Abstract
We compared the BD Phoenix automated microbiology system to the Bruker Biotyper (version 2.0) matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) system for identification of Gram-negative bacilli, using biochemical testing and/or genetic sequencing to resolve discordant results. The BD Phoenix correctly identified 363 (83%) and 330 (75%) isolates to the genus and species level, respectively. The Bruker Biotyper correctly identified 408 (93%) and 360 (82%) isolates to the genus and species level, respectively. The 440 isolates were grouped into common (308) and infrequent (132) isolates in the clinical laboratory. For the 308 common isolates, the BD Phoenix and Bruker Biotyper correctly identified 294 (95%) and 296 (96%) of the isolates to the genus level, respectively. For species identification, the BD Phoenix and Bruker Biotyper correctly identified 93% of the common isolates (285 and 286, respectively). In contrast, for the 132 infrequent isolates, the Bruker Biotyper correctly identified 112 (85%) and 74 (56%) isolates to the genus and species level, respectively, compared to the BD Phoenix, which identified only 69 (52%) and 45 (34%) isolates to the genus and species level, respectively. Statistically, the Bruker Biotyper overall outperformed the BD Phoenix for identification of Gram-negative bacilli to the genus (P < 0.0001) and species (P = 0.0005) level in this sample set. When isolates were categorized as common or infrequent isolates, there was statistically no difference between the instruments for identification of common Gram-negative bacilli (P > 0.05). However, the Bruker Biotyper outperformed the BD Phoenix for identification of infrequently isolated Gram-negative bacilli (P < 0.0001).
INTRODUCTION
Traditional bacterial identification in the clinical microbiology laboratory is done by biochemical and phenotypic analysis, using both manual and automated systems, in addition to molecular methods (17). While some of these techniques are rapid, the majority depend on microbial growth and utilization of biochemicals, requiring hours to days for identification.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) is a technology used to identify and analyze proteins. While the analysis of whole-cell bacteria for identification by MALDI-TOF MS was demonstrated a number of years ago (12, 13), not until recently has its potential for routine use been assessed for bacterial identification in the clinical laboratory (2, 5, 22, 27). The premise of bacterial identification by the MALDI-TOF MS approach is the generation of a spectral profile of abundant bacterial proteins, the majority of which are likely ribosomal (19). The spectral profile is referenced to a compiled database, allowing for comparison and differentiation of bacterial isolates by their protein profiles. Protein analysis by MALDI-TOF MS does not require lengthy biochemical reactions, making it a more rapid identification strategy than traditional methods (reviewed in references 4 and 20). Furthermore, costs associated with MALDI-TOF MS are significantly lower than those of commonly used techniques (5, 22).
Significant effort has focused on MALDI-TOF MS analyses of specific bacterial genera (9, 18, 21, 23, 25, 28). Additional studies analyzed general groups of bacteria (6, 16) or yeast (15, 24) or assessed its utility for routine bacterial identification in the clinical laboratory (1, 2, 5, 22, 27). Numerous reports focused on comparison of MALDI-TOF MS to automated systems, in particular the Vitek system (2, 5, 22, 27). Additionally, comparisons to traditional biochemical or commercial phenotypic identification assays (6) and 16S rRNA gene sequencing (16) have been reported. Despite this, comparisons of MALDI-TOF MS to the BD Phoenix automated microbiology system (Becton Dickinson, Franklin Lakes, NJ) (1, 10), and in particular analysis of Gram-negative bacilli, are limited. In studies comparing MALDI-TOF MS to the BD Phoenix, one report focused on coagulase-negative staphylococci (10) and the other compared a MALDI-TOF/TOF tandem MS system to identification by the BD Phoenix and API strips (1).
In this study, we compared the BD Phoenix to the Bruker Biotyper MALDI-TOF MS system (Bruker Daltonics, Billerica, MA) for the identification of Gram-negative bacilli, strictly using the manufacturer-provided reference databases for each instrument. Our study is unique in that we included an analysis of both common Gram-negative bacilli encountered in the clinical laboratory and those infrequently isolated. Identification achieved solely on the manufacturer-provided database demonstrates the performance of the instrument if implemented directly into the clinical laboratory.
(This work was presented, in part, at the 48th Annual Meeting of the Infectious Diseases Society of America, Vancouver, British Columbia, Canada, 21 to 24 October 2010.)
MATERIALS AND METHODS
Bacterial isolates.
Four hundred forty aerobic Gram-negative bacilli collected from multiple clinical sources, including blood, tissue, urine, stool, wound, cerebrospinal fluid, and respiratory and other sources, were studied. Isolates were categorized as either common or infrequent by their relative frequency of occurrence in daily laboratory identification. Commonly encountered species were defined as those encountered at least weekly in our laboratory (as assessed over a 2-week period). In order to obtain a representative sample of pathogens, we limited the number of common isolates analyzed, biasing toward infrequently isolated bacteria. All isolates were cultured overnight, or until visible growth was observed, on 5% sheep blood agar in 5% CO2 at 35°C, with the exception of Haemophilus spp. (chocolate agar), Legionella spp. (ambient atmosphere, buffered charcoal yeast extract [BCYE] agar), and Campylobacter spp. (capnophilic atmosphere, 42°C, chocolate agar).
Automated identification.
All isolates were analyzed by the BD Phoenix automated identification system by following the manufacturer's recommendations and as previously reported (8, 14). Briefly, isolated colonies subcultured and incubated overnight, as outlined above, were used to inoculate manufacturer-provided diluent to a McFarland standard of 0.5 to 0.6. Subsequently, BD Phoenix Gram-negative identification cassettes (negative identification [NID] panel 448007) were inoculated with the bacterial suspension and placed in the BD Phoenix system. Valid isolate identification required a score greater then 90%; otherwise, no identification was reported.
MALDI-TOF MS.
Bacterial isolates were cultured overnight as described above. Bacteria were applied as a thin film to a 24-spot steel plate (Bruker Daltonics) and allowed to visibly dry at room temperature (referred to as the direct colony technique). Subsequently, 2 μl of MALDI matrix (a saturated solution of α-cyano-4-hydroxycinnamic acid [HCCA; Bruker Daltonics] in 50% acetonitrile and 2.5% trifluoroacetic acid) was applied to the colony and dried in a fume hood. MALDI-TOF MS was performed with a MicroFlex LT system (Bruker Daltonics) tabletop mass spectrometer using the manufacturer's suggested settings. Briefly, ions generated with a 337-nm nitrogen laser were captured in the positive linear mode in a mass range of 2 to 20 kDa. Captured spectra were analyzed using MALDI Biotyper automation control and Bruker Biotyper 2.0 software (Bruker Daltonics, Bremen, Germany). The MALDI Biotyper database contained 3,740 spectra from 319 genera and 1,946 species. For each 24-spot plate, a standard (bacterial test standard; Bruker Daltonics) was included to calibrate the instrument and validate the run.
Identification criteria used in our analysis, outlined by the manufacturer, were as follows: a score of ≥2.000 indicated species level identification, a score of 1.700 to 1.999 indicated identification to the genus level, and a score of <1.700 was interpreted as no identification. Additionally, in order for the above criteria to be valid, a minimum difference of 10% between the top score and next closest scores was required for individual isolates (referred to as the 10% differential rule) (6). Isolates which had conflicting species or genus identifications that failed the 10% rule were considered incorrect. (This was not a manufacturer recommendation.)
After initial analysis on the Bruker Biotyper, any isolate that failed to produce a score of ≥2.000 or generated a score of ≥2.000 but failed the 10% differential rule was reanalyzed by the direct colony technique. Isolates which failed to generate a score of ≥2.000 were subsequently extracted prior to analysis. For extraction, a loopful of bacteria was suspended in 300 μl of molecular grade water (Sigma-Aldrich, St. Louis, MO) and vortexed. Next, 900 μl of 100% ethanol (Sigma-Aldrich) was added, vortexed, and centrifuged at top speed (20,800 × g) for 2 min. Supernatant was decanted, and the pellet was again centrifuged at top speed. Residual supernatant was removed, and the pellet was dried at room temperature. To the pellet, 50 μl of 70% formic acid (Fluka [Sigma-Aldrich], St. Louis, MO) was added and mixed by pipetting. Subsequently, 50 μl of acetonitrile (Fluka) was added and thoroughly mixed, followed by centrifugation at top speed for 2 min. One microliter of supernatant was spotted on the 24-spot plate and allowed to visibly dry at room temperature before the addition of 1 μl matrix.
Discrepant analysis.
Discrepant results generated by the two systems were resolved by traditional biochemical analysis, partial 16S rRNA gene sequencing (Microseq ID version 2.0 AB_bacterial500LIB_2.1; Applied Biosystems, Carlsbad, CA), or gyrB gene sequencing (7, 11, 17). Isolates for sequencing were lysed in PrepMan Ultra (Applied Biosystems). When definitive species identification was not achieved, isolates were classified to the genus level only.
Data analysis.
Subsets of closely related species are routinely grouped into complexes because they are difficult to separate biochemically or phenotypically. We observed similar challenges in this study with the BD Phoenix and Bruker Biotyper. When such isolates were analyzed, occasional differences in species identification between instruments was observed. Despite these differences, we considered the result correct if the species belonged to the Burkholderia cepacia complex, Citrobacter freundii complex, Enterobacter cloacae complex, or Klebsiella pneumoniae complex. Additionally, both systems had databases with heterotypic synonyms or outdated nomenclature, which would have resulted in misidentification (see Discussion). In these instances, the identification was considered correct.
Statistical analysis.
Comparison of the BD Phoenix with the Bruker Biotyper for the identification of genus or species was made using McNemar's test, a test of paired proportions. P values of <0.05 were considered statistically significant. Statistical analysis was performed using SAS version 9.1 (SAS Institute).
RESULTS
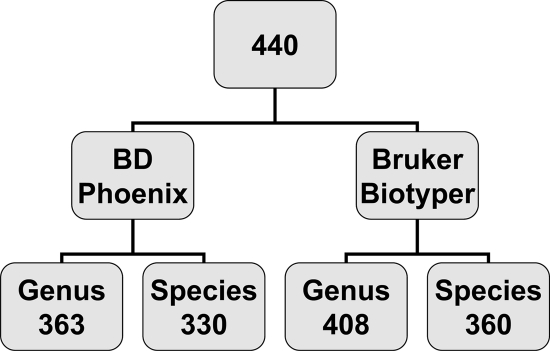
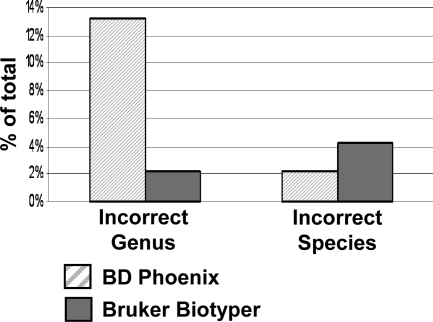
The BD Phoenix correctly identified 363 (83%) isolates to the genus level, of which 330 (75% of the total isolates) were correctly identified to the species level. In comparison, the Bruker Biotyper correctly identified 408 (93%) isolates to the genus level, and 360 (82%) to the species level (Fig. 1), statistically performing better then the BD Phoenix for both genus (P < 0.0001) and species (P = 0.0005) identification. Incorrect genus identification was provided for 58 (13%) and 10 (2%) isolates by the BD Phoenix and Bruker Biotyper, respectively (Fig. 2). Correct genus but incorrect species identification was provided for 10 (2%) and 19 (4%) isolates by the BD Phoenix and Bruker Biotyper, respectively (Fig. 2).
Fig. 1.
Identification of Gram-negative bacilli by BD Phoenix and Bruker Biotyper MALDI-TOF MS systems. Four hundred forty Gram-negative bacilli were analyzed by the BD Phoenix and Bruker Biotyper MALDI-TOF MS systems. The numbers of correct genus and species level identifications are shown for each instrument.
Fig. 2.
Incorrect genus or species level identifications, determined by additional biochemical or molecular methods, are shown for the BD Phoenix and Bruker Biotyper MALDI-TOF MS for the 440 isolates analyzed.
We observed that both the BD Phoenix and Bruker Biotyper performed well in identifying the 308 common isolates (Table 1), achieving genus level identification for 294 (95%) and 296 (96%) isolates, respectively. Additionally, both systems achieved species level identification for 93% of the common isolates (285 and 286, respectively). Statistically, there was no difference between the two systems for identification of common isolates (P > 0.05). In contrast, the BD Phoenix was outperformed by the Bruker Biotyper for both genus and species identification (P < 0.0001) (Table 1) of infrequent isolates. For these 132 isolates, the BD Phoenix correctly identified to the genus and species level 69 (52%) and 45 (34%) isolates, respectively, whereas the Bruker Biotyper correctly identified to the genus and species level 112 (85%) and 74 (56%) isolates, respectively.
Table 1.
Common and infrequent isolates identified by the BD Phoenix and Bruker Biotyper
| Isolate type and name | No. of isolates | No. of isolates identified to the indicated level by the: |
|||
|---|---|---|---|---|---|
| BD Phoenix |
Bruker Biotyper |
||||
| Genus | Species | Genus | Species | ||
| Common | |||||
| Acinetobacter sp. | 7 | 6 | 5 | 7 | 4 |
| Alcaligenes faecalis | 2 | 2 | 2 | 2 | 2 |
| Citrobacter freundii complex | 27 | 25 | 24 | 27 | 27 |
| Citrobacter koseri | 5 | 5 | 5 | 5 | 5 |
| Escherichia coli | 36 | 35 | 35 | 36 | 36 |
| Enterobacter aerogenes | 12 | 12 | 11 | 12 | 12 |
| Enterobacter cloacae complex | 23 | 20 | 20 | 23 | 23 |
| Klebsiella oxytoca | 16 | 14 | 13 | 8 | 8 |
| Klebsiella pneumoniae complex | 35 | 34 | 34 | 35 | 34 |
| Klebsiella/Raoultella spp. | 1 | 1 | 0 | 1 | 0 |
| Klebsiella pneumoniae/Raoultella planticola | 1 | 1 | 1 | 1 | 1 |
| Morganella morganii | 6 | 6 | 6 | 6 | 6 |
| Pantoea agglomerans | 4 | 3 | 3 | 0 | 0 |
| Proteus mirabilis | 22 | 22 | 21 | 22 | 22 |
| Pseudomonas aeruginosa | 69 | 67 | 64 | 69 | 69 |
| Serratia marcescens | 23 | 22 | 22 | 23 | 18 |
| Stenotrophomonas maltophilia | 19 | 19 | 19 | 19 | 19 |
| Total no. of isolates (% correct identification) | 308 | 294 (95) | 285 (93) | 296 (96) | 286 (93) |
| Infrequent | |||||
| Achromobacter denitrificans | 1 | 1 | 0 | 1 | 0 |
| Achromobacter piechaudii | 2 | 2 | 0 | 2 | 0 |
| Achromobacter xylosoxidans | 8 | 8 | 0 | 8 | 0 |
| Actinobacillus ureae | 1 | 1 | 1 | 1 | 1 |
| Aeromonas hydrophila | 1 | 1 | 1 | 1 | 0 |
| Aeromonas caviae complex | 2 | 2 | 2 | 2 | 0 |
| Aggregatibacter actinomycetemcomitans | 1 | 0 | 0 | 1 | 0 |
| Aggregatibacter aphrophilus | 2 | 0 | 0 | 2 | 2 |
| Azorhizobium caulinodans | 1 | 0 | 0 | 0 | 0 |
| Bordetella holmesii | 1 | 0 | 0 | 0 | 0 |
| Bordetella petrii | 1 | 0 | 0 | 0 | 0 |
| Brevundimonas diminuta/bullata | 1 | 1 | 1 | 1 | 1 |
| Brevundimonas diminuta/vullata | 1 | 1 | 1 | 1 | 1 |
| Burkholderia cepacia complex | 8 | 4 | 4 | 8 | 8 |
| Burkholderia gladioli | 1 | 1 | 1 | 1 | 1 |
| Burkholderia spp. | 1 | 0 | 0 | 0 | 0 |
| Campylobacter jejuni | 2 | 0 | 0 | 2 | 2 |
| Cardiobacterium hominis | 1 | 1 | 1 | 1 | 0 |
| Cedecea spp. | 1 | 1 | 0 | 1 | 0 |
| Chryseobacterium indologenes/gleum | 2 | 1 | 1 | 1 | 1 |
| Chryseobacterium spp. | 3 | 0 | 0 | 2 | 0 |
| Citrobacter amalonaticus | 1 | 1 | 1 | 1 | 1 |
| Citrobacter farmeri | 2 | 2 | 2 | 2 | 2 |
| Comamonas kersterii | 1 | 1 | 0 | 1 | 1 |
| Delftia acidovorans | 1 | 1 | 1 | 1 | 0 |
| Eikenella corrodens | 3 | 3 | 3 | 3 | 1 |
| Haemophilus influenzae | 9 | 0 | 0 | 9 | 9 |
| Haemophilus parainfluenzae | 8 | 0 | 0 | 8 | 7 |
| Herbaspirillum sp. | 1 | 0 | 0 | 1 | 0 |
| Inquilinus limosus | 1 | 0 | 0 | 1 | 1 |
| Legionella pneumophila | 1 | 0 | 0 | 1 | 1 |
| Moraxella atlantae | 1 | 0 | 0 | 0 | 0 |
| Moraxella nonliquefaciens | 1 | 1 | 0 | 1 | 1 |
| Moraxella osloensis | 4 | 1 | 0 | 3 | 0 |
| Moraxella sp. | 1 | 1 | 0 | 1 | 0 |
| Neisseria bacilliformis | 1 | 0 | 0 | 0 | 0 |
| Neisseria elongata/bacilliformis | 2 | 0 | 0 | 0 | 0 |
| Neisseria elongata | 1 | 0 | 0 | 1 | 0 |
| Neisseria weaveri | 3 | 2 | 0 | 3 | 3 |
| Oligella urethralis | 5 | 4 | 4 | 5 | 4 |
| Paracoccus sp. | 2 | 0 | 0 | 0 | 0 |
| Paracoccus yeei | 1 | 1 | 1 | 1 | 1 |
| Pasteurella canis | 4 | 1 | 0 | 4 | 4 |
| Pasteurella dagmatis | 1 | 0 | 0 | 0 | 0 |
| Pasteurella multocida | 6 | 5 | 5 | 6 | 6 |
| Providencia rettgeri | 2 | 2 | 2 | 2 | 2 |
| Providencia stuartii | 3 | 3 | 3 | 3 | 3 |
| Pseudomonas aeruginosa/fluorescens/putida | 1 | 1 | 1 | 1 | 1 |
| Pseudomonas fluorescens/putida | 3 | 2 | 2 | 3 | 2 |
| Pseudomonas luteola | 2 | 2 | 2 | 2 | 2 |
| Pseudomonas oryzihabitans | 2 | 1 | 1 | 2 | 1 |
| Pseudomonas sp. | 2 | 2 | 0 | 2 | 0 |
| Psychrobacter phenylpyruvicus | 1 | 0 | 0 | 0 | 0 |
| Psychrobacter sp. | 4 | 0 | 0 | 0 | 0 |
| Ralstonia mannitolilytica | 1 | 1 | 0 | 1 | 0 |
| Rhizobium radiobacter | 3 | 2 | 2 | 3 | 2 |
| Roseomonas mucosa | 1 | 0 | 0 | 1 | 1 |
| Salmonella sp. (not Salmonella typhi) | 2 | 2 | 0 | 2 | 0 |
| Shigella flexneri | 1 | 1 | 1 | 0 | 0 |
| Weeksella virosa | 1 | 1 | 1 | 1 | 1 |
| Total no. of isolates (% correct identification) | 132 | 69 (52) | 45 (34) | 112 (85) | 74 (56) |
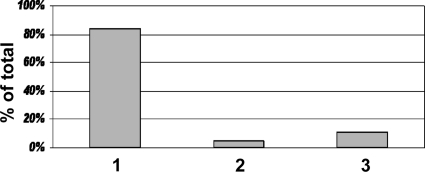
Using the Bruker Biotyper system, we found that 370 (84%) of the isolates were identified with high confidence to the species level following single direct colony analysis (Fig. 3). Four yielded a wrong identification, seven an identification to the genus level only (despite having a score of ≥2.000), and 26 scored ≥2.000 but failed the 10% differential rule. We reanalyzed the 26 isolates that failed the 10% rule by a second direct colony analysis which resolved five isolates to correct species identification. The remaining 21 isolates had no improvement, thus still failing the 10% differential rule. The remaining 70 isolates yielded an initial score of <2.000 and were reanalyzed by the direct colony technique. Repeat direct colony testing yielded a score of ≥2.000 for 21 (5%) isolates (Fig. 3). Of these, 14 were resolved to the species level, 3 were resolved to the genus level (despite having a score of ≥2.000), and 4 had a score of ≥2.000 but failed the 10% differential rule. The remaining 49 (11%) isolates which scored <2.000 after a second direct colony analysis were extracted (Fig. 3). This resulted in 8 identifications to the species level, 19 identifications to the genus level, and 22 nonidentifications (score, <1.700).
Fig. 3.
Percentage of isolates identified by direct colony testing or requiring extraction for high confidence scores by MALDI-TOF MS. Eighty-nine percent of 440 isolates generated a high confidence score (≥2.000) after a single (84%) (bar 1) or secondary (5%) (bar 2) direct colony test. Those which did not generate a score of ≥2.000 (11%) were extracted (bar 3).
DISCUSSION
Rapid and accurate identification of pathogens is essential to ensure ideal patient outcomes. Achieving bacterial identifications within a matter of minutes after culture, as opposed to hours or days, is possible with MALDI-TOF MS. Using the approach described herein with single direct colony testing, 23 isolates can be identified in less than 1 h. In comparison, isolate identification by the BD Phoenix can take several hours. Previous reports assessed MALDI-TOF MS performance in comparison to biochemical, automated, and molecular methodologies (2, 5, 6, 16, 22, 27). However, to date no study has specifically compared the performances of the BD Phoenix automated microbiology system and the Bruker Biotyper MALDI-TOF MS system for identification of Gram-negative bacilli.
We found overall that the Bruker Biotyper exhibited better performance for identifying isolates to the genus and species level (93% and 82%, respectively) than the BD Phoenix (83% and 75%, respectively). This difference can be attributed to the improved performance of the Bruker Biotyper in identifying isolates that are infrequently isolated in the clinical laboratory (Table 1). Even when isolates which would not normally be analyzed on the BD Phoenix (e.g., Campylobacter jejuni or Legionella pneumophila) were excluded from analysis, the Bruker Biotyper still overall outperformed the BD Phoenix (P < 0.0001; data not shown) for this sample set. Statistically equivalent performances between the instruments were observed for common Gram-negative bacilli (Table 1). Because of the open platform design, one could expect that the ability to upload additional spectra to the Bruker Biotyper library database would continue to improve the superior performance of the Bruker Biotyper over that of the BD Phoenix, achieving, in many cases, bacterial identification previously made possible only with 16S rRNA gene sequencing.
We observed that the majority of the Gram-negative isolates, 362 of 440 (82%), were correctly identified with high confidence (score, ≥2.000) using the direct colony technique. Considering all isolates with a score of ≥2.000, whether or not they gave a correct identification or passed the imposed 10% differential criteria, 391 of 440 (89%) yielded scores of ≥2.000. Further extraction of the remaining 49 isolates generated only a species level identification in eight cases, suggesting that extraction does not appreciably improve the overall level of identification by the system analyzed, at least in regards to Gram-negative bacilli. These observations are consistent with other reports that analyzed Gram-negative bacteria (5, 22, 27) and suggest that for these bacteria, extraction is not needed for routine diagnostic use.
For our analysis, we observed instances in which an analyzed isolate generated a score very close to another but represented a different genus or species, making interpretation difficult. Despite numerous studies using MALDI-TOF MS, few have addressed this issue (6, 27). The authors of one report required a score difference of 10% to consider the analysis correct (6). We also used this criterion. Therefore, while the top MALDI-TOF score may have given the correct identification, if a different genus or species score was within 10% of this top score, we considered the result incorrect (in the case of different genera) or identification to genus level only (in the case of different species). In total, 25 of the 440 isolates (6%) failed the 10% differential rule. This was highlighted by 6 of the 10 incorrect MALDI-TOF MS results, in which Klebsiella oxytoca and Raoultella ornithinolytica failed to be differentiated by more then 10%. Resolution of these observations regarding scores close to one another is imperative before implementation in the clinical laboratory.
We considered particular bacteria to belong to complexes; an example is the E. cloacae complex, which includes E. cloacae, Enterobacter hormaechei, Enterobacter kobei, and others. For closely related species within complexes, laboratories should consider reporting to the complex level, and ideally the database software should provide an alert to proceed as such. In some cases, (e.g., isolation of B. cepacia complex organisms from cystic fibrosis patients), species identification within a complex may provide clinically useful information. Whether the Bruker Biotyper can achieve this level of identification requires further study.
We identified instances where library updates were required, either as the result of heterotypic synonyms or absence of spectra within the database, consistent with previous reports (reviewed in reference 3). Specifically regarding the Bruker Biotyper, Stenotrophomonas maltophilia was in some instances reported as Pseudomonas beteli, Pseudomonas geniculata, or Pseudomonas hibiscicola. This may result in end user confusion if clinicians or laboratorians are unfamiliar with these pseudonyms.
The absence of some species' spectra within the library is also a concern (e.g., Shigella species and Vibrio cholerae). The absence of Shigella entries (likely due to their indistinguishable peptidic spectra compared to those of Escherichia coli) resulted in misidentification of an isolate of Shigella flexneri as E. coli in this study. Exclusion of Shigella species by additional tests would be required by the user to differentiate possible Shigella species from E. coli. Finally, the absence of spectral profiles from organisms considered select agents is not only a limitation of the current database but importantly poses a risk to laboratory staff. Rapid identification of these organisms reduces the chance of exposure to these potentially dangerous pathogens. A separate database (Security library 1.0; Bruker Daltonics) is needed to identify such agents. This database is subject to export authorization from Germany and is sold separately from the reference library used for this study.
Culture conditions may affect MALDI-TOF MS analysis (2, 16, 26, 29). We evaluated a limited number of media with one isolate of Klebsiella pneumoniae prior to MALDI-TOF MS. We found that analysis of the isolate grown on eosin methylene blue, MacConkey, or Hektoen enteric agar required extraction to generate a high confidence score (data not shown), an observation previously noted (2). In addition, the same isolate cultured for more than 2 days required extraction to obtain high-confidence species identification (data not shown). Nevertheless, incorrect identification did not appear to be associated with various culture conditions.
In conclusion, the Bruker Biotyper MALDI-TOF MS outperformed the BD Phoenix automated microbiology system for Gram-negative bacteria that are infrequently cultured in the clinical laboratory. However, for common isolates, the systems performed equivalently. With the improved turnaround time and cost-effectiveness of the Bruker Biotyper system, MALDI-TOF MS technology provides an advance in bacterial identification in the clinical microbiology laboratory.
ACKNOWLEDGMENTS
We thank the clinical microbiology laboratory technologists for their help in collecting and analyzing isolates for this study.
Funding for this study was provided by the Mayo Clinic.
Footnotes
Published ahead of print on 5 January 2011.
REFERENCES
- 1. Bessede E., et al. 27 May 2010, posting date Matrix-assisted laser-desorption/ionization BIOTYPER: experience in the routine of a University hospital. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2010.03274 [DOI] [PubMed] [Google Scholar]
- 2. Bizzini A., Durussel C., Bille J., Greub G., Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bizzini A., Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619 [DOI] [PubMed] [Google Scholar]
- 4. Carbonnelle E., et al. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:164–169 [DOI] [PubMed] [Google Scholar]
- 5. Cherkaoui A., et al. 2010. Comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Degand N., et al. 2008. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delmas J., Breysse F., Devulder G., Flandrois J. P., Chomarat M. 2006. Rapid identification of Enterobacteriaceae by sequencing DNA gyrase subunit B encoding gene. Diagn. Microbiol. Infect. Dis. 55:263–268 [DOI] [PubMed] [Google Scholar]
- 8. Donay J. L., et al. 2004. Evaluation of the automated Phoenix system for potential routine use in the clinical microbiology laboratory. J. Clin. Microbiol. 42:1542–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubois D., et al. 2010. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dupont C., et al. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 16:998–1004 [DOI] [PubMed] [Google Scholar]
- 11. Hall L., Doerr K. A., Wohlfiel S. L., Roberts G. D. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holland R. D., et al. 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:1227–1232 [DOI] [PubMed] [Google Scholar]
- 13. Krishnamurthy T., Ross P. L. 1996. Rapid identification of bacteria by direct matrix-assisted laser desorption/ionization mass spectrometric analysis of whole cells. Rapid Commun. Mass Spectrom. 10:1992–1996 [DOI] [PubMed] [Google Scholar]
- 14. Liu Z. K., Ling T. K., Cheng A. F. 2005. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of common clinical isolates. Med. Princ. Pract. 14:250–254 [DOI] [PubMed] [Google Scholar]
- 15. Marklein G., et al. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mellmann A., et al. 2008. Evaluation of matrix-assisted laser desorption ionization–time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. (ed.). 2007. Manual of clinical microbiology, 9th ed., vol. 1 ASM Press, Washington, DC [Google Scholar]
- 18. Pennanec X., Dufour A., Haras D., Rehel K. 2010. A quick and easy method to identify bacteria by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 24:384–392 [DOI] [PubMed] [Google Scholar]
- 19. Ryzhov V., Fenselau C. 2001. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73:746–750 [DOI] [PubMed] [Google Scholar]
- 20. Sauer S., Kliem M. 2010. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 8:74–82 [DOI] [PubMed] [Google Scholar]
- 21. Seibold E., Maier T., Kostrzewa M., Zeman E., Splettstoesser W. 2010. Identification of Francisella tularensis by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry: fast, reliable, robust, and cost-effective differentiation on species and subspecies levels. J. Clin. Microbiol. 48:1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 23. Stephan R., Ziegler D., Pfluger V., Vogel G., Lehner A. 2010. Rapid genus and species specific identification of Cronobacter spp. by MALDI-TOF mass spectrometry. J. Clin. Microbiol. 48:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stevenson L. G., Drake S. K., Shea Y. R., Zelazny A. M., Murray P. R. 2010. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szabados F., Woloszyn J., Richter C., Kaase M., Gatermann S. 2010. Identification of molecularly defined Staphylococcus aureus strains using matrix-assisted laser desorption/ionization time of flight mass spectrometry and the Biotyper 2.0 database. J. Med. Microbiol. 59:787–790 [DOI] [PubMed] [Google Scholar]
- 26. Valentine N., Wunschel S., Wunschel D., Petersen C., Wahl K. 2005. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 71:58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Veen S. Q., Claas E. C., Kuijper E. J. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vargha M., Takats Z., Konopka A., Nakatsu C. H. 2006. Optimization of MALDI-TOF MS for strain level differentiation of Arthrobacter isolates. J. Microbiol. Methods 66:399–409 [DOI] [PubMed] [Google Scholar]
- 29. Walker J., Fox A. J., Edwards-Jones V., Gordon D. B. 2002. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J. Microbiol. Methods 48:117–126 [DOI] [PubMed] [Google Scholar]