Abstract
Antibodies to the hepatitis delta virus (HDV) were found in 17.6% of 233 hepatitis B virus surface antigen-positive subjects in Cameroon. Phylogenetic analyses showed the presence of HDV-1, HDV-5, HDV-6, and HDV-7 genotypes. These results enrich the limited data on HDV prevalence and molecular diversity in Cameroon.
Hepatitis delta virus (HDV) is associated with hepatitis B virus (HBV) infection and is frequently related to more severe disease than that due to the underlying HBV monoinfection (5, 10). HDV infection has a worldwide distribution, but its frequency varies greatly throughout different geographic regions. It is highly endemic in the Middle East, in the Mediterranean area, in the Amazonian region, and in several African countries (3). Genomic analysis of HDV isolates from different regions of the world reported at least eight phylogenetically distinct genotypes with dissimilar geographic distributions. Apart from HDV genotype 1 (HDV-1), which is ubiquitous, each virus clade is geographically localized: HDV-2 is found in Japan, Taiwan, and Russia; HDV-4 is found in Taiwan and Japan; HDV-3 is found in the Amazonian region; and HDV-5, HDV-6, HDV-7, and HDV-8 are found in Africa (6). The role of these HDV genotypes is not yet well determined, but some studies have suggested an association between the severity of disease and infection with different HDV genotypes (4, 8). The two studies conducted 2 decades ago on HDV infection in Cameroon have reported the prevalence of antibodies against HDV (HDV-Ab) of 6.5% and 27.3%, respectively (9, 12). In spite of this high prevalence, no data on HDV genotype diversity in Cameroon are available. Therefore, in the present study, we investigated the current HDV seroprevalence and the genotype diversity in Cameroonian patients with chronic HBV infection.
This study was performed on plasma samples from 233 hepatitis B virus surface antigen (HBsAg) carriers (mean age, 34.5 years; 79 women and 154 men) seen at two medical care centers in Yaounde, Cameroon, from May 2008 to May 2009. As recommended by the Cameroonian Society of Gastroenterology, in addition to the HBV DNA quantification and the assessment of biochemical liver enzyme, HDV screening should be part of the screening of HBsAg carriers, considering the endemicity of this infection in Central Africa. The analyses included in HDV screening are HDV antibody (HDV-Ab) and HDV RNA quantification for HDV-Ab-positive patients. During the medical consultation, HBsAg carriers were screened for HDV infection and were asked to participate in this study. For carriers who agreed to participate, written informed consent was obtained for HDV genotyping, since this analysis is not part of the original protocol in a routine workup of liver disease.
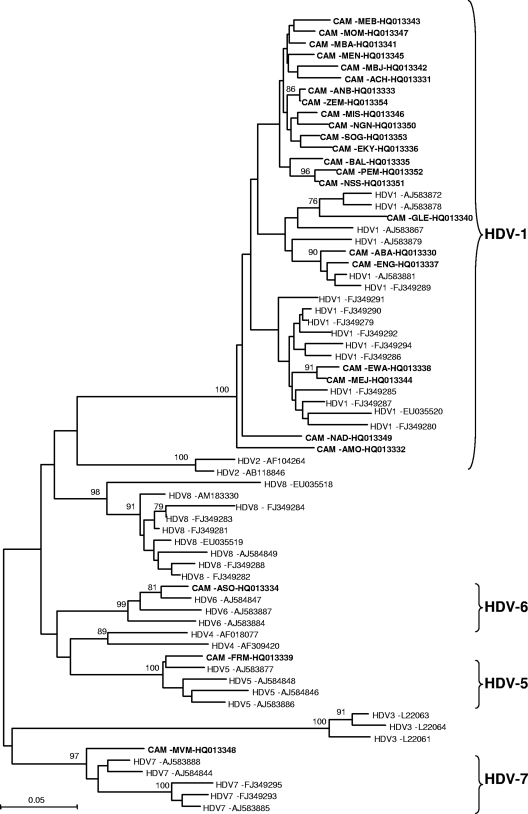
The presence of HDV-Ab was determined by the Murex anti-delta assay (Abbott, Wiesbaden, Germany), and HBV DNA viral load was determined by the Abbott RealTime HBV quantification kit (Abbott Molecular Inc., Rungis, France) according to the manufacturer's protocol. During routine workup of the liver disease, the biochemical liver enzyme alanine aminotransferase (ALT) (upper normal limit, 45 IU/liter) was also assessed by standard laboratory tests. In order to detect HDV, RNA was extracted from 140 μl HDV-Ab-positive plasma using the QIAamp RNA minikit (Qiagen, Courtaboeuf, France) and HDV RNA was detected by reverse transcription (RT)-PCR amplification of a 360-bp fragment coding for the small hepatitis delta antigen (HDV-sHD) gene as described previously (4). HDV-Ab was found in 41 (17.6%; 95% confidence interval [CI], 13.1 to 23.2%) of the 233 tested plasma specimens, and then HDV RNA was detected in 25 (61%; 95% CI: 44.5 to 75.4%) of the 41 HDV-Ab-positive plasma specimens. The percentage of patients with abnormal serum ALT levels was significantly higher in HBsAg carriers with HDV-Ab than in those without HDV-Ab. Remarkably; the proportion of HBsAg carriers with undetectable HBV DNA was higher in HDV-Ab-positive than in HDV-Ab-negative patients (Table 1). The phylogenetic analyses, performed as described elsewhere (11), revealed that 22 (88%) of the 25 HDV Cameroonian strains were classified as genotype 1 (HDV-1), with a 100% bootstrap value. The other three strains clustered with the previously described HDV-5, HDV-6, and HDV-7 genotypes, with bootstrap values of 100%, 99%, and 97%, respectively (Fig. 1). Although they were not supported by significant bootstrap values (less than 70%), the phylogenetic analyses indicated splitting of the Cameroonian HDV-1 genotypes into five probable “subgenotypes” (Fig. 1). The average of nucleotide differences between the 25 Cameroonian HDV genotypes was 15.5%, whereas the average of nucleotide differences within the 22 Cameroonian HDV-1 strains was 12.9% (data not shown). The average of nucleotide differences among the Cameroonian HDV-1 strains seems to be higher than the 11% average reported among other worldwide HDV-1 strains (6). This result highlights the extent of intragenotypic variation for the Cameroonian HDV-1 isolates.
Table 1.
Demographic and laboratory characteristics of the 233 HBV carriers according to HDV antibody (HDV-Ab) status
| Parameter | Specimen |
Pa | |
|---|---|---|---|
| HDV-Ab positive (n = 41) | HDV-Ab negative (n = 192) | ||
| Age (mean, yr) | 35.7 | 35.4 | >0.05 |
| Gender (% male) | 65.9 | 66.1 | >0.05 |
| Serum ALT level (median, IU/liter) | 96.75 | 58.5 | 0.0004 |
| HBV DNA level (% undetectable) | 61 | 35.4 | 0.002 |
Statistical differences were evaluated using the chi-square test or Fisher's exact test to compare qualitative data. The Mann–Whitney U test was used to compare quantitative variables. Differences were considered statistically significant for P values less than 0.05.
Fig. 1.
Phylogenetic analysis of a 360-bp fragment of the HDV-sHD gene from different HDV strains using the neighbor-joining (NJ) method. Twenty-five HDV sequences from Cameroonian patients (highlighted in boldface; name of the isolate and GenBank accession number are indicated) were aligned with 41 previously published HDV sequences of clades 1 to 8 (HDV-1 to HDV-8) available from GenBank as reference genes (Genotype and GenBank accession number are indicated) by using the CLUSTAL X v1.81 software program and imported into MEGA v.4.0 software for phylogenetic analysis. Numbers next to the nodes of the tree represent bootstrap values (1,000 replicates). Only bootstrap values above 70% are presented. Genetic distances were calculated using the Kimura two-parameter method.
The prevalence of HDV-Ab obtained in this study is as high as that previously reported by other groups from Cameroon (9, 12). HDV RNA was found in 61% of the HDV-Ab patients, indicating the low prevalence of HDV clearance in this population. The natural history of HDV infection shows that coinfection evolves to chronicity in only a small number of patients and patients recover from both hepatitis B and hepatitis D, while superinfection of HDV leads to progressive disease and cirrhosis in approximately 60 to 80% of cases (1, 2). It can thus be suggested that most of our patients acquired the infection as a superinfection on hepatitis B. It is known that coinfection or superinfection of HBV and HDV may cause severe liver diseases (1). Our observations showed that the median serum ALT level was significantly higher in carriers with HDV-Ab than in those without HDV-Ab (Table 1). In our study, the proportion of HBsAg-positive patients with undetectable HBV DNA was higher in patients with HDV-Ab than in those without HDV-Ab (61% versus 35.4%; P < 0.05). This result indicates, as reported by previous studies (2), the suppression/inhibition of HBV replication by HDV infection and highlights the importance of screening for HDV infection in HBV carriers. We showed the presence of four different genotypes: HDV-1, HDV-5, HDV-6, and HDV-7. HDV-1 was found to be the predominant genotype. Three new strains from Cameroon were closely related to HDV-5, HDV-6, and HDV-7 genotypes previously described from African patients (6), indicating that these genotypes are in circulation in Africa. However, although recently reported to originate and to be endemic in Central Africa (7), the newly described HDV-8 was not found in our study. As claimed by other authors (6), the great genetic diversity of HDV found in Cameroon, with at least four different genotypes, suggests a wide and ancient spread of African HDVs. In contrast to previous studies (13), our results showed no significantly well-defined subgenotypes within genotype 1. Further studies, with full genome characterization of these strains, are required.
In conclusion, our results point to a high endemicity of HDV in Cameroonian populations, which is in sharp accordance with data reported from Cameroon 20 years ago and with the situation in neighboring Central African countries. Therefore, practitioners and health care managers should be made aware of the risk of dual infection with HBV and HDV, especially when the HBV viral load is low or undetectable with abnormal serum ALT levels. HDV infection in Cameroon is characterized by a wide genetic diversity, with the circulation of four different genotypes (1, 5, 6, and 7), HDV-1 being predominant.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the new HDV strains isolated from the population in this study are HQ013330 to HQ013354.
Acknowledgments
We are grateful to the HBsAg carriers who agreed to participate in this study. We are also grateful to Jean-Marc Reynes for critical reading of the manuscript.
This work was supported by funds from the Centre Pasteur of Cameroon.
Footnotes
Published ahead of print on 5 January 2011.
REFERENCES
- 1. Govindarajan S., De Cock K., Redeker A. 1986. Natural course of delta superinfection in chronic hepatitis B virus-infected patients: histopathologic study with multiple liver biopsies. Hepatol. Res. 6:640–644 [DOI] [PubMed] [Google Scholar]
- 2. Heidrich B., et al. 2009. Virological and clinical characteristics of delta hepatitis in Central Europe. J. Viral Hepat. 16:883–894 [DOI] [PubMed] [Google Scholar]
- 3. Husa P., Linhartová A., Nemecek V., Husová L. 2005. Hepatitis D. Acta Virol. 49:219–225 [PubMed] [Google Scholar]
- 4. Ivaniushina V., et al. 2001. Hepatitis delta virus genotypes I and II cocirculate in an endemic area of Yakutia, Russia. J. Gen. Virol. 82:2709–2718 [DOI] [PubMed] [Google Scholar]
- 5. Karayiannis P. 1998. Hepatitis D virus. Rev. Med. Virol. 8:13–24 [DOI] [PubMed] [Google Scholar]
- 6. Le Gal F., et al. 2006. Eighth major clade for hepatitis delta virus. Emerg. Infect. Dis. 12:1447–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Makuwa M., et al. 2008. Prevalence and genetic diversity of hepatitis B and delta viruses in pregnant women in Gabon: molecular evidence that hepatitis delta virus clade 8 originates from and is endemic in central Africa. J. Clin. Microbiol. 46:754–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakano T., et al. 2001. Characterization of hepatitis D virus genotype III among Yucpa Indians in Venezuela. J. Gen. Virol. 82:2183–2189 [DOI] [PubMed] [Google Scholar]
- 9. Ndumbe P. 1991. Hepatitis D in Yaounde, Cameroon. APMIS 99:196–198 [DOI] [PubMed] [Google Scholar]
- 10. Niro G., et al. 1997. The predominance of hepatitis delta virus genotype I among chronically infected Italian patients. Hepatol. Res. 25:728–734 [DOI] [PubMed] [Google Scholar]
- 11. Njouom R., et al. 2009. Predominance of hepatitis C virus genotype 4 infection and rapid transmission between 1935 and 1965 in the Central African Republic. J. Gen. Virol. 90:2452–2456 [DOI] [PubMed] [Google Scholar]
- 12. Poveda J., et al. 1986. Carriage of HBs antigen and infection by delta agent in Cameroon. Bull. Soc Pathol. Exot. Filiales 79:785–787 [In French.] [PubMed] [Google Scholar]
- 13. Zhang Y., Tsega E., Hansson B. 1996. Phylogenetic analysis of hepatitis D viruses indicating a new genotype I subgroup among African isolates. J. Clin. Microbiol. 34:3023–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]