistance to ciprofloxacin (12). The equivalents of these genes in the genome of isolate 39016 either are incomplete or are pseudogenes, which may explain the resistance phenotype.
Prophages and genomic islands.
The genome of strain 39016 contains four putative prophages, including prophages sharing some similarity with F10 (31) and D3 (27). In addition, there are prophages sharing extensive similarity with the Mu-like phage B3 (6) and a group of generalized transducing phages, including MP38, DMS3 (7), MP22, and D3112 (19) (Table 2). The genome of strain 39016, however, lacks the genes encoding the filamentous phage Pf1 (21), which is carried by strain PAO1 and many other P. aeruginosa strains (Table 6).
The genome of strain 39016 also contains a number of genomic islands, some of which have been reported for other P. aeruginosa strains. For example, in contrast to the reference strain PAO1, the genome of strain 39016 carries the incomplete version of the flagellin glycosylation island (2), the exoU island A (28), PAGI-1 (36), the O11 O-antigen biosynthesis genes, and type II pyoverdine biosynthesis genes. However, genomic islands sharing little or no similarity with previously reported P. aeruginosa genomes also were identified. These include a large genomic island (>110 kb) sharing some putative ORFs with PAGI-2 and P. aeruginosa strain 2192 but with regions of high similarity to clusters of genes from the genomes of diverse bacteria, including Bordetella petrii, Acidovorax sp. strain JS42, and Delftia acidovorans (Table 5). As with PAGI-2 and related islands (26), many ORFs within this island have putative roles in heavy-metal resistance or transport. Other shorter islands also contained putative ORFs with transport, sensing, or regulatory functions (Table 5).
Distribution of representative sequences of novel regions.
To address the question of whether novel components of the accessory genome of strain 39016 are widespread among the collection of keratitis isolates, we designed a number of indicative PCR amplification assays. The distribution data obtained suggested clear linkage between the accessory genome and O11 serotype, but variations were evident within the whole O11 group and the clone D subgroup (Table 7), suggesting considerable diversity with respect to the carriage or composition of genomic islands or prophages. There was little evidence for the widespread sharing of common accessory genes across serotypes. To examine the distribution of the strain 39016 variant of pilA among the keratitis isolates, the variable region of the novel pilA gene also was targeted in PCR assays. The novel pilA was detected in 12 of the keratitis isolates, including 46% of serotype O11 isolates and 63% of clone D isolates (Tables 1 and 7).
Table 7.
Distribution of ORFs within 39016 ROD according to PCR assaysa
| Serotype/clone | n | ROD1 (265) | ROD2 (624) | ROD6 (2240) | ROD7 (2549) | ROD11 (3034) | ROD15 (4339) | ROD16 (5228) | ROD17 (5375) | ROD17 (5388) | ROD18 (6116) | Novel PilA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O1 | 15 | 0 | 0 | 3 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 |
| O2 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| O3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| O4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| O6 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| O8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| O10 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 |
| O11 | 24 | 6 | 1 | 3 | 3 | 2 | 8 | 8 | 0 | 7 | 10 | 11 |
| NT | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 |
| Clone D | 8 | 5 | 1 | 2 | 1 | 1 | 5 | 6 | 1 | 4 | 6 | 5 |
Primer pair names are indicated in parentheses beneath the ROD they were designed to target. NT, nontypeable by serotyping.
A dendrogram of the different PilA sequences is shown in Fig. 2. There is evidence for the association of specific type IV pilin allele types with isolates from a particular source (group I pilins in cystic fibrosis) (29). Although our numbers are insufficient for statistical significance to be adequately assessed, we did find a predominance of particularly O11, exoU-positive, novel pilA type isolates with good twitching motility in our keratitis collection.
Fig. 2.
Dendrogram based on PilA amino acid sequences. The figure shows a phylogenetic tree comparing 28 database PilA sequences, including those of the commonly used strains PAO1 (P04739) and PA14 (AAL12242), to the PilA sequence of strain 39016, indicating the considerable diversity in P. aeruginosa PilA sequences.
DISCUSSION
Previous studies have suggested that although dominant clones are a feature of P. aeruginosa populations, these clones are distributed widely among collections from diverse geographical, environmental, or clinical sources (42, 57). We present evidence that a specific subpopulation of P. aeruginosa is associated with corneal infections (keratitis). Other approaches have been used to study populations of P. aeruginosa from various sources. Multilocus sequence typing indicates the divergence of oceanic P. aeruginosa isolates from the general population (25), whereas an alternative genotyping approach suggests that diversity among river water isolates broadly reflects global diversity (45). Our observations strongly indicate an association between a particular group of related P. aeruginosa clones and keratitis. Although this keratitis-associated subpopulation is in equilibrium with the global P. aeruginosa population for many of the markers analyzed in this study, including common bacteriophage-related genes like PA0722 and PA0728, or the pyoverdine receptors, there is a strong disequilibrium for some specific characteristics, with the keratitis group being characterized by oriC allele type 1, the presence of exoU, glycosylated but unmodified flagellin, the absence of the nonheme catalase gene katN, and the lack of tRNA(Lys)-integrated genomic islands like pKLC102 or PAPI-1. Although there was variation within the group with respect to SNP patterns, the majority of keratitis isolates fell within a cluster of highly related strains, suggesting common ancestry.
The cluster including the majority of keratitis isolates includes 7 of the 16 most common clones reported in a previous study (56). Within this cluster lie eight keratitis clone D isolates, representing the largest single clone grouping. All but two of these isolates have been identified as serotype O11. Although most isolates sharing common clone designations also shared serotypes, we found some evidence for variable serotypes within single clone types, mostly due to isolates being either O1 or O11. This suggests that P. aeruginosa clones can undergo serotype conversion. Particular bacteriophage-mediated examples of this have been reported previously (30).
Serotype O11 isolates have been reported at prevalence levels of 15 to 18% in a number of surveys of clinical and environmental isolates (5, 43, 44). We (37% [58]) and others (28% [62]) have reported higher prevalences among collections of isolates associated with keratitis. There is also a significant association between serotype O11 isolates and the presence of exoU (5, 59, 62). We also have noted previously that the two dominant serotypes among the keratitis collection (O1 and O11) both exclusively carried type a flagellin genes (59).
Despite previous observations that particular clones of P. aeruginosa are characterized by specific repertoires comprising their accessory genomes (56), genotyping using the AT system and indicative PCR assays aimed at surveying the distribution of genes from the accessory genome of strain 39016 indicated that there is little evidence for the widespread sharing of genes from the accessory genome among keratitis isolates. Bacteriophages can contribute to virulence (60), but there was no evidence for the common carriage of particular prophages among the keratitis isolates. As has been reported previously for other genome-sequenced P. aeruginosa isolates (40, 47, 60), the genome of isolate 39016 harbored a number of genomic islands, some closely related to previously sequenced islands, others more novel. Beyond some associations with the O11 serotype, however, there was little evidence to suggest the presence of a particular genomic island associated with keratitis infections. It is difficult to make direct links between the genomic content of the keratitis-associated subpopulation of P. aeruginosa and specific roles in corneal infections. The keratitis isolates tend to lack KatN, a nonheme catalase. Active oxygen species, such as peroxide, can play a role in corneal ulceration (8), and P. aeruginosa tolerance to hydrogen peroxide may be an advantage. The lack of a catalase therefore may seem counterproductive. There are, however, other catalases that could compensate for this (9).
The majority of clone D isolates, and some other O11 isolates, carry an unusual group II pilA gene (29). It has been demonstrated that mutants of P. aeruginosa defective in twitching motility show a reduced ability to colonize the cornea of mice (63), suggesting an important role for type IV pili in corneal infection. We reported previously that 90% of the 63 keratitis isolates used in this study exhibited better twitching motility than strain PA14 (59). Given the importance of type IV pili in twitching motility and the potential additional role in adhesion, our observations suggest that the carriage of certain pilA allele types confer some advantage to P. aeruginosa strains in relation to corneal infection.
Organism virulence is only one contributor to outcome in infectious diseases. We have shown previously in bacterial keratitis, using a general linear multivariate model, a weak (14%) but significant association between the MIC of the antimicrobial prescribed and clinical outcome, defined by the ratio of healing time to ulcer size (24). We also demonstrated the importance of the bacterial type and particular antimicrobial used for treatment (24, 52). The importance of this relationship is underlined by the finding that although 99% of P. aeruginosa isolates were found, for example (using systemic breakpoint data), to be susceptible to ceftazidime, ciprofloxacin, levofloxacin, and moxifloxacin, the association between clinical outcome and MIC for P. aeruginosa was 10% (24, 52). Hence, although clinical outcome in P. aeruginosa-associated keratitis is dependent in part on the MIC of the prescribed antimicrobial (10% for fluoroquinolones), the majority of the clinical outcome relates to other factors. The finding therefore of novel genotypic factors in P. aeruginosa isolates, some of which are associated with prolonged healing or poor outcome, such as that described in this study for isolate 39016, suggests that these are important for virulence. The identification of such genotypes therefore potentially is important and may help both the microbiologist and clinician in determining prognosis and possible treatment modifications.
In conclusion, our study suggests that there is a subgroup of P. aeruginosa isolates that is more likely to be found among keratitis isolates. Although such a group may represent a novel pathotype, it is clear that other P. aeruginosa strains also can cause corneal infections, and the biological basis of the clustering has yet to be elucidated. Associations between specific gene profiles and host niche preferences have been demonstrated in other pathogens (4, 23), including the plant pathogen Pseudomonas syringae (48). Previous studies generally have found little evidence for such associations in populations of P. aeruginosa (10, 61). It is known, however, that a greater prevalence of carriage of exoU is a feature among keratitis isolates (37, 59). Here, we report further evidence for specialization within P. aeruginosa populations associated with infections of the cornea.
ACKNOWLEDGMENTS
The members of the Microbiology Ophthalmic Group are Stephen Tuft, Stephen Kaye, Timothy Neal, Derek Tole, John Leeming, Peter McDonnell, Timothy Weller (deceased), Francisco Figueiredo, Steven Pedler, Andrew Tullo, and Malcolm Armstrong.
We acknowledge the support of the Royal Liverpool and Broad Green University Hospitals NHS Trust. The development of Artemis and ACT was funded by the Wellcome Trust's Beowulf Genomics initiative through its support of the Pathogen Sequencing Unit, Sanger Centre, United Kingdom.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1. Andrews J. M. 2009. BSAC standardized disc susceptibility testing method (version 8). J. Antimicrob. Chemother. 64:454–489 [DOI] [PubMed] [Google Scholar]
- 2. Arora S. K., Wolfgang M. C., Lory S., Ramphal R. 2004. Sequence polymorphism in the glycosylation island and flagellins of Pseudomonas aeruginosa. J. Bacteriol. 186:2115–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Battle S. E., Rello J., Hauser A. R. 2009. Genomic islands of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 290:70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ben Zakour N. L., Guinane C. M., Fitzgerald J. R. 2008. Pathogenomics of the staphylococci: insights into niche adaptation and the emergence of new virulent strains. FEMS Microbiol. Lett. 289:1–12 [DOI] [PubMed] [Google Scholar]
- 5. Berthelot P., et al. 2003. Genotypic and phenotypic analysis of type III secretion system in a cohort of Pseudomonas aeruginosa bacteremia isolates: evidence for a possible association between O serotypes and exo genes. J. Infect. Dis. 188:512–518 [DOI] [PubMed] [Google Scholar]
- 6. Braid M. D., Silhavy J. L., Kitts C. L., Cano R. J., Howe M. M. 2004. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 186:6560–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Budzik J. M., Rosche W. A., Rietsch A., O'Toole G. A. 2004. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 186:3270–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carubelli R., Nordquist R. E., Rowsey J. J. 1990. Role of active oxygen species in corneal ulceration. Effect of hydrogen peroxide generated in situ. Cornea 9:161–169 [PubMed] [Google Scholar]
- 9. Choi Y. S., et al. 2007. Identification of Pseudomonas aeruginosa genes crucial for hydrogen peroxide resistance. J. Microbiol. Biotechnol. 17:1344–1352 [PubMed] [Google Scholar]
- 10. Curran B., Jonas D., Grundmann H., Pitt T., Dowson C. G. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Vos D., et al. 1997. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J. Clin. Microbiol. 35:1295–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dötsch A., et al. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:2522–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleiszig S. M., Arora S. K., Van R., Ramphal R. 2001. FlhA, a component of the flagellum assembly apparatus of Pseudomonas aeruginosa, plays a role in internalization by corneal epithelial cells. Infect. Immun. 69:4931–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fleiszig S. M., Evans D. J. 2002. The pathogenesis of bacterial keratitis: studies with Pseudomonas aeruginosa. Clin. Exp. Optom. 85:271–278 [DOI] [PubMed] [Google Scholar]
- 15. Gal-Mor O., Finlay B. B. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 8:1707–1719 [DOI] [PubMed] [Google Scholar]
- 16. Hart C. A., Winstanley C. 2002. Persistent and aggressive bacteria in the lungs of cystic fibrosis children. Br. Med. Bull. 61:81–96 [DOI] [PubMed] [Google Scholar]
- 17. Hayden H. S., et al. 2008. Large-insert genome analysis technology detects structural variation in Pseudomonas aeruginosa clinical strains from cystic fibrosis patients. Genomics 91:530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He J., et al. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U. S. A. 101:2530–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heo Y. J., Chung I. Y., Choi K. B., Lau G. W., Cho Y. H. 2007. Genome sequence comparison and superinfection between two related Pseudomonas aeruginosa phages, D3112 and MP22. Microbiology 153:2885–2895 [DOI] [PubMed] [Google Scholar]
- 20. Hertle R., Mrsny R., Fitzgerald D. J. 2001. Dual-function vaccine for Pseudomonas aeruginosa: characterization of chimeric exotoxin A-pilin protein. Infect. Immun. 69:6962–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hill D. F., Short N. J., Perham R. N., Petersen G. B. 1991. DNA sequence of the filamentous bacteriophage Pf1. J. Mol. Biol. 218:349–364 [DOI] [PubMed] [Google Scholar]
- 22. Kang P. J., et al. 1997. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 24:1249–1262 [DOI] [PubMed] [Google Scholar]
- 23. Kaper J. B., Nataro J. P., Mobley H. L. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 24. Kaye S., et al. 2010. Bacterial susceptibility to topical antimicrobials and clinical outcome in bacterial keratitis. Investig. Ophthalmol. Vis. Sci. 51:362–368 [DOI] [PubMed] [Google Scholar]
- 25. Khan N. H., et al. 2008. Multilocus sequence typing and phylogenetic analyses of Pseudomonas aeruginosa isolates from the ocean. Appl. Environ. Microbiol. 74:6194–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klockgether J., Wurdemann D., Reva O., Wiehlmann L., Tummler B. 2007. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J. Bacteriol. 189:2443–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kropinski A. M. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kulasekara B. R., et al. 2006. Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 188:4037–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kus J. V., Tullis E., Cvitkovitch D. G., Burrows L. L. 2004. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 150:1315–1326 [DOI] [PubMed] [Google Scholar]
- 30. Kuzio J., Kropinski A. M. 1983. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J. Bacteriol. 155:203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kwan T., Liu J., Dubow M., Gros P., Pelletier J. 2006. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 188:1184–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lam D. S., et al. 2002. Incidence and risk factors for microbial keratitis in Hong Kong: comparison with Europe and North America. Eye 16:608–618 [DOI] [PubMed] [Google Scholar]
- 33. Larbig K. D., et al. 2002. Gene islands integrated into tRNA(Gly) genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee D. G., et al. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee E. J., Cowell B. A., Evans D. J., Fleiszig S. M. 2003. Contribution of ExsA-regulated factors to corneal infection by cytotoxic and invasive Pseudomonas aeruginosa in a murine scarification model. Investig. Ophthalmol. Vis. Sci. 44:3892–3898 [DOI] [PubMed] [Google Scholar]
- 36. Liang X., Pham X. Q., Olson M. V., Lory S. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lomholt J. A., Poulsen K., Kilian M. 2001. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect. Immun. 69:6284–6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lyczak J. B., Cannon C. L., Pier G. B. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 39. Mainz J. G., et al. 2009. Concordant genotype of upper and lower airways Pseudomonas aeruginosa and Staphylococcus aureus isolates in cystic fibrosis. Thorax 64:535–540 [DOI] [PubMed] [Google Scholar]
- 40. Mathee K., et al. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U. S. A. 105:3100–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morlet N., Minassian D., Butcher J. 1999. Risk factors for treatment outcome of suspected microbial keratitis. Ofloxacin Study Group. Br. J. Ophthalmol. 83:1027–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pirnay J. P., et al. 2009. Pseudomonas aeruginosa population structure revisited. PLoS One 4:e7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pirnay J. P., et al. 2002. Pseudomonas aeruginosa displays an epidemic population structure. Environ. Microbiol. 4:898–911 [DOI] [PubMed] [Google Scholar]
- 44. Pirnay J. P., et al. 2003. Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone. J. Clin. Microbiol. 41:1192–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pirnay J. P., et al. 2005. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbiol. 7:969–980 [DOI] [PubMed] [Google Scholar]
- 46. Rakhimova E., et al. 2009. Pseudomonas aeruginosa population biology in chronic obstructive pulmonary disease. J. Infect. Dis. 200:1928–1935 [DOI] [PubMed] [Google Scholar]
- 47. Roy P. H., et al. 2010. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5:e8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sarkar S. F., Gordon J. S., Martin G. B., Guttman D. S. 2006. Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174:1041–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smart C. H., Walshaw M. J., Hart C. A., Winstanley C. 2006. Use of suppression subtractive hybridization to examine the accessory genome of the Liverpool cystic fibrosis epidemic strain of Pseudomonas aeruginosa. J. Med. Microbiol. 55:677–688 [DOI] [PubMed] [Google Scholar]
- 50. Spencer D. H., et al. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sueke H., et al. 2010. An in vitro investigation of synergy or antagonism between antimicrobial combinations against isolates from bacterial keratitis. Investig. Ophthalmol. Vis. Sci. 51:4151–4155 [DOI] [PubMed] [Google Scholar]
- 52. Sueke H., et al. 2010. Minimum inhibitory concentrations of standard and novel antimicrobials for isolates from bacterial keratitis. Investig. Ophthalmol. Vis. Sci. 51:2519–2524 [DOI] [PubMed] [Google Scholar]
- 53. Tam C., et al. 2007. Mutation of the phospholipase catalytic domain of the Pseudomonas aeruginosa cytotoxin ExoU abolishes colonization promoting activity and reduces corneal disease severity. Exp. Eye Res. 85:799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thomas P. A., Geraldine P. 2007. Infectious keratitis. Curr. Opin. Infect. Dis. 20:129–141 [DOI] [PubMed] [Google Scholar]
- 55. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wiehlmann L., et al. 2007. Functional genomics of Pseudomonas aeruginosa to identify habitat-specific determinants of pathogenicity. Int. J. Med. Microbiol. 297:615–623 [DOI] [PubMed] [Google Scholar]
- 57. Wiehlmann L., et al. 2007. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8101–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Willcox M. D., et al. 2008. Role of quorum sensing by Pseudomonas aeruginosa in microbial keratitis and cystic fibrosis. Microbiology 154:2184–2194 [DOI] [PubMed] [Google Scholar]
- 59. Winstanley C., et al. 2005. Genotypic and phenotypic characteristics of Pseudomonas aeruginosa isolates associated with ulcerative keratitis. J. Med. Microbiol. 54:519–526 [DOI] [PubMed] [Google Scholar]
- 60. Winstanley C., et al. 2009. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res. 19:12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wolfgang M. C., et al. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:8484–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhu H., et al. 2006. Type III secretion system-associated toxins, proteases, serotypes, and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis. Curr. Eye Res. 31:297–306 [DOI] [PubMed] [Google Scholar]
- 63. Zolfaghar I., Evans D. J., Fleiszig S. M. 2003. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 71:5389–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]