Abstract
In eight successive seasons (2001 to 2009), a total of 726 human respiratory syncytial virus (HRSV) infections from a total of 1,560 children with acute lower respiratory tract illness were identified. Molecular analysis of the attachment (G) protein gene confirmed that 52 (7.8%) children were infected more than once with any of the 3 genotypes of HRSV-A (genotypes GA5, NA1, and NA2) and/or 6 genotypes of HRSV-B (genotypes BA4, BA5, and BA7 to BA10). Repeated infections in 46 cases (82.1%) occurred in the next season, and only one case occurred in the same season (10-day interval). First infections were 33 (63.5%) HRSV-A cases and 19 (36.5%) HRSV-B cases, whereas second infections occurred in 35 (67.3%) HRSV-A cases and 17 (32.7%) HRSV-B cases. Third infections were attributed to 4 (100.0%) HRSV-A cases. Homologous subgroup reinfections were detected in 28 cases, 23 HRSV-A cases and 5 HRSV-B cases (P = 0.005), whereas homologous genotype reinfections were detected only for 5 HRSV-A cases (2GA5 and 3NA2) but not any HRSV-B case. Heterologous subgroup reinfections were detected in 28 cases, 12 cases from HRSV-A-to-HRSV-B reinfections and 16 cases from HRSV-B-to-HRSV-A reinfections. Genotypes NA1 and NA2 had higher numbers of heterologous genotype infections than did other genotypes. Our observations suggest that repeated infections occur more frequently in HRSV-A strains than in HRSV-B strains, and heterologous genotype reinfections occur more frequently than homologous genotype reinfections, especially in the case of the emerging genotypes NA1 and NA2 of HRSV-A strains that circulated in the community during our study period.
INTRODUCTION
Human respiratory syncytial virus (HRSV), a member of the genus Pneumovirus within the family Paramyxoviridae, is the most common cause of acute lower respiratory tract disease among infants and young children (3). In spite of maternal antibodies, 50% to 70% of infants are infected with HRSV in the first year of life, and almost all children are infected by 2 years of age (3). After that period, HRSV causes repeated infections throughout life, but infections in older children and adults are usually associated with milder disease, indicating that HRSV infections induce local immunity (10, 13).
The ability of the virus to reinfect may be due to an inadequate immune response and/or to strain variability allowing evasion of the immune response (25). HRSV has been classified into antigenic subgroups A and B (HRSV-A and HRSV-B, respectively), initially on the basis of the reactivity of the virus with monoclonal antibodies against the attachment glycoprotein (G protein) (1, 5, 14, 18), and have now been classified into genotypes in each subgroup by genetic analyses (8, 16, 23, 27, 29). The G protein is one of the main targets for inducing neutralizing and protective antibodies, and extensive antigenic and genetic diversity within this protein may facilitate repeated HRSV infections (2–4, 8, 15, 21–23, 26). Reinfections with homologous and heterologous groups of HRSV have been shown, but most studies of HRSV reinfection analyzed HSRV by groups and not by genotypes (17, 28). However, the relative contribution of antigenic diversity and/or inadequate immune responses to the establishment of reinfections has yet to be elucidated.
We previously reported that the newly identified genotype NA2 of HRSV-A caused the outbreak in the 2006-2007 season, while emerging HRSV-B genotypes did not cause an outbreak in the community (7, 26). In conjunction with data from previous molecular epidemiological studies, we undertook the present study to analyze the characteristics of reinfections in relation to HRSV genetic diversity among children with repeated HRSV infections in Japan.
MATERIALS AND METHODS
Study population and clinical samples.
This study was conducted for eight successive seasons from November 2001 through April 2009 at one pediatric outpatient clinic in Niigata City, Japan. Niigata City is the prefectural capital of Niigata Prefecture, and its total population is approximately 0.8 million. Children under 5 years of age with acute lower respiratory tract illness, such as wheezing, cough, rhinorrhea, and fever, were enrolled in this study. We retained written informed consent from their guardians and obtained nasopharyngeal aspirate samples or nasal swabs from the patients. Basic patient data such as registration number in the clinic, sex, age, date of clinic visit, and date of fever onset were recorded by a pediatrician at the clinic. The first infection was defined as the first episode of HRSV infection in children who subsequently had one or more HRSV infections. Repeated infection was identified by the unique registration number of the patient. This study was approved by the Medical Faculty Ethics Committee of Niigata University. Clinical specimens were kept at 4°C in the clinic; were transported to the Department of Public Health, Graduate School of Medical and Dental Sciences, Niigata University, within 5 days of sampling; and were then kept frozen at −80°C for further examination.
RT-PCR and nucleotide sequencing.
Briefly, viral RNA was extracted from 100-μl samples of the nasopharyngeal aspirate samples or swabs by using an Extragen II kit (Kainos, Tokyo, Japan) according to the manufacturer's instructions. Reverse transcription (RT) to create cDNA was performed by using random primers and Moloney murine leukemia virus reverse transcriptase (Invitrogen Corp., Carlsbad, CA) (23).
In accordance with a method previously reported (23), first and heminested PCRs targeting a 270-nucleotide segment (330-nucleotide segment in the case of HRSV-B) of the G-protein gene's second hypervariable region were performed. Amplified PCR products were purified by using MSB Spin PCRapace (Invitek GmbH, Berlin, Germany), labeled with a BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions, and then analyzed with an ABI 3100 automatic DNA sequencer.
Phylogenetic analysis.
Multiple-sequence alignments of G genes for HRSV-A and -B were compiled by using BioEdit, versions 7.0 and 9.0, respectively (12). Phylogenetic trees of the G-protein gene's second hypervariable region were generated by using the neighbor-joining method with MEGA software, version 4.0 (30). Bootstrap probabilities for 1,000 iterations were calculated to evaluate confidence estimates. All sequences for both subgroup A and B viruses of the repeated infection cases were included in the phylogenetic analysis. Reference sequences (BA/100/04, BA/524/04, BA/354/04, and BE/46/03) were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/). The G-protein gene sequences newly reported in this paper were deposited in the DDBJ database, and others were reported previously (7, 23).
Evaluation of repeated infections and statistical analysis.
The sequential infection pattern by subtype or genotype was evaluated between “previous” and “subsequent” infections. Thus, the first infection was compared to the second, and the second infection was compared to the third, to evaluate the influence of immunity from the most recent infections.
Fisher's exact test was used to evaluate statistical significance for 2-by-2 tables in homologous and heterologous subtype infections and that among homologous and heterologous genotype infections (SPSS, version 11.0). A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
The G-protein gene sequences newly reported in this paper were deposited in the DDBJ database under accession numbers AB603443 to AB603484 (http://www.ddbj.nig.ac.jp/index-e.html).
RESULTS
Study population and demographic details.
For eight successive seasons, from November 2001 through April 2009, 1,560 cases were screened in Niigata City, Japan. A total of 726 HRSV samples were positive by PCR. All HRSV-positive samples were sequenced and further analyzed for genotypes by the gene analysis of the attachment (G) protein gene of HRSV. Three genotypes of HRSV-A, GA5, NA1, and NA2, and 6 genotypes of HRSV-B, BA4, BA5, BA7, BA8, BA9, and BA10, were identified (7, 23, 26). The yearly distribution of positive cases varied from 33 to 154 depending on the season, with the highest positivity rate being observed for the 2006-2007 season (Table 1).
Table 1.
Seasonal genotype distributions of HRSV-A and -B strains in repeated infections from the 2002-2003 to 2008-2009 seasons
| Infection and season (mo/yr) | No. (%) of cases with repeated infectionsa | No. of isolatesb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HRSV-A |
HRSV-B |
|||||||||
| GA5 | NA1 | NA2 | BA4 | BA5 | BA7 | BA8 | BA9 | BA10 | ||
| 1st | ||||||||||
| 2002–2003 | 7/60 (11.7) | 1 | — | — | — | 6 | — | — | — | — |
| 2003–2004 | 3/75 (4.0) | 3 | — | — | — | — | — | — | — | — |
| 2004–2005 | 6/45 (13.3) | 1 | 4 | — | 1 | — | — | — | — | — |
| 2005–2006 | 18/121 (14.9) | — | 8 | 5 | — | — | 5 | — | — | — |
| 2006–2007 | 12/154 (7.8) | — | — | 9 | — | — | — | 2 | 1 | — |
| 2007–2008 | 6/80 (7.5) | 1 | — | 1 | — | — | 1 | — | — | 3 |
| 2nd | ||||||||||
| 2003–2004 | 3/75 (4.0) | 3 | — | — | — | — | — | — | — | — |
| 2004–2005 | 3/45 (6.7) | 2 | — | — | 1 | — | — | — | — | — |
| 2005–2006 | 8/121 (6.6) | — | 4 | 2 | 1 | — | — | — | 1 | — |
| 2006–2007 | 20/154 (13.0) | — | — | 14 | — | — | — | 5 | 1 | — |
| 2007–2008 | 9/80 (11.3) | 1 | — | — | — | — | — | — | 1 | 7 |
| 2008–2009 | 9/79 (11.4) | — | 6 | 3 | — | — | — | — | — | — |
| 3rd | ||||||||||
| 2006–2007 | 2/154 (1.3) | — | — | 2 | — | — | — | — | — | — |
| 2008–2009 | 2/79 (2.5) | — | 2 | — | — | — | — | — | — | — |
Percentages were calculated from number of patients who experienced repeated infections divided by the total number of RSV cases detected for the season.
—, no cases.
State of repeat infections.
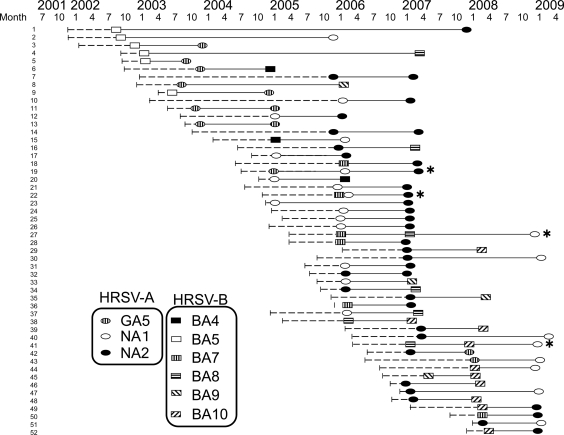
Typing of HRSV confirmed that 52 children (7.8%) had repeated HRSV infections. First infections included 33 (63.5%) HRSV-A cases and 19 (36.5%) HRSV-B cases, whereas second infections occurred in 35 (67.3%) HRSV-A cases and 17 (32.7%) HRSV-B cases. Third infections were attributed to 4 (100.0%) HRSV-A cases. Genotypes were comprised of genotypes GA5, NA1, and NA2 for HRSV-A and/or genotypes BA4, BA5, and BA7 to BA10 for HRSV-B (Fig. 1 and 2 and Table 1). Four of these patients were infected with HRSV three times (Fig. 1). All patients had no underlying conditions and did not show any serious conditions for hospitalization.
Fig. 1.
Repeat HRSV infection cases divided by genotypes shown in a timeline from 2001 to 2009. Dashed lines indicate birth to first infection. Ellipses indicate HRSV-A infections, while rectangles indicate HRSV-B infections. Solid lines mark the interval of infections. Lines with asterisk are cases with three infections.
The average ages of the first (primary), second, and third infections were 10.6 ± 7.9, 25.8 ± 11.5, and 33.6 ± 7.6 months, respectively (Table 2). The average age upon the second infections was higher for patients with genotypes NA1, NA2, BA8, BA9, and BA10 than for patients with other genotypes. Repeated infections in 46 (82.1%) cases occurred in the next season, and only one case had a repeated infection in the same season (10-day interval). The average interval period for reinfection was 14.6 ± 8.8 months (range, 10 days to 50 months).
Table 2.
Numbers and average ages of patients with HRSV-A and HRSV-B infections divided by genotypea
| Genotype | Total |
1st infection |
2nd infection |
3rd infection |
||||
|---|---|---|---|---|---|---|---|---|
| No. of cases | Mean avg age (mo) ± SD | No. of cases | Mean avg age (mo) ± SD | No. of cases | Mean avg age (mo) ± SD | No. of cases | Mean avg age (mo) ± SD | |
| HRSV-A | ||||||||
| GA5 | 12 | 11.6 ± 6.4 | 6 | 8.3 ± 6.1 | 6 | 14.9 ± 4.8 | — | |
| NA1 | 24 | 19.0 ± 12.5 | 12 | 12.1 ± 8.1 | 10 | 27.5 ± 11.8 | 2 | 38.6 ± 7.7 |
| NA2 | 36 | 21.2 ± 13.6 | 15 | 12.2 ± 9.8 | 19 | 28.2 ± 13.0 | 2 | 28.7 ± 3.4 |
| HRSV-B | ||||||||
| BA4 | 3 | 17.0 ± 6.5 | 1 | 9.7 | 2 | 20.8 ± 4.8 | — | |
| BA5 | 6 | 6.8 ± 7.9 | 6 | 6.8 ± 7.9 | — | — | ||
| BA7 | 6 | 9.6 ± 8.3 | 6 | 9.6 ± 8.3 | — | — | ||
| BA8 | 7 | 26.5 ± 12.2 | 2 | 17.0 ± 7.0 | 5 | 30.4 ± 12.0 | — | |
| BA9 | 4 | 22.8 ± 10.4 | 1 | 8.4 | 3 | 27.6 ± 7.2 | — | |
| BA10 | 10 | 16.8 ± 12.5 | 3 | 11.3 ± 4.9 | 7 | 23.3 ± 11.4 | — | |
| Total | 108 | 18.5 ± 12.5 | 52 | 10.6 ± 7.9 | 52 | 25.8 ± 11.5 | 4 | 33.6 ± 7.6 |
—, no cases.
Upon the first infection, 33 cases were infected with 3 genotypes of HRSV-A strains, genotypes GA5 (6 cases), NA1 (12 cases), and NA2 (15 cases), while 19 cases were infected with 6 genotypes of HRSV-B strains, genotypes BA4 (1 case), BA5 (6 cases), BA7 (6 cases), BA8 (2 cases), BA9 (1 case), and BA10 (3 cases) (Table 1 and Fig. 2). Genotypes GA5 and BA5 were the predominant genotypes in the repeated infections from the 2002-2003 season to the 2003-2004 season, and the predominance shifted to genotypes NA1 and NA2 from the 2004-2005 season (Table 1). Furthermore, the number of repeated infection cases increased in the 2005-2006 and 2006-2007 seasons, perhaps due to the circulation of the emerging genotypes NA1, NA2, and BA7 (7, 26).
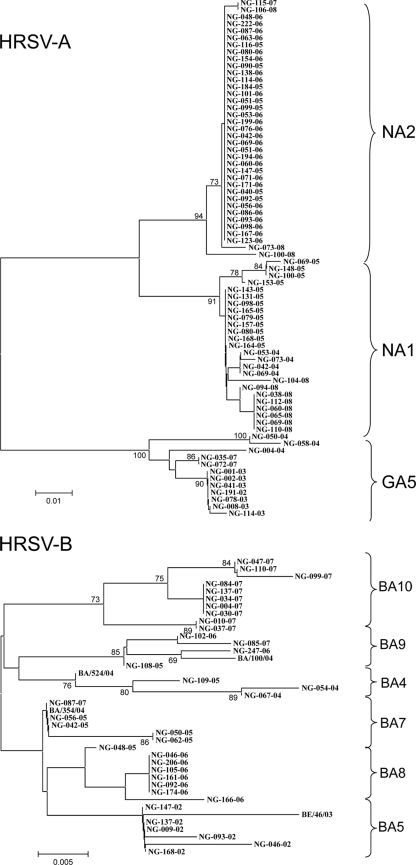
Fig. 2.
Phylogenic trees of HRSV-A and HRSV-B nucleotide sequences based on the second variable region of the G-protein gene (270 and/or 330 bp) using the neighbor-joining method with MEGA software, version 4.0. Reference sequences of group B genotypes from other studies were included in the analysis of respective group comparisons. The scale bars show the proportions of nucleotide substitutions per site, and the values at the nodes are bootstrap values for 1,000 iterations. Only bootstrap values greater than 70% are shown.
Upon the second infection, 35 cases were infected with 3 genotypes of HRSV-A strains, genotypes GA5 (6 cases), NA1 (10 cases), and NA2 (19 cases), while 17 cases were infected with 4 genotypes of HRSV-B strains, genotypes BA4 (2 cases), BA8 (5 cases), BA9 (3 cases), and BA10 (7 cases) (Table 2). The proportion of reinfected cases over total HRSV cases had increased since the 2006-2007 season, due mainly to emerging genotypes NA1, NA2, BA8, and BA10 (Table 1 and Fig. 1).
Upon the third infection, 4 cases were reinfected with 2 strains each of genotypes NA1 and NA2 (Table 2 and Fig. 2). Overall, for three-time infections one patient was infected with HRSV-A of different genotypes, and one was infected once with HRSV-B followed twice by infection with HRSV-A. The other two cases were repeatedly infected with HRSV-B, and it changed to HRSV-A in the third infection (Table 3). There were no homologous genotype infections with the four cases.
Table 3.
Genotypes attributed to HRSV infections three times
| Patient | Genotype |
||
|---|---|---|---|
| 1st infection | 2nd infection | 3rd infection | |
| 19 | GA5 | NA1 | NA2 |
| 22 | BA7 | NA1 | NA2 |
| 27 | BA7 | BA8 | NA1 |
| 41 | BA8 | BA10 | NA1 |
Throughout all seasons, homologous subgroup reinfections were detected in 28 cases: 23 HRSV-A cases and 5 HRSV-B cases. Homologous HRSV-A infections occurred more frequently (65.7%) than homologous HRSV-B infections (23.8%), with statistical significance (P = 0.005) (Table 4 and Fig. 2). Homologous genotype reinfections were detected in 2 repeated genotype GA5 cases (interval, 14.4 and 14.9 months) and 3 repeated genotype NA2 cases (interval, 11.9, 12.7, and 13.7 months) with HRSV-A but not any case with HRSV-B, including the third infection. Among HRSV-A strains, heterologous genotype reinfections were detected in 18 cases: 2 cases (genotype GA5 to genotype NA1), 11 cases with a change from genotype NA1 to genotype NA2, 1 case with a change from genotype NA2 to genotype GA5, and 4 cases with a change from genotype NA2 to genotype NA1 (Tables 3 and 5). Among HRSV-B strains, heterologous genotype reinfections were detected in 5 cases: 1 case with a change from genotype BA5 to genotype BA8, 1 case with a change from genotype BA7 to genotype BA8, 2 cases with a change from genotype BA8 to genotype BA10, and 1 case with a change from genotype BA9 to genotype BA10.
Table 4.
Total number of repeated HRSV infection cases divided by subtypes of HRSV-A and -B
| Previous infection | No. (%) of cases of subsequent infection witha: |
|
|---|---|---|
| HRSV-A | HRSV-B | |
| HRSV-A (n = 35) | 23 (65.7)* | 12 (34.3) |
| HRSV-B (n = 21) | 16 (76.2) | 5 (23.8)* |
An asterisk indicates that a statistically significant difference in the number of homologous subtype reinfection cases was shown between HRSV-A and -B over hetrologous subtype infection (P = 0.005). The P value was tested by a two-tailed Fisher's exact probability test.
Table 5.
Total number of repeated HRSV infection cases divided by genotypes of HRSV-A and -B in first and second infections
| Genotype of previous infection | Total no. of cases of previous infection | No. of cases of subsequent infectiona |
No. (%) of homologous genotype reinfection cases | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HRSV-A |
HRSV-B |
||||||||
| GA5 | NA1 | NA2 | BA4 | BA8 | BA9 | BA10 | |||
| HRSV-A | |||||||||
| GA5 | 6 | 2 | 2 | 0 | 1 | 0 | 1 | 0 | 2 (33.3) |
| NA1 | 14 | 0 | 0 | 11 | 1 | 1 | 1 | 0 | 0 (0) |
| NA2 | 15 | 1 | 4 | 3 | 0 | 2 | 1 | 4 | 3 (20.0) |
| Subtotal | 35 | 3 | 6 | 14 | 2 | 3 | 3 | 4 | 5* (14.3) |
| HRSV-B | |||||||||
| BA4 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 (0) |
| BA5 | 6 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 0 (0) |
| BA7 | 6 | 0 | 1 | 4 | 0 | 1 | 0 | 0 | 0 (0) |
| BA8 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 (0) |
| BA9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 (0) |
| BA10 | 4 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 (0) |
| Subtotal | 21 | 3 | 6 | 7 | 0 | 2 | 0 | 3 | 0* (0) |
| Total | 56 | 6 | 12 | 21 | 2 | 5 | 3 | 7 | 5 (8.9) |
An asterisk indicates that statistical significance was not shown for the number of homologous genotype reinfection cases between HRSV-A and -B over heterologous genotype infections (P = 0.145). The P value was tested by a two-tailed Fisher's exact probability test.
Heterologous subgroup reinfections were detected in 28 cases: 12 cases of HRSV-A-to-HRSV-B reinfections and 16 cases of HRSV-B-to-HRSV-A reinfections (Tables 3 and 4 and Fig. 1). For the HRSV-A-to-HRSV-B group, heterologous genotype reinfections were detected in one case each of a change of genotype GA5 to genotype BA4 and genotype GA5 to genotype BA9; one case each of genotype NA1 to genotype BA4, genotype NA1 to genotype BA8, and genotype NA1 to genotype BA9; 2 cases of genotype NA2 to genotype BA8; one case of genotype NA2 to genotype BA9; and 4 cases of genotype NA2 to genotype BA10 (Table 5). For the HRSV-B-to-HRSV-A group, heterologous genotype reinfections were detected in one case of a change of genotype BA4 to genotype NA1; 3 cases of genotype BA5 to genotype GA5, one case each of genotype BA5 to genotype NA1 and genotype BA5 to genotype NA2, one case of genotype BA7 to genotype NA1, 4 cases of genotype BA7 to genotype NA2, one case of genotype BA8 to genotype NA1, and 2 cases each of genotype BA10 to genotype NA1 and genotype BA10 to genotype NA2 (Table 5).
Overall, we observed more homologous genotype infections with HRSV-A (14.3%) than with HRSV-B (0%) over heterologous genotype infections but without statistical significance (Table 5).
DISCUSSION
We found that the number of reinfections with the HRSV-A subgroup was significantly higher than that with the HRSV-B subgroup. This finding is different from data from other reports showing that children initially infected with an HRSV-A strain appeared more likely to experience reinfection with an HRSV-B strain (17). The higher prevalence of reinfections among HRSV-A subgroups may indicate that the subgroup has sufficient antigenic divergence to allow escape from an individual's immune response. Another possible explanation for the more frequent occurrence of homologous HRSV-A reinfections is that HRSV-B strains elicit a more complete and/or long-lasting subgroup-specific immune response than do HRSV-A strains (17, 32), and thus, HRSV-B may cause repeated infections less often.
We previously reported several genotypes for each HRSV subgroup, genotypes GA5 and GA7 and emerging genotypes NA1 and NA2 for HRSV-A and genotypes GB3 and SAB3, and emerging genotypes BA4 to BA10 for HRSV-B, during eight successive seasons from 2001 to 2009 (7, 23, 26). Large outbreaks of HRSV in the community were observed upon the emergence of newly identified HRSV-A genotypes (genotypes NA1 and NA2) but not upon the emergence of newly identified HRSV-B genotypes. Our study showed that the repeated infections were attributed to 3 genotypes of HRSV-A (genotypes GA5, NA1, and NA2) and 6 genotypes of HRSV-B (genotypes BA4, BA5, and BA7 to BA10). The overall distribution of genotypes of HRSV in the first and repeated infections in each season was similar to the prevalence of genotypes that circulated in the season (7, 26).
Among the reinfection episodes, repeat infections in 46 cases (82.1%) occurred in the succeeding season, and only one case was a heterologous genotype reinfection in the same season (10-day interval). This case eventually had another genotype reinfection with HRSV-A in the following season. It is known that following primary infection, some infants lose strain-specific immunity after 7 to 9 months (between epidemics) and group-specific immunity in 2 to 4 months (within an epidemic period) (24). This information is relevant in trying to elucidate the nature of immunity to reinfection by HRSV in children in relation to the primary infecting and reinfecting variants.
Protective immunity to the virus of the homologous subgroup can be short-lived in some individuals. Our study clearly indicated that homologous subgroup reinfections occurred more frequently in HRSV-A than in HRSV-B strains (23 versus 5 events). Furthermore, we demonstrated that homologous genotype reinfections were detected in 5 HRSV-A cases but not were detected in any case of HRSV-B infection. These findings strongly suggest that a stronger immune response against HRSV-B itself might had led to a lower rate of homologous reinfections with HRSV-B in general.
Our study clearly indicated that homologous (28 cases) and heterologous (28 cases) subgroup reinfections occurred with the same frequency, although previous studies indicated that children infected with one antigenic group were more likely to be reinfected with the heterologous group than with the homologous group, although the numbers examined were small (17). In the case of heterologous subgroup infections, HRSV-B-to-HRSV-A reinfections occurred more frequently than did HRSV-A-to-HRSV-B reinfections. These findings may be the result of the higher rate of circulation of emerging genotype NA1 and NA2 strains than of other strains in the community at the time of infection. However, during our study period, we found several emerging genotypes for each HRSV group: genotypes NA1 and NA2 for HRSV-A and genotypes BA4, BA5, and BA7 to BA10 for HRSV-B (7, 26). The genetic diversity in the second hypervariable region of these strains was within the range of what we reported previously (7, 26). Of note, genotype NA2 was associated with an increase of infection and reinfection cases in the 2006-2007 season (26), whereas newly identified genotype BA10, as the most prevalent type of HRSV-B in the 2007-2008 season, did not increase the number of reinfections in the 2007-2008 season. Furthermore, the proportion of reinfections among the circulating strains in our study(7.8%) was higher than those reported previously (19, 20, 23, 25), probably due to the newly identified genotypes NA1 and NA2. Thus, our observations lead us to surmise that even newly identified genotypes, such as genotype NA2, which played an important role in increased infection and reinfection, may not elicit a complete and/or long-lasting genotype-specific immune response compared to other genotypes. This remains to be confirmed by serological studies, and further investigation of the role of genotype NA2 in repeat infections is warranted. We also consider the point of view that the observed overall prevalence of HRSV-A over HRSV-B strains during the epidemic could be due to more transient subgroup A-specific immune protection rather than to higher HRSV-A genetic diversity, as was previously suggested (6, 32).
We found cases infected with HRSV three times during the study period. To our knowledge, this is the first report to elaborate the shift of HRSV genotypes in multiple infections, although one paper in the past reported three-time infections with serotype HRSV F and G proteins (31). Of note, homologous subgroup infections with HRSV-A occurred three times in one patient, but no other patients had serial HRSV-B infections three times, which also suggests a short-lived immunity to HRSV-A.
This study was implemented at one outpatient clinic where children consistently visited for years. Samples were collected mainly from children less than 5 years of age with acute lower respiratory tract symptoms, such as wheezing, cough, rhinorrhea, and fever. It is well known that there is a reduction in the severity of clinical illness upon reinfection (9–11, 13). Since we could not obtain disease or severity scores in this study, we could not discuss the differences in the clinical pictures at the time of repeated infections. In addition, a limitation of the study is that there is a possibility of missing cases due to no visit to the clinic because of mild symptoms.
We demonstrated the antigenic variability of the G protein between and within the two major HRSV antigenic groups that may be related to reinfection by evading the preexisting host immune response. Further study will be required to characterize the relationship between genomic variants and primary and/or repeat infections for the development of effective HRSV vaccines.
ACKNOWLEDGMENTS
We thank the staff at Sano Pediatric Clinic who assisted with the collection of the clinical information and samples and Akinori Miyashita and Ryozo Kuwano at the Center for Bioresources, Brain Research Institute, Niigata University, for utilization of the DNA sequencer. We thank Naohito Tanabe, Hassan Zaraket, Tatiana Baranovich, and Taeko Oguma for reviewing the manuscript; Akemi Watanabe for technical assistance in virus isolation; and Yoshiko Kato for intensive secretarial work.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1. Anderson L., et al. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626–633 [DOI] [PubMed] [Google Scholar]
- 2. Cane P. 1997. Analysis of linear epitopes recognised by the primary human antibody response to a variable region of the attachment (G) protein of respiratory syncytial virus. J. Med. Virol. 51:297–304 [DOI] [PubMed] [Google Scholar]
- 3. Cane P. 2001. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 11:103–116 [DOI] [PubMed] [Google Scholar]
- 4. Cane P., Pringle C. 1995. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J. Virol. 69:2918–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coates H., Alling D., Chanock R. 1966. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am. J. Epidemiol. 83:299–313 [DOI] [PubMed] [Google Scholar]
- 6. Coggins W., Lefkowitz E., Sullender W. 1998. Genetic variability among group A and group B respiratory syncytial viruses in a children's hospital. J. Clin. Microbiol. 36:3552–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dapat I., et al. 2010. New genotypes within respiratory syncytial virus group B genotype BA in Niigata, Japan. J. Clin. Microbiol. 48:3423–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. García O., et al. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 68:5448–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glezen W., Paredes A., Allison J., Taber L., Frank A. 1981. Risk of respiratory syncytial virus infection for infants from low-income families in relationship to age, sex, ethnic group, and maternal antibody level. J. Pediatr. 98:708–715 [DOI] [PubMed] [Google Scholar]
- 10. Glezen W., Taber L., Frank A., Kasel J. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140:543–546 [DOI] [PubMed] [Google Scholar]
- 11. Hall C., Walsh E., Long C., Schnabel K. 1991. Immunity to and frequency of reinfection with respiratory syncytial virus. J. Infect. Dis. 163:693–698 [DOI] [PubMed] [Google Scholar]
- 12. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp. Ser. 41:95–98 [Google Scholar]
- 13. Henderson F., Collier A., Clyde W. J., Denny F. 1979. Respiratory-syncytial-virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N. Engl. J. Med. 300:530–534 [DOI] [PubMed] [Google Scholar]
- 14. Hendry R., et al. 1986. Concurrent circulation of antigenically distinct strains of respiratory syncytial virus during community outbreaks. J. Infect. Dis. 153:291–297 [DOI] [PubMed] [Google Scholar]
- 15. Johnson P., Spriggs M., Olmsted R., Collins P. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. U. S. A. 84:5625–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Melero J., García-Barreno B., Martínez I., Pringle C., Cane P. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 78(Pt. 10):2411–2418 [DOI] [PubMed] [Google Scholar]
- 17. Mufson M., Belshe R., Orvell C., Norrby E. 1987. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J. Clin. Microbiol. 25:1535–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mufson M., Orvell C., Rafnar B., Norrby E. 1985. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 66(Pt. 10):2111–2124 [DOI] [PubMed] [Google Scholar]
- 19. Nokes D., et al. 2004. Respiratory syncytial virus epidemiology in a birth cohort from Kilifi district, Kenya: infection during the first year of life. J. Infect. Dis. 190:1828–1832 [DOI] [PubMed] [Google Scholar]
- 20. Parveen S., Broor S., Kapoor S., Fowler K., Sullender W. 2006. Genetic diversity among respiratory syncytial viruses that have caused repeated infections in children from rural India. J. Med. Virol. 78:659–665 [DOI] [PubMed] [Google Scholar]
- 21. Peret T., Hall C., Schnabel K., Golub J., Anderson L. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79(Pt. 9):2221–2229 [DOI] [PubMed] [Google Scholar]
- 22. Rueda P., Delgado T., Portela A., Melero J., García-Barreno B. 1991. Premature stop codons in the G glycoprotein of human respiratory syncytial viruses resistant to neutralization by monoclonal antibodies. J. Virol. 65:3374–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sato M., et al. 2005. Molecular epidemiology of respiratory syncytial virus infections among children with acute respiratory symptoms in a community over three seasons. J. Clin. Microbiol. 43:36–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scott P., et al. 2006. Molecular analysis of respiratory syncytial virus reinfections in infants from coastal Kenya. J. Infect. Dis. 193:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scott P., et al. 2007. Comparison of strain-specific antibody responses during primary and secondary infections with respiratory syncytial virus. J. Med. Virol. 79:1943–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shobugawa Y., et al. 2009. Emerging genotypes of human respiratory syncytial virus subgroup A among patients in Japan. J. Clin. Microbiol. 47:2475–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sullender W., Mufson M., Anderson L., Wertz G. 1991. Genetic diversity of the attachment protein of subgroup B respiratory syncytial viruses. J. Virol. 65:5425–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sullender W., Mufson M., Prince G., Anderson L., Wertz G. 1998. Antigenic and genetic diversity among the attachment proteins of group A respiratory syncytial viruses that have caused repeat infections in children. J. Infect. Dis. 178:925–932 [DOI] [PubMed] [Google Scholar]
- 29. Sullender W., Sun L., Anderson L. 1993. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J. Clin. Microbiol. 31:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 31. Wagner D. K., et al. 1989. Serum immunoglobulin G antibody subclass response to respiratory syncytial virus F and G glycoproteins after first, second, and third infections. J. Clin. Microbiol. 27:589–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zlateva K., Vijgen L., Dekeersmaeker N., Naranjo C., Van Ranst M. 2007. Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J. Clin. Microbiol. 45:3022–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]