Abstract
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry is a rapid and accurate tool for the identification of many microorganisms. We assessed this technology for the identification of 103 Haemophilus parainfluenzae, Aggregatibacter aphrophilus, Aggregatibacter actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella kingae (HACEK) clinical isolates and 20 Haemophilus influenzae clinical isolates. Ninety-three percent of HACEK organisms were identified correctly to the genus level using the Bruker database, and 100% were identified to the genus level using a custom database that included clinical isolates.
Clinical microbiology laboratories strive to identify infectious organisms from patient samples in the most-accurate yet time- and cost-efficient manner possible. This process may be impeded by rare or difficult-to-identify organisms such as those of the HACEK group (Haemophilus parainfluenzae, Aggregatibacter aphrophilus, Aggregatibacter actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella kingae). HACEK organisms are fastidious Gram-negative rods found in the healthy oropharynx or upper respiratory tract and can cause endocarditis, especially in young children and patients with heart defects (3). HACEK organisms are also involved in a wide array of serious infections involving the head and neck, bone, joints, lungs, and other soft tissues (17).
Numerous reports have recently been published showing the successful integration of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) in the identification of microorganisms routinely encountered in clinical microbiology labs (1, 2, 12, 15). These reports have focused largely on easy-to-culture bacteria and yeasts, including Staphylococcus aureus and nonfermenting Gram-negative rods (6, 14). To date, three genera of fastidious Gram-negative rods (Legionella, Bartonella, and Francisella) have been extensively evaluated by the newer MALDI-TOF MS identification systems (4, 7, 10, 11). Several studies have also investigated subsets of organisms, such as difficult-to-identify Nocardia spp., anaerobes, and oral flora, as well as fastidious upper respiratory flora (e.g., Neisseria spp., Moraxella spp., Haemophilus spp.) (5, 9, 12, 13, 15, 16). Haemophilus spp., including H. influenzae, were previously shown to be distinguishable by MALDI-TOF MS (5), and since H. influenzae and HACEK organisms have similar growth requirements, this suggested that MALDI-TOF MS may be useful in their identification. HACEK organisms are not readily identified on most automated bacterial identification systems, and most require either time-consuming biochemical profiling or genetic analysis, such as 16S rRNA gene sequencing, for definitive identification (8). This study aimed to determine whether MALDI-TOF MS could accurately identify a large panel of HACEK clinical isolates.
HACEK and H. influenzae isolates were grown on chocolate agar plates (Hardy Diagnostics, Santa Maria, CA), and 24- to 48-h-old cultures were used for identification. Organisms were previously identified by partial 16S rRNA gene sequencing in our laboratory (n = 103) or by outside laboratories (n = 20). The collection of organisms investigated in this study included 24 H. parainfluenzae, 20 A. aphrophilus, 5 A. actinomycetemcomitans, 14 C. hominis, 35 E. corrodens, 5 K. kingae, and 20 H. influenzae isolates. The complete panel of HACEK organisms was extracted using the standard ethanol-formic acid (FA) extraction procedure recommended by Bruker Daltonics (Billerica, MA). Approximately 5 mg of cells from pure cultures was extracted, and 1 μl of bacterial extract was spotted and allowed to air dry on an MSP 96 polished steel target (Bruker Daltonics). Samples were overlaid with 1.5 μl of matrix (saturated α-cyano-4-hydroxy-cinnamic acid in 50% acetonitrile–2.5% trifluoracetic acid), dried at room temperature, and analyzed immediately. Mass spectra were acquired on a microflex LT MALDI-TOF spectrometer (Bruker Daltonics) in the linear positive-ion mode using FlexControl 3.0 software (Bruker Daltonics). Sample data were collected over an m/z range of 2,000 to 20,000 Da, without gating, using a detector gain of 2,605 V. Each spot was measured by collecting 500 laser shots at 60 Hz in groups of 100 shots per locus, and spectra were combined into a sum spectrum. The Bruker bacterial test standard was used as a calibrator for each sample run. Organisms were identified by comparing their characteristic mass spectra with reference spectra from the integrated database provided by the manufacturer, using the MALDI Biotyper 2.0 software package (Bruker Daltonics). Scores greater than 1.7 were considered reliable for genus level identification, while scores greater than 1.9 were considered reliable for species level identification as described by Seng et al. (12). Scores lower than 1.7 were not considered reliable according to the algorithm. Samples resulting in scores lower than 1.9 were reextracted and reanalyzed to ensure proper sampling.
Using the default database, 93% of isolates (96) were identified to at least the genus level, and 66% (68 isolates) were correctly identified to the species level (Table 1). Six isolates were not identified according to our scoring algorithm (e.g., scores of <1.7); however, it is interesting to note that the top Biotyper organism identification (irrespective of score) was correct for all but 1 isolate in the study. Figure 1 illustrates that overall, H. parainfluenzae, A. actinomycetemcomitans, E. corrodens, K. kingae, and H. influenzae isolates were readily identified to the species level; however, several isolates of C. hominis and A. aphrophilus yielded relatively low scores. A. aphrophilus isolates produced mainly genus level identifications, and one isolate was incorrectly identified as H. influenzae with a score of 1.860.
Table 1.
Score distribution of 103 formic acid-extracted HACEK isolates
| Score | No. (%) of isolates |
|---|---|
| >1.9 | 68 (66.0) |
| 1.7–1.9 | 28 (27.2) |
| <1.7 | 6 (5.8) |
| Incorrect IDa | 1 (0.97) |
ID, identification.
Fig. 1.
Organism-specific breakdown of identification scores achieved for HACEK organisms and H. influenzae using the standard Biotyper database. Bars represent the percentages of the total numbers of isolates for each species.
A recent report by van Veen et al. showed that a high proportion (98%) of 51 HACEK isolates were identified to the species level by MALDI-TOF MS (15). This study differed from ours in the sample preparation method, the use of replicate samples, and the instrument settings, which could have affected the final results. In addition, the van Veen et al. study specifically mentioned testing only H. parainfluenzae and A. aphrophilus, so it is possible that their cohort was composed primarily of the easily identified H. parainfluenzae. This could explain the higher proportion of species level identifications by van Veen et al. As our data show, inclusion of all members of the HACEK group resulted in variable performances among the different species.
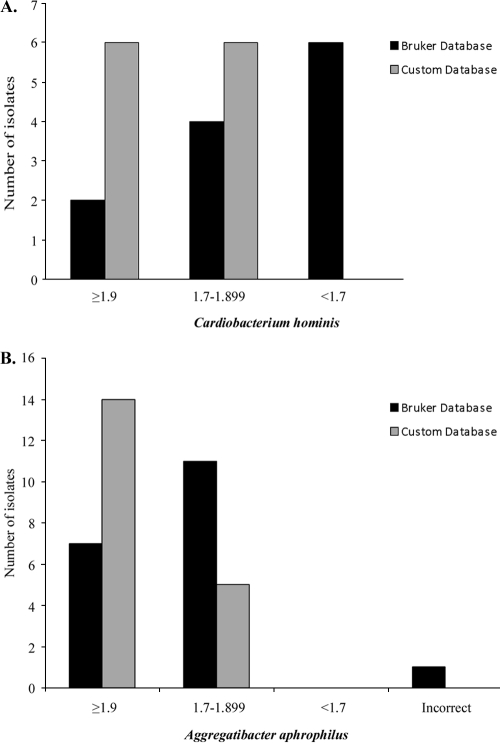
Specific C. hominis and A. aphrophilus isolates showed notable spectral differences compared to the single-reference database entries, which suggested that a representative clinical isolate for each species would increase the score quality. Custom database spectra were created by plating a sequenced clinical isolate (not included in our analysis) on three separate chocolate agar plates. Growth from each plate was extracted individually and spotted in eight replicates, and a composite reference spectrum was created by adding 500 shots per individual spot. Student's t test was used to analyze the mean score differences between standard and custom databases (GraphPad Prism v5.03; GraphPad Software, Inc.). The average scores for C. hominis increased from 1.759 to 1.92 (P = 0.0011), and those for A. aphrophilus increased from 1.814 to 2.153 (P < 0.0001), using the Bruker and custom databases, respectively. These increased scores resulted in all C. hominis isolates being identified to the genus or species level and the majority of A. aphrophilus isolates achieving species level identification, including the isolate previously misidentified as H. influenzae (Fig. 2). Using the custom database, all of the isolates were identified to at least the genus level, and 79% were correctly identified to the species level. One explanation for this improvement is that the single reference spectrum for each organism in the manufacturer's database was not representative of a typical clinical isolate from our laboratory. This could be a limitation of the system for any organism with only one reference spectrum. In fact, Seng et al. showed that higher numbers of reference spectra in the database resulted in more reliable identification (12). It is also possible that the strain diversity within a given species may lead to lower scores for some isolates. A third possible explanation is that our custom database spectra were more similar to our other patient samples simply because the isolates used were grown, extracted, and analyzed in the same manner.
Fig. 2.
Score comparison of C. hominis (A) and A. aphrophilus (B) identified using the Bruker database and a custom database that included representative clinical isolates for each species.
Overall, these data show that customization of the default database can allow for improved identification of clinical isolates. Unlike public nucleic acid sequence databases which may be populated with poor-quality entries, the custom database feature of MALDI-TOF MS allows the user to control the quality of spectra in the database and can also allow a laboratory to populate its database according to the representative isolates that it encounters through routine practice without relying on the manufacturer to release database updates. Despite the fastidious nature of HACEK organisms, this report shows that they can be accurately identified using MALDI-TOF MS. These organisms currently require identification methods that are time and/or cost intensive, and therefore MALDI-TOF MS provides an attractive alternative for clinical microbiology laboratories looking to simplify and accelerate their HACEK identification algorithm.
Acknowledgments
We thank Gary Kruppa and Gongyi Shi (Bruker Daltonics) for expert assistance on MALDI-TOF MS setup and analysis.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1. Bessede E., et al. 27 May 2010, posting date Matrix-assisted laser-desorption/ionization biotyper: experience in the routine of a University hospital. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2010.03274.x [DOI] [PubMed] [Google Scholar]
- 2. Bizzini A., Durussel C., Bille J., Greub G., Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferrieri P., et al. 2002. Unique features of infective endocarditis in childhood. Pediatrics 109:931–943 [DOI] [PubMed] [Google Scholar]
- 4. Fournier P. E., Couderc C., Buffet S., Flaudrops C., Raoult D. 2009. Rapid and cost-effective identification of Bartonella species using mass spectrometry. J. Med. Microbiol. 58:1154–1159 [DOI] [PubMed] [Google Scholar]
- 5. Haag A. M., Taylor S. N., Johnston K. H., Cole R. B. 1998. Rapid identification and speciation of Haemophilus bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 33:750–756 [DOI] [PubMed] [Google Scholar]
- 6. Mellmann A., et al. 2009. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 47:3732–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moliner C., et al. 2010. Rapid identification of Legionella species by mass spectrometry. J. Med. Microbiol. 59:273–284 [DOI] [PubMed] [Google Scholar]
- 8. Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. (ed.). 2007. Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC [Google Scholar]
- 9. Nagy E., Maier T., Urban E., Terhes G., Kostrzewa M. 2009. Species identification of clinical isolates of Bacteroides by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 15:796–802 [DOI] [PubMed] [Google Scholar]
- 10. Pennanec X., Dufour A., Haras D., Rehel K. 2010. A quick and easy method to identify bacteria by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 24:384–392 [DOI] [PubMed] [Google Scholar]
- 11. Seibold E., Maier T., Kostrzewa M., Zeman E., Splettstoesser W. 2010. Identification of Francisella tularensis by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry: fast, reliable, robust, and cost-effective differentiation on species and subspecies levels. J. Clin. Microbiol. 48:1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 13. Stingu C. S., Rodloff A. C., Jentsch H., Schaumann R., Eschrich K. 2008. Rapid identification of oral anaerobic bacteria cultivated from subgingival biofilm by MALDI-TOF-MS. Oral Microbiol. Immunol. 23:372–376 [DOI] [PubMed] [Google Scholar]
- 14. Szabados F., Woloszyn J., Richter C., Kaase M., Gatermann S. 2010. Identification of molecularly defined Staphylococcus aureus strains using matrix-assisted laser desorption/ionization time of flight mass spectrometry and the Biotyper 2.0 database. J. Med. Microbiol. 59:787–790 [DOI] [PubMed] [Google Scholar]
- 15. van Veen S. Q., Claas E. C., Kuijper E. J. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verroken A., et al. 2010. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Nocardia species. J. Clin. Microbiol. 48:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winn W. C., Koneman E. W. 2006. Koneman's color atlas and textbook of diagnostic microbiology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]