Abstract
Human enterovirus 71 (EV71) is the major etiological agent of hand, foot, and mouth disease (HFMD), which is a common infectious disease in young children and infants. EV71 can cause various clinical manifestations and has been associated with severe neurological complications; it has resulted in fatalities during recent outbreaks in Asian-Pacific regions since 1997. The early and rapid detection is critical for prevention and control of EV71 infection, since no vaccine or antiviral drugs are currently available. In this study, a simple and sensitive reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay was developed for rapid detection of EV71. The detection limit of the RT-LAMP assay was approximately 0.01 PFU per reaction mixture, and no cross-reactive amplification with other enteroviruses was observed. The assay was evaluated further with 40 clinical specimens and exhibited 92.9% sensitivity and 100% specificity. This RT-LAMP assay may become a useful alternative in clinical diagnosis of EV71, especially in resource-limited hospitals or rural clinics of China and other countries in the Asian-Pacific region.
INTRODUCTION
Hand, foot, and mouth disease (HFMD) is a common infectious disease in young children and infants and is characterized by fever, ulcers in the mouth, and vesicles on the hands and feet. This disease is mainly caused by two enteroviruses: human enterovirus 71 (EV71) and coxsackievirus A16 (CVA16). The epidemics of HFMD pose serious public health threats among infants throughout the world (19, 22). Since 1997, large HFMD epidemics with severe neurological complications were found to be caused by EV71 infection, and a large number of fatalities occurred in the Asian-Pacific region, including Malaysia (34), Taiwan (8, 12), Singapore (1, 2), Vietnam (7), Australia (23), and Japan (13). Recently, since 2008, there has been a significant increase in epidemics of EV71 in mainland China (38), and millions of cases and hundreds of deaths among infants have been reported every year (9). There is a great demand for the rapid detection and differentiation of EV71 infection in the acute phase of illness in order to provide timely clinical treatment and disease control, especially considering the fact that no vaccine or specific antiviral drugs are currently available for severe EV71 infections.
EV71, first isolated from an infant with encephalitis in California in 1969 (31), is a member of the Enterovirus genus of the family Picornaviridae. The genome of EV71 is a positive-sense single-stranded RNA of approximately 7.5 kb in length. It contains a single open reading frame (ORF) flanked by the 5′ and 3′ untranslated regions (UTRs). The single ORF encodes a polyprotein that can be processed into four capsid proteins (VP1, VP2, VP3, and VP4) and seven nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) (22). Based on the highly variable sequences of the VP1 gene, EV71 can be classified into four genogroups (A, B, C, and D) (6, 10). Genogroups B and C both consist of five lineages (B1 to B5 and C1 to C5). The predominant genotypes currently circulating in Asia are C1, C4, C5, and B5 (5).
Presently, laboratory diagnosis of EV71 includes virus isolation, detection of virus-specific antibodies, and detection of viral genomic sequences by nucleic acid amplification techniques. EV71 can be isolated from a variety of clinical specimens, which always requires 1 to 2 weeks to obtain results. IgM assays based on enzyme immunological techniques have been developed (35, 37), but these require specialized equipment and need serum samples, which are difficult to collect from infant patients. Reverse transcription-PCR (RT-PCR) and real-time RT-PCR (rRT-PCR) assays for detection of EV71 have been reported (11, 14, 33, 36), and they exhibited high sensitivities of at least 5 viral RNA copies/ml or 0.1 50% tissue culture infectious doses (TCID50)/ml. However, these methods require specific, expensive equipment and skilled technicians, which limits their applications in resource-limited laboratories such as in community clinics, especially rural clinics.
The loop-mediated isothermal amplification (LAMP) method is a cheap, rapid, and simple method that was first described in 2000 (24). This novel detection method works on the principle of a strand displacement reaction with the specific stem-loop structures, and the whole amplification reaction takes place continuously under isothermal conditions. Previously, LAMP methods for detection of different viruses have been developed, including West Nile virus (27), dengue virus serotypes 1 to 4 (26), Japanese encephalitis virus (20, 28), avian H5 and H7 influenza viruses (29, 32), the 2009 pandemic H1N1 influenza virus (15, 21), Marburg virus (16), Ebola virus (17), foot-and-mouth disease virus (25), and herpes simplex virus 1 (30). All these assays showed high specificity, efficiency, and sensitivity that were similar to or higher than PCR assays. These previous studies exhibited the potential of LAMP as a routine diagnostic method for viral infectious diseases in a hospital laboratory.
In this study, an RT-LAMP assay targeting the VP3 gene of EV71 was developed and further evaluated with clinical specimens. The results demonstrated that the RT-LAMP assay was sensitive and accurate, and it may become a useful alternative in clinical diagnosis of EV71, especially in resource-limited hospital laboratories and rural clinics.
MATERIALS AND METHODS
Clinical samples.
A total of 40 clinical stool samples were collected from suspected HFMD patients according to the guidelines for HFMD diagnosis from the Chinese Center for Disease Control (http://www.chinacdc.cn/n272442/n272530/n3479265/n3479308/31860.html) during the epidemics of HFMD in Guangxi Province of China in 2009. All samples were collected between 2 days and 7 days after the onset of illness. The clinical samples used in this study were appropriately coded to be anonymous. Local ethical approval was obtained, and guidelines were followed for use of clinical material and access to diagnostic results.
A 10% stool suspension was made by adding 1 g of stool sample to 10 ml of phosphate-buffered saline (PBS; 0.02 mol/liter; pH 7.2). The suspension was then centrifuged at 13,000 × g for 5 min. The upper suspension was filtered and then subsequently processed.
Cell culture.
Human RD cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, CA) supplemented with 10% fetal bovine serum (FBS; Excell, Shanghai, China) plus 2 mM l-glutamine, 100 IU of penicillin, and 100 μg of streptomycin per ml at 37°C in the presence of 5% CO2.
Virus.
Six EV71 strains (HN0803, HN0804, HN0808, AH0806, SD18, and SD24) isolated from clinical throat swab or stool samples of confirmed HFMD cases during outbreaks in Henan, Anhui, and Shandong provinces of China were used in this study. The EV71 protype strain BrCr was kindly provided by the Beijing Institute of Biological Products. Poliovirus type 1 (PV1), CVA16, coxsackievirus B3 (CVB3), and CVB5 were used to assay the specificity. Seven other viruses that can cause flu-like illness or encephalitis, such as the 2009 pandemic H1N1 influenza virus, H5N1 avian influenza virus, seasonal influenza viruses H1N1 and H3N2, parainfluenza virus type 2, adenovirus type 3, dengue 2 virus, Japanese encephalitis virus, and West Nile virus, were also used to for the specificity tests. All these viruses were stored at −80°C in our laboratory.
A monolayer of RD cells was infected by EV71 and then incubated at 36°C until 75% to 100% cytopathic effect was observed. The culture supernatant was collected and stored at −80°C until use. The titer of virus stock was determined by standard plaque assay as previously described (3).
RNA extraction.
Total RNA was extracted from 200-μl aliquots of the clinical samples or viral culture supernatant using the RNeasy minikit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The RNA was eluted in a final volume of 50 μl of RNase-free water and stored at −80°C until use.
rRT-PCR.
rRT-PCR was carried out with the published set of specific primers and probe for the detection of EV71 (36). The rRT-PCR was performed with the One-Step PrimeScript RT-PCR kit (Takara, Dalian, China) in the LightCycler 2.0 system (Roche, Mannheim, Germany) in a 20-μl mixture containing 2 μl total RNA, 10 μl 2× One Step RT-PCR buffer III, 0.4 μl ExTaq, 0.4 μl PrimeScript RT enzyme mix II, 0.25 μM forward primer, 0.25 μM reverse primer, and 0.25 μM probe. The reaction was performed for 5 min at 42°C, followed by 20 s at 95°C, with a subsequent 40 cycles of amplification (95°C for 5 s and 60°C for 20 s). Fluorescence was recorded at 60°C.
Design of EV71-specific RT-LAMP primers.
The nucleotide sequences of the complete genomes of EV71 strains were retrieved from the GenBank database and aligned by using the ClustalX multiple sequence alignment program (18). The alignment file was input into the LAMP primer designing software PrimeExplorer (http://primerxplorer.p/e/). Primer sets comprising two outer primers (F3 and B3), two inner primers (FIP and BIP), and two loop primers (LF and LB), which recognize eight distinct regions on the target sequence, were designed on the basis of the sequence of the Henan10-08 virus (GenBank accession no. GU366191). The feasibility of all sets of primers was then subsequently validated by using the BLAST program (http://blast/ncbi.nlm.nih.gov/blast.cgi) and tested with different EV71 isolates from China. The best set was selected. All the primers were synthesized by Shanghai Invitrogen Company.
RT-LAMP assay.
The RT-LAMP reaction was carried out using the Loopamp RNA amplification kit (Eiken Chemical, Tokyo, Japan). The reaction system contained 2.5 μl total RNA, 40 pmol each of the primers FIP and BIP, 5 pmol each of the outer primers F3 and B3, 20 pmol each of LF and LB, 12.5 μl 2× reaction mix, 1 μl enzyme mix, and 1 μl fluorescent regent. The reaction mixture was incubated at 63°C for 35 min in a Loopamp real-time turbidimeter LA-320 (Eiken Chemical, Tokyo, Japan), followed by heating at 80°C for 5 min to terminate the reaction. For the real-time monitoring of the amplification of the RT-LAMP reaction, the optical density data of each reaction were recorded every 6 s by the Loopamp real-time turbidimeter. The threshold of turbidity for positive samples was defined as 0.1. The time of positivity (Tp) was determined as the time when the turbidity value increased above the threshold.
End point visual detection was performed by using a fluorescent detection reagent (Eiken Chemical, Tokyo, Japan). The color change under normal light and UV light was recorded.
Sensitivity and specificity of the RT-LAMP assay.
Viral RNA was extracted from EV71-infected RD cell cultures. The sensitivity of the RT-LAMP assay was analyzed using 10-fold serial dilutions of viral RNA. The final concentrations of viral RNA were between 1 PFU and 0.000001 PFU per reaction mixture, respectively. The specificity of the assay was evaluated further by cross-reactivity tests with RNA extracted from virus stock of seven EV71 isolates as well as PV1, CVB3, CVB5, CVA16, and seven other viral pathogens that may cause flu-like illness and encephalitis.
RESULTS
Design of EV71-specific RT-LAMP primers.
The nucleotide sequences of EV71 strain Henan10-08 were retrieved from the GenBank database and aligned with other EV71 isolates from mainland China between 2008 and 2010 by using the ClustalX multiple sequence alignment program. The most conserved regions in the VP3 gene were selected as the target for primer design. Four sets of primers of EV71-specific RT-LAMP were designed based on the aligned sequences. The possible mismatches between all sets of primers and EV71 were checked by using the BLAST program. The efficiency and sensitivity of the different sets of primers were evaluated with different EV71 isolates, and the best set was selected. The details for each primer and the positions of the primers in the viral genomic sequence are shown in Table 1. The sequence homology of the primers and EV71 Chinese isolates and other EV71 strains of circulating genotypes available from GenBank were analyzed. There were only one or two mismatches between the selected set of primers and EV71 isolates, and all mismatches were located in the 5′-terminal part.
Table 1.
RT-LAMP primer sets designed for rapid detection of EV71
| Primer | Genome positiona | Length (bp) | Sequenceb (5′–3′) |
|---|---|---|---|
| F3 | 1921–1938 | 18 | CTTCCCGGTCTCAGCACA |
| B3 | 2104–2123 | 20 | GCGGTGTATAGGCTATGAGC |
| FIP | (1999–2019) + (1939–1956) | 43 | TGACCCAGCAAGGTGGATTGCTTTTAGCAGGGAAAGGTGAGCT |
| BIP | (2024–2044) + (2074–2092) | 44 | TGCGGGTACTACACCCAATGGTTTTAGCCATGAAGGACCCAGTA |
| LF | 1957–1976 | 20 | CGGCTCTGAACACCGCACAC |
| LB | 2048–2072 | 25 | GGATCATTGGAAGTCACCTTCATGT |
EV71 strain Henan10-08 (GenBank accession no. GU366191). FIP and BIP are long primers containing two recognition sequences (positions indicated with parentheses) with a TTTT linker.
Underlined portions indicate a TTTT linker.
Sensitivity and specificity of the EV71-specific RT-LAMP assay.
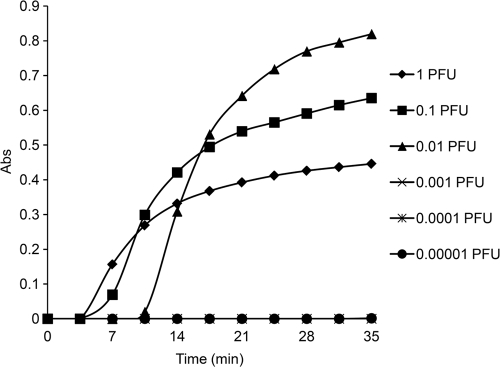
The sensitivities of the RT-LAMP assay and rRT-PCR assay were analyzed using RNA extracted from 10-fold dilution series of viral cultures. The detection limit of the RT-LAMP assay was 0.01 PFU per reaction mixture (Fig. 1), which was equal to that of the real-time RT-PCR assay. Furthermore, the specificity of the RT-LAMP assay was also analyzed with different EV71 strains and other common enteroviruses and respiratory viruses. The RT-LAMP assay could detect all the EV71 strains tested, and no cross-reaction with other enteroviruses, respiratory viruses, or encephalitis viruses was observed.
Fig. 1.
Sensitivity and dynamic range of EV71-specific RT-LAMP assay with 10-fold serial dilutions of viral RNA (from 1 to 0.00001 PFU per reaction mixture). The detection limit is 0.01 PFU per reaction mixture.
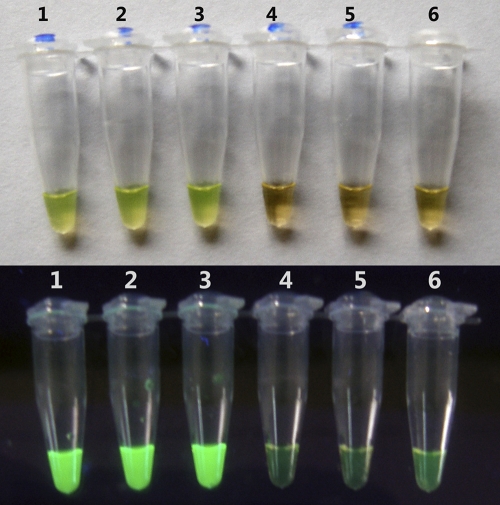
The RT-LAMP assay could be completed in 35 min. In addition, by using a fluorescent detection regent, the inspection for amplification was also performed by the observation of color change. In cases of positive amplification, the color of the reaction mixture changed from orange to green under natural light, and bright green fluorescence was observed under UV light. For negative control and negative samples, the color of reaction mixture did not change under either normal light or UV light (Fig. 2).
Fig. 2.
Visual detection of EV71 by RT-LAMP with a fluorescent detection reagent. Ten-fold serial dilutions of viral RNA (lanes 1 to 6, 1 to 0.00001 PFU per reaction mixture, respectively) were detected. (A) Evaluation under normal light. The color of positive samples changed from orange to green, whereas the color of negative samples and the negative control remained orange. (B) Evaluation under UV light. Positive samples were bright green under UV light, while negative samples or the negative control remained weakly green.
Evaluation of the RT-LAMP assay with clinical samples.
The RT-LAMP assay for EV71 was further evaluated with clinical samples from suspected HFMD cases. Of the 40 samples, 28 were found to be positive for EV71 and 7 were positive for CVA16 by rRT-PCR assay, respectively. A Tp value of 35 min was set as the cutoff for a positive result by the EV71-specific RT-LAMP assay. The results of the RT-LAMP assay in comparison with the rRT-PCR assay for 40 clinical stool samples are shown in Table 2. Twenty-six positive samples (92.9%) were confirmed by RT-LAMP assay, while 28 samples tested positive by rRT-PCR methods. Two positive samples by rRT-PCR were not detected by the RT-LAMP assay. Nonspecific amplification was not observed in the detection of 40 clinical samples. None of the negative samples by rRT-PCR tested positive by the RT-LAMP assay, and seven samples positive for CVA16 also tested negative by the RT-LAMP assay. The sensitivity and the specificity of the RT-LAMP assay in comparison with the rRT-PCR method were 92.9% and 100%, respectively.
Table 2.
Comparison of detection of EV71 from clinical specimensa by RT-LAMP assay and rRT-PCR assay
| Result | No. of samples with indicated result by: |
Correlation (%) | |
|---|---|---|---|
| RT-LAMP assayb | rRT-PCR | ||
| Positive | 26 | 28 | 92.9 |
| Negative | 14 | 12 | |
| Total | 40 | 40 | |
A total of 40 clinical fecal samples were collected between 2 days and 7 days after the onset of symptoms from suspected HFMD infant patients during epidemics in Guangxi, China (2009 to 2010).
In comparison with the rRT-PCR assay, the RT-LAMP assay exhibited a sensitivity of 92.9% and a specificity of 100%.
DISCUSSION
In comparison with mild cases of CVA16 infection, neurotropic EV71 can cause severe neurological disease with rapid progression and high mortality, and most EV71 cases occur in rural areas, where the level of sanitation is lower and laboratory equipments or skilled technicians are lacking (9). For inexperienced doctors in community or rural clinics, it is difficult to distinguish the EV71 infection from other diseases with fever, rash, or encephalitis. Therefore, there is great demand for simple, rapid, and sensitive diagnostic methods or kits for rapid detection of EV71 to provide timely clinical treatment and disease control.
Here, we developed a one-step RT-LAMP assay for detection of EV71 from clinical samples. The sensitivity of the EV71-specific RT-LAMP is 0.01 PFU per reaction mixture, which is similar to the rRT-PCR assay. It is also similar to the other rRT-PCR system described in previous studies, which had a detection limit of 0.1 TCID50/ml, or five viral copies (33, 36). Previously, an RT-LAMP system was developed for detection of enterovirus (4), but this system only exhibited a high sensitivity for the detection of poliovirus and human enterovirus C and was less sensitivite to human enterovirus B and human enterovirus A, including EV71 and CVA16. The detection limit of EV71 BrCr-TR strain was only 7.8 TCID50 (7, 400 copies of the viral genome), which limited use of this RT-LAMP system in diagnosis of EV71 infection.
Based on 40 clinical samples from suspected HFMD cases, 92.9% diagnostic sensitivity was achieved with the RT-LAMP assay. Two samples tested negative by the RT-LAMP assay but positive by the rRT-PCR assay. The cross-points (Cp) of two samples obtained by rRT-PCR with the LightCycler system were above 33. This may indicate that the RT-LAMP assay is slightly less sensitive than the rRT-PCR assay for specimens with very low viral titers. The RT-LAMP assay employs six primers that recognize eight distinct regions of the target, which prevents cross-reactions (24). Our data showed that there was no cross-reactive amplification with the RNA of other enteroviruses, such as CVA16, CVB3, CVB5, and PV1. The high specificity of the RT-LAMP assay was also exhibited in 40 clinical samples, including seven CVA16-positive samples.
The RT-LAMP assay is simple and easy to perform. The assay employs regular primers without need for probe-labeled dye. The reaction mixture, which amplifies at a constant temperature (63°C), could be incubated in a regular laboratory water bath. The assay could be achieved in 35 min, which is rapid, even including the time required for RNA extraction from the clinical specimens. These advantages would make such a diagnostic tool very useful in the early determination of EV71.
In conclusion, a simple, rapid, highly sensitive and specific one-step EV71-specific RT-LAMP assay was developed and evaluated for detection of EV71 in clinical samples. The RT-LAMP assay is rapid, accurate, and feasible without need for specific equipment and trained operators. It has the potential as a routine diagnostic method for EV71 infection in resource-limited laboratories, such as community or rural clinics. The EV71-specific RT-LAMP is useful for the early diagnosis of suspected patients, especially patients in rural areas, which will contribute to the prevention and control of EV71 epidemics in China and other countries in the Asian-Pacific region.
ACKNOWLEDGMENTS
This work was supported by the Major Special Program of Na-tional Science and Technology of China (2009ZX10004-204 and 2008ZX10004-001).
We thank Li-Juan Liu and Wei Liu for sample collection.
We declare that we have no potential conflicts of interest relevant to this article.
Footnotes
Published ahead of print on 22 December 2011.
REFERENCES
- 1. Ahmad K. 2000. Hand, foot and mouth disease outbreak reported in Singapore. Lancet 356:1338. [DOI] [PubMed] [Google Scholar]
- 2. Ang L. W., et al. 2009. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001–2007. Ann. Acad. Med. Singapore. 38:106–112 [PubMed] [Google Scholar]
- 3. Arita M., et al. 2005. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J. Gen. Virol. 86:1391–1401 [DOI] [PubMed] [Google Scholar]
- 4. Arita M., et al. 2009. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) system for a highly sensitive detection of enterovirus in the stool samples of acute flaccid paralysis cases. BMC Infect. Dis. 9:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bek J. E., McMinn P. C. 2010. Recent advances in research on human enterovirus 71. Future Virol. 5:453–468 [Google Scholar]
- 6. Brown B. A., Oberste M. S., Alexander J. P., Jr., Kennett M. L., Pallansch M. A. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cardosa M. J., McMinn P. C. 2007. Epidemiologic and virologic investigation of hand, foot, and mouth disease, southern Vietnam, 2005. Emerg. Infect. Dis. 11:1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang L. Y. 2008. Enterovirus 71 in Taiwan. Pediatr. Neonatol. 49:103–112 [DOI] [PubMed] [Google Scholar]
- 9. China Ministry of Health 2010. Information on prevention and control of HFMD. Http://www.moh.gov.cn/publicfiles/business/htmlfiles/mohbgt/s3582/201006/47871.htm (In Chinese)
- 10. Deshpande J. M., Nadkarni S. S., Francis P. P. 2003. Enterovirus 71 isolated from a case of acute flaccid paralysis in India represents a new genotype. Curr. Sci. 84:1350–1353 [Google Scholar]
- 11. Fujimoto T., et al. 2008. Detection and quantification of enterovirus 71 genome from cerebrospinal fluid of an encephalitis patient by PCR applications. Jpn. J. Infect. Dis. 61:497–499 [PubMed] [Google Scholar]
- 12. Ho M., et al. 1999. An epidemic of enterovirus 71 infection in Taiwan. N. Engl. J. Med. 341:929–935 [DOI] [PubMed] [Google Scholar]
- 13. Hosoya M., et al. 2006. Genetic diversity of enterovirus 71 associated with hand, foot and mouth disease epidemics in Japan from 1983 to 2003. Pediatr. Infect. Dis. J. 25:691–694 [DOI] [PubMed] [Google Scholar]
- 14. Kapusinszky R., Szomor N. K., Farkas A., Takacs M., Berencsi G. 2010. Detection of non-polio enteroviruses in Hungary 2000–2008 and molecular epidemiology of enterovirus 71, coxsackievirus A16, and echovirus 30. Virus Genes 40:163–173 [DOI] [PubMed] [Google Scholar]
- 15. Kubo T., et al. 2010. Development of a reverse transcription-loop-mediated isothermal amplification assay for detection of pandemic (H1N1) 2009 virus as a novel molecular method for diagnosis of pandemic influenza in resource-limited settings. J. Clin. Microbiol. 48:728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kurosaki Y., Grolla A., Fukuma A., Feldmann H., Yasuda J. 2010. Development and evaluation of a simple assay for Marburg virus detection using a reverse transcription-loop-mediated isothermal amplification method. J. Clin. Microbiol. 48:2330–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kurosaki Y., et al. 2007. Rapid and simple detection of Ebola virus by reverse transcription-loop-mediated isothermal amplifi cation. J. Virol. Methods 141:78–83 [DOI] [PubMed] [Google Scholar]
- 18. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 19. Lee T. C., et al. 2009. Diseases caused by enterovirus 71 infection. Pediatr. Infect. Dis. J. 28:904–910 [DOI] [PubMed] [Google Scholar]
- 20. Liu Y., Chuang C. K., Chen W. J. 2009. In situ reverse-transcription loop-mediated isothermal amplification (in situ RT-LAMP) for detection of Japanese encephalitis viral RNA in host cells. J. Clin. Virol. 46:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma X. J., et al. 2010. Visual detection of pandemic influenza A H1N1 virus 2009 by reverse-transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. J. Virol. Methods 167:214–217 [DOI] [PubMed] [Google Scholar]
- 22. McMinn P. C. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107 [DOI] [PubMed] [Google Scholar]
- 23. McMinn P. C., Stratov I., Nagarajan L., Davis S. 2001. Neurological manifestations of enterovirus 71 infection in children during an outbreak of hand, foot, and mouth disease in Western Australia. Clin. Infect. Dis. 32:236–242 [DOI] [PubMed] [Google Scholar]
- 24. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paraguison R. C., Flores E. B., Cruz L. C. 2010. Possible use of RNA isolate from inactivated vaccine for external positive control in reverse transcription-based detection of foot-and-mouth disease virus in bull semen. Biochem. Biophys. Res. Commun. 392:557–560 [DOI] [PubMed] [Google Scholar]
- 26. Parida M., et al. 2005. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 43:2895–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parida M., Posadas G., Inoue S., Hasebe F., Morita K. 2004. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 42:257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parida M., et al. 2006. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J. Clin. Microbiol. 44:4172–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Postel A., et al. 2010. Evaluation of two commercial loop-mediated isothermal amplification assays for detection of avian influenza H5 and H7 hemagglutinin genes. J. Vet. Diagn. Invest. 22:61–66 [DOI] [PubMed] [Google Scholar]
- 30. Reddy A. K., Balne P. K., Reddy R. K., Mathai A., Kaur I. 2010. Loop-mediated isothermal amplification assay for the diagnosis of retinitis caused by herpes simplex virus-1. Clin. Microbiol. Infect. 17:210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt N. J., Lennette E. H., Ho H. H. 1974. An apparently new enterovirus isolated from patients with disease of the central nervous system. J. Infect. Dis. 129:304–309 [DOI] [PubMed] [Google Scholar]
- 32. Shivakoti S., et al. 2010. Development of reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay for detection of avian influenza viruses in field specimens. J. Vet. Med. Sci. 72:519–523 [DOI] [PubMed] [Google Scholar]
- 33. Tan E. L., Chow V. T., Quak S. H., Yeo W. C., Poh C. L. 2008. Development of multiplex real-time hybridization probe reverse transcriptase polymerase chain reaction for specific detection and differentiation of enterovirus 71 and coxsackievirus A16. Diagn. Microbiol. Infect. Dis. 61:294–301 [DOI] [PubMed] [Google Scholar]
- 34. Tee K. K., Takebe Y., Kamarulzaman A. 2009. Emerging and re-emerging viruses in Malaysia, 1997–2007. Int. J. Infect. Dis. 13:307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang S. Y., Chin T. L., Chen H. Y., Lin T. S. 2004. Early and rapid detection of enterovirus 71 infection by an IgM-capture ELISA. J. Virol. Methods 119:37–43 [DOI] [PubMed] [Google Scholar]
- 36. Xiao X. L., et al. 2009. Simultaneous detection of human enterovirus 71 and coxsackievirus A16 in clinical specimens by multiplex real-time PCR with an internal amplification control. Arch. Virol. 154:121–125 [DOI] [PubMed] [Google Scholar]
- 37. Xu F., et al. 2010. Performance of detecting IgM antibodies against enterovirus 71 for early diagnosis. PLoS One 5:e11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang F., et al. 2009. Enterovirus 71 outbreak in the People's Republic of China in 2008. J. Clin. Microbiol. 47:2351–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]