Abstract
Real-time PCR has the potential to streamline detection and identification of Cryptosporidium spp. in human clinical samples. In the present article, we report the first such assay to allow not only detection and differentiation of the most common human pathogens, Cryptosporidium hominis and Cryptosporidium parvum, but also simultaneous amplification of a region of the small subunit (SSU) rRNA gene, permitting direct sequence analysis to identify any Cryptosporidium species. An internal control is incorporated to identify the presence of PCR inhibitors. Analytical sensitivity was determined to be as low as 200 oocysts per gram of feces processed, equivalent to 2 oocysts per PCR. The C. hominis and C. parvum PCRs specifically detected only species/genotypes in their respective target clades. Diagnostic sensitivity and specificity, evaluated against a widely used conventional nested SSU rRNA gene PCR as a nominated gold standard using a panel of 258 (151 positive and 107 negative) samples, were 100% and 99.1%, respectively. The assay agreed with PCR-restriction fragment length polymorphism analysis of the Cryptosporidium oocyst wall protein gene for 134 of 136 (98.5%) samples tested prospectively and typed two additional isolates. The real-time PCR assay was sensitive, specific, and reproducible and significantly improved laboratory work flow and turnaround times.
INTRODUCTION
Cryptosporidium is a genus of parasitic protozoa causing diarrheal illness in humans and animals. Currently more than 20 species of Cryptosporidium have been identified, infecting a wide range of hosts. In humans, the majority of cryptosporidiosis cases in most countries are caused by C. hominis or C. parvum (39). However, a number of other species and genotypes have been detected in human stools, especially in developing countries, with Cryptosporidium meleagridis, Cryptosporidium felis, Cryptosporidium canis, and Cryptosporidium ubiquitum being the most commonly identified, but others have been reported, including Cryptosporidium muris, Cryptosporidium andersoni, Cryptosporidium suis, Cryptosporidium cuniculus, Cryptosporidium chipmunk genotype I, and horse, skunk, and C. hominis monkey genotypes (40).
Routine clinical diagnosis is by microscopy, immunoassay, or PCR to ascertain the presence/absence of the genus. For the identification of a Cryptosporidium species or genotype, required for epidemiological and outbreak investigations, PCR followed by restriction fragment length polymorphism (RFLP) analysis or sequencing has been widely employed. These assays have targeted different regions of the genome, including the small subunit (SSU) rRNA, Cryptosporidium oocyst wall protein (COWP), thrombospondin-related adhesive proteins, 70-kDa heat shock protein (HSP70), and actin genes (14, 19, 29, 32, 33, 35). In our laboratory, COWP PCR-RFLP, supported by SSU rRNA gene analysis, has been used routinely for typing an annual average of 2,000 isolates for epidemiological purposes in the United Kingdom since January 2000 (7–9). This work has shown that 96% of cases here are caused by C. hominis or C. parvum but that other species/genotypes may be involved in human infection and disease.
To improve work flow and efficiency for large-scale molecular surveillance, a real-time PCR which would identify the main human pathogenic species and also detect and allow timely identification of other Cryptosporidium spp. was sought. The lack of a requirement for downstream processing, such as restriction digests and agarose gel electrophoresis, was a perceived benefit, improving turnaround times, particularly for outbreak investigations. Real-time PCRs for detection of Cryptosporidium spp. and genotypes in human clinical samples have been described (2, 17, 18, 20, 22, 36, 38). However, none identify C. hominis and C. parvum and detect other Cryptosporidium spp. by amplifying a region which can be directly sequenced to identify species/genotype. We describe the development, evaluation, and clinical validation of a real-time PCR targeting C. hominis and C. parvum alleles of a locus of unknown function, LIB13 (36), while simultaneously detecting all known Cryptosporidium species and genotypes by amplification of a region of the SSU rRNA gene allowing identification by direct sequence analysis (26). The assay also incorporates an exogenous internal control (IC) for the identification of inhibited reactions. The accuracy of the assay is assessed against nested, conventional PCR analysis of the SSU rRNA gene and compared with that of COWP PCR-RFLP.
MATERIALS AND METHODS
Cryptosporidium sources, DNA preparation, and species identification.
Unless indicated otherwise, Cryptosporidium genomic DNA originated with and was extracted from oocysts isolated from microscopy-positive diarrheic fecal samples referred by local diagnostic laboratories for identification to the species level using COWP and SSU rRNA PCR-RFLP and sequencing as described previously (8).
Design and development of real-time PCR for detection and characterization of Cryptosporidium DNA.
Real-time PCR was carried out in two duplex reactions: (i) a genus-specific PCR amplifying ∼300 bp of the Cryptosporidium SSU rRNA gene, duplexed with a C. parvum-specific PCR amplifying 166 bp of the LIB13 locus, and (ii) a C. hominis-specific PCR amplifying 169 bp of the LIB13 locus, duplexed with a commercial noncompetitive (primer-limited) IC PCR (PrimerDesign, Southampton, United Kingdom). PCR primers and probes (Table 1) were designed using the BioEdit software program, version 7.0.9.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) to create Clustal W (37) alignments of representative Cryptosporidium sequences from GenBank (National Center for Biotechnology Information [NCBI]; http://www.ncbi.nlm.nih.gov/GenBank/). The Primer Express software program (Applied Biosystems, Warrington, United Kingdom) was used to calculate melting temperatures and check for undesirable inter- and intramolecular binding. Primer and probe sequences were then checked for cross-reactions with nontarget sequences on the GenBank database using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Table 1.
Real-time PCR primers and probes used in this study
| Target locus | Primer/probe | Position (nt)a | Sequence (5′–3′)b |
|---|---|---|---|
| Cryptosporidium species SSU rRNA gene | CRU18SFc | 440–468 | GAGGTAGTGACAAGAAATAACAATACAGG |
| CRU18SRc | 710–738 | CTGCTTTAAGCACTCTAATTTTCTCAAAG | |
| CRU18STM | 587–609 | FAM-TACGAGCTTTTTAACTGCAACAA MGB-NFQ | |
| C. parvum LIB13 | CRULib13F | 228–253 | TCCTTGAAATGAATATTTGTGACTCG |
| CRULib13RCp | 370–393 | TTAATGTGGTAGTTGCGGTTGAAC | |
| CRULib13TMCp | 339–358 | VIC-TATCTCTTCGTAGCGGCGTA MGB-NFQ | |
| C. hominis LIB13 | CRULib13F | 228–253 | As above |
| CRULib13RCh | 374–396 | AAATGTGGTAGTTGCGGTTGAAA | |
| CRULib13TMCh | 340–357 | VIC-CTTACTTCGTGGCGGCGT MGB-NFQ |
Each 25-μl reaction mixture contained 12.5 μl of TaqMan environmental master mix 2.0 (Applied Biosystems). All primers (Integrated DNA Technologies, Glasgow, United Kingdom) were included at 900 nM except CRULib13RCh, which was at 300 nM. The minor groove binding (MGB) TaqMan probes (Applied Biosystems) CRU18STM (6-carboxyfluorescein [FAM] labeled) and CRULIB13Cp and CRULIB13Ch (both VIC labeled) were at 100 nM, 150 nM, and 100 nM, respectively. The C. hominis LIB13-IC tube contained 1 μl of primer/probe (FAM labeled) mix and 5 μl of a 1:20 dilution of the IC DNA (PrimerDesign). To each tube, 2 μl of DNA was added. C. hominis, C. parvum, and no-template PCR controls were included in each run.
Reactions were prepared using a CAS1200 automated pipetting instrument (Corbett Research, Cambridge, United Kingdom) and run on a Rotorgene 6000 real-time PCR instrument (Corbett Research) using the 72-well Rotor-Disc format (Qiagen, Crawley, United Kingdom). Thermocycling conditions were as follows: 95°C for 10 min, followed by 55 cycles of 95°C for 15 s and 60°C for 60 s. Data were collected from the green (FAM), yellow (VIC), and orange (ROX normalization dye) channels during each 60°C annealing/extension phase, and postrun analysis was performed using the Rotorgene 6000 software program, version 1.7 (Corbett Research). Threshold cycles (CTs) for each reaction were determined by the cycle at which the fluorescence plot crossed a standardized threshold of 0.02 and 0.05 normalized fluorescence units for the green and yellow channels, respectively. IC reactions were considered inhibited if the fluorescence failed to cross the threshold. Initial evaluation (data not shown) showed the assay was capable of detecting the cloned SSU rRNA gene, C. hominis LIB13 locus, and C. parvum LIB13 locus diluted down to 10, 8, and 4 copies per PCR, respectively.
DNA sequence analysis of real-time PCR products.
To assess the suitability of the real-time PCR products for sequencing, these were purified using the QIAquick cleanup kit (Qiagen). Bidirectional sequencing was carried out by Source BioScience (Cambridge, United Kingdom) using the BigDye Terminator cycle sequencing kit on an ABI3730 automated sequencer (Applied Biosystems). Consensus sequences were created (ChromasPro 1.4a; Technelysium Ltd., Tewantin, Australia), and compared with published sequences in the GenBank database using BLAST.
Analytical sensitivity and specificity of real-time PCR.
For estimation of C. hominis analytical sensitivity, Cryptosporidium-negative stools were seeded with 1,000 and 200 oocysts per gram (opg) (5 replicates each). The concentration of oocysts in each seeding suspension was obtained from 10 counts of immunofluorescence-labeled (CryptoCel; Cellabs, Brookvale, Australia) oocysts fixed on microscope slides according to the manufacturer's instructions. Genomic DNA was extracted from 0.5 g of seeded stool, as described previously (8).
For assessment of C. parvum analytical sensitivity, genomic DNA was from flow cytometer-counted oocysts in nuclease-free water seeded at a concentration of 2 oocysts/μl, extracted by a freeze-thaw method (13).
Analytical specificity was assessed by testing DNA from 14 different Cryptosporidium spp./genotypes, a range of C. hominis and C. parvum isolates, and non-Cryptosporidium spp.: Eimeria tenella, Toxoplasma gondii, Cyclospora cayetanensis, and Homo sapiens (NEQAS 0703; National External Quality Assessment Service, Watford, United Kingdom). To identify variation in C. hominis and C. parvum and ensure this was represented in the panel, subtype analysis of isolates at the 60-kDa glycoprotein (GP60) gene was performed as described previously (1, 5, 34). As many other isolates as possible from each Cryptosporidium species examined were included, but many are rare in the United Kingdom.
Diagnostic sensitivity and specificity of real-time PCR.
To compare the sensitivity and specificity of the real-time PCR and COWP PCR-RFLP, a blind-coded panel of 258 DNA extracts (151 Cryptosporidium positive and 107 negative), derived from anonymous routine diagnostic stools from the local microbiology laboratory and the UK Cryptosporidium Reference Unit (UKCRU), was tested, and the results were compared with a nominated gold standard, nested SSU rRNA PCR (19). Ninety-five percent confidence intervals were calculated using the Confidence Interval Analysis software program (2000; http://www.som.soton.ac.uk/cia/download.asp). At least 250 samples were needed to ensure that differences in sensitivities and specificities in excess of 10% had an 80% chance of being detected at a 5% significance level. The positive samples were 80 for C. hominis, 67 for C. parvum, 2 for C. felis, 1 for C. ubiquitum, and 1 for C. meleagridis.
Validation of real-time PCR by prospective testing of clinical samples.
To validate performance in clinical practice, the real-time assay was prospectively compared with the routinely used COWP PCR-RFLP assay for typing of 136 Cryptosporidium-positive stools received at the UKCRU during January through March 2010. To evaluate the repeatability of the real-time PCR typing results, 29 of the Cryptosporidium-positive DNA extracts were tested three times each in separate runs by the same operator using the same equipment. To evaluate reproducibility of the real-time PCR, 62 of the samples were tested by three different operators, all experienced at performing PCR.
Nucleotide sequence accession numbers.
DNA sequences were deposited in the GenBank sequence database under accession numbers HQ149020 to HQ149041.
RESULTS
Analytical sensitivity and specificity.
Analytical sensitivity for detection of C. hominis genomic DNA from feces was very close to 200 opg feces, which equates to 2 oocysts per PCR (Table 2). A lower limit of detection was found for genomic DNA extracted from flow cytometer-counted C. parvum oocysts in aqueous suspension, with all replicates of 1 oocyst per PCR being detected (Table 2).
Table 2.
Detection of low copy numbers of genomic Cryptosporidium DNA by real-time PCR
| Real-time PCR and target | No. (%) of samples positivea |
||||
|---|---|---|---|---|---|
| 1,000 opg feces (10 opPCR) | 800 opg feces (8 opPCR)* | 400 opg feces (4 opPCR)* | 200 opg feces (2 opPCR) | 100 opg feces (1 opPCR)† | |
| C. hominis | |||||
| LIB13 | 25/25 (100) | 10/10 (100) | 9/10 (90) | 16/25 (64) | NT |
| SSU rRNA | 25/25 (100) | 10/10 (100) | 9/10 (90) | 24/25 (96) | NT |
| C. parvum | |||||
| LIB13 | NT | NT | 10/10 (100) | 9/10 (90) | 10/10 (100) |
| SSU rRNA | NT | NT | 10/10 (100) | 8/10 (80) | 8/10 (80) |
DNA dilutions made from 1,000 and 200 oocyst per gram samples in nuclease-free water are shown by “*” and “†,” respectively. NT, not tested; opPCR, oocytes per PCR.
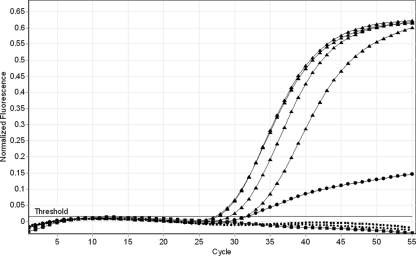
Evaluation of analytical specificity revealed no cross-reactions with other genera, and all Cryptosporidium isolates tested were detected by the SSU rRNA real-time PCR (Table 3). The C. hominis LIB13 real-time PCR specifically amplified C. hominis and the very closely related C. cuniculus and C. hominis monkey genotype. The C. parvum LIB13 PCR specifically amplified C. parvum; the Cryptosporidium horse genotype was also amplified, but it gave a differentiating amplification curve (Fig. 1) due to mismatches between the probe and horse genotype genomic DNA. The characteristic flattened curve shape was observed for 4/4 confirmed Cryptosporidium horse genotype samples tested. Proof of principle for sequencing the SSU rRNA real-time PCR product for identification of Cryptosporidium spp. was provided in vitro by sequence analysis of C. meleagridis, C. felis, C. cuniculus, Cryptosporidium horse genotype, and C. hominis monkey genotype. All were identified correctly, showing 100% identity with the expected GenBank entry. Financial constraints prevented in vitro analysis of more isolates, but in silico analysis revealed no unexpected identical sequences over the amplified region among GenBank entries representing 24 Cryptosporidium spp. and 37 genotypes (data not shown). The closely related ruminant species, Cryptosporidium xiaoi and Cryptosporidium bovis, are generally indistinguishable over this region, having as few as 1 bp of difference over the gene (19). This confirmed the suitability of the region for the purpose of detection and differentiation of Cryptosporidium spp. from human clinical samples by sequencing.
Table 3.
Detection and amplification of a panel of Cryptosporidium species, genotypes, and subtypes and non-Cryptosporidium DNA by real-time PCR
| Description (no. of samples) | Host | Real-time PCR result |
||
|---|---|---|---|---|
| C. hominis LIB13 | C. parvum LIB13 | SSU rRNA | ||
| C. hominis clade | ||||
| C. hominis GP60 subtype | ||||
| IaA15R3 (1) | Human | + | − | + |
| IbA9G3 (1) | Human | + | − | + |
| IbA10G2 (1) | Human | + | − | + |
| IdA16 (1) | Human | + | − | + |
| IeA11G3T3 (1) | Human | + | − | + |
| IfA12G1 (1) | Human | + | − | + |
| C. hominis monkey genotype (1) | Human | + | − | + |
| C. cuniculus (5) | Human | + | − | + |
| C. parvum GP60 subtype | ||||
| IIaA15G2R1 (1) | Human | − | + | + |
| IIaA17G1 (1) | Human | − | + | + |
| IIaA19G2R1 (1) | Human | − | + | + |
| IIbA15G2R1 (1) | Human | − | + | + |
| IIcA5G3 (1) | Human | − | + | + |
| IIdA22G1 (1) | Human | − | + | + |
| Cryptosporidium horse genotype (4) | Human | − | +a | + |
| C. andersoni (2) | Cow | − | − | + |
| C. muris (1) | Mouse | − | − | + |
| C. meleagridis (6) | Human | − | − | + |
| C. felis (4) | Human | − | − | + |
| C. ubiquitum (5) | Human | − | − | + |
| C. bovis (1) | Cow | − | − | + |
| C. baileyi (1) | Chicken | − | − | + |
| C. canis (1) | Human | − | − | + |
| C. xiaoi (3) | Sheep/goat | − | − | + |
| Eimeria tenella (1) | Chicken | − | − | − |
| Cyclospora cayetanensis (1) | Human | − | − | − |
| Toxoplasma gondii (1) | Human | − | − | − |
| Homo sapiens (1) | − | − | − | |
Differentiating amplification curve.
Fig. 1.
Amplification plots from the C. parvum LIB13 real-time PCR showing the characteristic curve produced by the Cryptosporidium horse genotype (circles) compared with the typical C. parvum curves (triangles). Also shown: C. hominis (dashed lines) and no-template control (squares).
Diagnostic specificity and sensitivity.
There was no significant difference between the diagnostic sensitivity and specificity for the detection of Cryptosporidium by the real-time PCR (100% and 99.1%, respectively) and COWP PCR-RFLP (98.0% and 100%, respectively) compared with the nominated gold standard (Table 4). The real-time PCR resulted in one false-positive (C. parvum) result; this was attributed to an isolated cross-contamination event, since this sample was retested twice and was found to be negative. Of three COWP PCR-RFLP false negatives, all were detected by the real-time PCR, one identified as C. hominis and two as Cryptosporidium sp., C. felis on sequencing. All other typing results were in agreement with the gold standard.
Table 4.
Diagnostic sensitivities and specificities of real-time PCR and COWP PCR-RFLP for 259 samples compared with a nominated gold standard
| Assay | % sensitivity (95% confidence interval) | % specificity (95% confidence interval) | No. of false negatives | No. of false positives |
|---|---|---|---|---|
| Real-time PCR | 100 (97.5 to 100) | 99.1 (94.9 to 99.8) | 0 | 1 |
| COWP PCR-RFLP | 98.0 (94.3 to 99.3) | 100 (96.5 to 100) | 3 | 0 |
IC PCR CT values for patient samples ranged from 31.8 to 47.9 cycles (mean, 37.2 cycles; standard deviation, 3.2 cycles), apart from one outlier at 52.6 cycles. This range of CTs was probably influenced by a combination of unquantified factors, including the presence of inhibitors and relative amounts of C. hominis and IC DNA in the tube. IC PCRs were negative for only two samples, one of which was a gold standard-negative sample also negative by the real-time assay and the other of which was a gold standard Cryptosporidium-positive sample in which real-time PCR detected C. hominis with a below-average LIB13 CT value, suggesting that the IC reaction was negative due to it being outcompeted by the C. hominis PCR rather than the action of PCR inhibitors. These results suggest efficient removal of inhibitors during DNA preparation or a high level of tolerance to inhibitors in the reactions. Fluorescence levels detected in the orange channel emitted from the ROX dye integral to the polymerase mix were constant.
Validation by prospective testing of clinical samples.
Prospective comparison of the real-time and COWP PCR-RFLP assays for typing of 136 Cryptosporidium-positive stools submitted for typing showed the two assays were in agreement for a total of 134 (98.5%) samples (Table 5). Overall, the real-time PCR was more sensitive than the COWP PCR-RFLP, detecting Cryptosporidium spp. in two more samples. Of 129 samples having C. hominis or C. parvum, the assays were in agreement for 128 (99.2%); 1 C. hominis sample was negative by COWP PCR-RFLP. Other Cryptosporidium spp. were detected in five and four samples by real-time PCR and COWP PCR-RFLP, respectively, with the species identified agreeing for four samples and one C. felis sample negative by COWP PCR-RFLP. Assay times for processing a batch of 30 samples by each assay (from receipt of DNA extract to identification of C. hominis or C. parvum) were approximately 6 h and 3 h, and hands-on times were approximately 100 min and 40 min for COWP-PCR-RFLP and real-time PCR, respectively.
Table 5.
Prospective analysis of 136 human clinical stool samples using real-time PCR and COWP PCR-RFLP
| Organism | No. of samples identified by: |
|
|---|---|---|
| Real-time PCR | COWP PCR-RFLP | |
| C. hominis | 81 | 80 |
| C. parvum | 48 | 48 |
| Other Cryptosporidium spp. | ||
| C. meleagridis | 2 | 2 |
| C. ubiquitum | 1 | 1 |
| Novela | 1 | 1 |
| C. felis | 1 | 0 |
| Total no. positive | 134 | 132 |
| No. negative | 2 | 4 |
Identity with C. cuniculus at SSU rRNA gene, 99.0%.
Repeatability and reproducibility.
Repeatability was demonstrated by a single operator achieving the same results for 28 of 29 (97%) samples when they were tested three times on separate runs. One sample was identified as mixed C. hominis and C. parvum in one run but as C. parvum only in two runs, which was the result obtained by COWP PCR-RFLP, possibly due to sampling variation from the single DNA extraction. The reproducibility of the real-time PCR was 100% for 62 samples when tested by three different operators.
DISCUSSION
Characterization of Cryptosporidium spp. by molecular methods has proven invaluable for the understanding of the epidemiology of cryptosporidiosis and for investigation of outbreaks (4, 9, 11, 12, 25, 28). Real-time PCR offers the potential to streamline the process through simultaneous amplification and amplicon detection, reducing assay times and contamination risk.
An assay should ideally be able to detect all Cryptosporidium spp. or at least detect and differentiate all species that infect humans (41). Although real-time-PCR-based assays have been described previously, there remained a need for a well-characterized assay for identification of the main human infective species, C. hominis and C. parvum, which also allows detection and timely sequence identification of other Cryptosporidium spp. The real-time PCR described here targets the SSU rRNA gene to detect all members of the Cryptosporidium genus. The region amplified was chosen because it covers the major region of interspecies/genotype variability in the gene, allowing identification of nearly all Cryptosporidium spp. and genotypes by sequence analysis of the PCR product while being relatively short to allow efficient amplification and real-time detection. The region has previously been used successfully to identify Cryptosporidium spp. by conventional PCR and sequencing (26). C. xiaoi and C. bovis are the only species that could not be differentiated from each other by sequencing the ∼300 bp of the SSU rRNA gene amplified in this assay and have as little as a 1-bp difference over the ∼830-bp nested PCR amplicon (19); differences are more marked at the HSP70 and actin genes (15). C. xiaoi has never been detected in humans and C. bovis only once, in a dairy farm worker in India (21), so this is unlikely to be problematic for human clinical samples.
Since 1 April 2010, the assay has been used for routine typing of confirmed Cryptosporidium-positive isolates referred to our laboratory (unpublished data). During the first 3 months of use, 681 samples were tested; 678 (99.6%) were typed by real-time PCR, 674 (99.4%) of these on the first test, compared to 88% using the previous routine assay, COWP PCR-RFLP (8). Of the positives, 123 (18.1%) were C. hominis, 533 (78.6%) were C. parvum, 4 (0.6%) were mixed C. hominis/C. parvum, and 18 (2.7%) were other Cryptosporidium spp., a distribution typical for the United Kingdom in the second quarter of the year (7–9). The numbers of undifferentiated C. cuniculus or monkey genotype cases among those with C. hominis profiles are likely to be very small; over the same time frame in 2007, only two C. cuniculus cases were detected by SSU rRNA PCR-RFLP; none were detected in 2008 (6) or 2009 (3), and no monkey genotype cases were found.
Although SSU rRNA gene PCR allows detection and identification of all Cryptosporidium spp., it is compromised in its ability to detect mixed species due to preferential amplification of the predominant species in a sample (4, 30), and the lack of suitable primer sites prevents species-specific targeting of the gene. The LIB13 locus has been used previously as an alternative to specifically detect C. hominis and C. parvum (20, 36). Separate PCRs were performed for these species to reduce amplification competition in mixed infections of C. hominis and C. parvum. However, a rate of only 0.6% mixed C. hominis/C. parvum infection was identified in our unpublished analysis of 681 Cryptosporidium-positive samples. This supports the evidence for mixed C. hominis/C. parvum infections being an uncommon event in England and Wales from studies by McLauchlin et al. (25), who found only 0.4% of 1,705 cases, and Chalmers et al. (8), who found only 0.5% of 7,758 cases using COWP PCR-RFLP. Jothikumar et al. (20) found 3.0% by species-specific real-time PCR in 67 cases from the United States and Botswana. These are all markedly lower than the 12.0% rate found by Mallon et al. (23) among 135 cryptosporidiosis cases in the northeast of Scotland using a multilocus fragment typing approach and the 11.9% rate reported by Nagamani et al. (27) in 59 cases from India using SSU rRNA PCR-RFLP. It is likely that the variation is linked to local prevalence of infection, epidemiology, and transmission. In some developing countries, mixed infections with other Cryptosporidium species have been reported (4, 16), for which different combinations of species-specific primers and probes would be required. However, infections with species other than C. hominis and C. parvum occur in the United Kingdom at a very low rate, and mixes with other species have not been detected even during enhanced testing (6).
MGB TaqMan probes were used for the two loci for two reasons. First, their increased melting temperatures allowed the use of shorter probes, simplifying probe design for the variable, A-T-rich SSU rRNA region. Second, they were used for their increased specificity due to their lower tolerance for mismatches than conventional TaqMan probes (42). Unexpectedly, the C. parvum LIB13 PCR detected the horse genotype in all four isolates tested despite three mismatched bases. However, the horse genotype could be consistently differentiated from C. parvum on the basis of markedly different amplification curves. The results are in accordance with the finding of Yao et al. (42) and Whiley and Sloots (39), who reported such effects even with single nucleotide mismatches. In our test algorithm, any samples producing unusual amplification curves in the C. parvum LIB13 PCR are sequenced at the SSU rRNA gene to confirm identity.
Potentially more problematic is the detection of C. cuniculus and the C. hominis monkey genotype by the C. hominis LIB13 PCR, although both are very closely related to C. hominis and form part of the same SSU rRNA gene clade (31) and only two human cases of C. hominis monkey genotype have been reported (24). To date there has been just one waterborne outbreak caused by C. cuniculus (10) and a sporadic case rate of 1.2% identified in the United Kingdom (6). Routine differentiation of C. cuniculus and C. hominis poses difficulties. The two species are not differentiated by COWP PCR-RFLP but are by SSU rRNA PCR-RFLP with extended electrophoresis times (10) and by sequencing. If required, the SSU rRNA gene real-time PCR product can be sequenced, since they differ by at least 3 bp, and this can be undertaken in outbreaks or where animal contact is reported. If identification is required in the initial analysis, a discriminatory real-time PCR has been developed (S. J. Hadfield and R. M. Chalmers, unpublished data) which may be incorporated into the test algorithm. It should also be noted that there is no assay available which differentiates all Cryptosporidium spp. and genotypes, except for sequencing of the SSU rRNA gene, which is impractical for most laboratories and for typing for epidemiological purposes.
The real-time PCR was able to detect Cryptosporidium in clinical samples with sensitivity and specificity comparable to those of nested SSU rRNA gene PCR, a test that has provided the benchmark for typing. The real-time PCR is capable of accurately identifying the two main species of Cryptosporidium infecting humans in a rapid semiautomated format which decreased handling times with costs comparable to those of conventional PCR-based methods. Unusual isolates could be identified by sequencing of the SSU rRNA gene PCR products after a simple cleanup procedure. The assay is well suited to processing of high numbers of samples, especially when an automated PCR set-up system is used, as was the case here. The widely applicable reagents utilized and the steady signal from the ROX dye, which is used for normalization on certain real-time systems (not required by the Rotorgene), suggests the method can be readily transferred to other real-time platforms. Further developments, including consolidation into a single-tube assay, with multiplexed PCRs for C. hominis, C. parvum, C. cuniculus, and Cryptosporidium spp., are under investigation. From results to date, it could be argued that the IC may not be necessary for epidemiological analysis of known Cryptosporidium-positive clinical samples, but it would be of value if the assay is being used for initial diagnosis.
Since replacing COWP PCR-RFLP for routine typing of clinical samples for epidemiological purposes in our laboratory, the real-time PCR assay has shown improved performance and efficiency, resulting in fewer retests. The absence of downstream processing required for most samples has enabled results to be reported much earlier in the working day, of particular importance in outbreak investigations. The real-time PCR has therefore provided significant improvements in laboratory work flow and service provision.
ACKNOWLEDGMENTS
We thank Nigel Crouch, Jonathon Goss, and Brian Campbell of the UKCRU for preparation of Cryptosporidium sample panels, Andre Charlett, Health Protection Agency Centre for Infection, London, United Kingdom, for advice on statistics, George Di Giovanni, Texas A&M University, for provision of flow cytometer-counted C. parvum and C. muris oocysts, Joaquin Quilez, University of Zaragoza, Zaragoza, Spain, for E. tenella oocysts, Janet Francis, Toxoplasma Reference Unit, Public Health Wales, Swansea, United Kingdom, for T. gondii DNA, and Michaela Giles, Veterinary Laboratories Agency, Weybridge, United Kingdom, for C. baileyi oocysts.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1. Alves M., et al. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41:2744–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amar C. F., Dear P. H., McLauchlin J. 2004. Detection and identification by real time PCR/RFLP analyses of Cryptosporidium species from human faeces. Lett. Appl. Microbiol. 38:217–222 [DOI] [PubMed] [Google Scholar]
- 3. Anonymous 2010. Investigation of the taxonomy and biology of the Cryptosporidium rabbit genotype. Final report DWI 70/2/241. Drinking Water Inspectorate, Department for Environment, Food & Rural Affairs, United Kingdom: http://www.dwi.gov.uk/research/completed-research/reports/DWI70_2_241.pdf [Google Scholar]
- 4. Cama V., et al. 2006. Mixed Cryptosporidium infections and HIV. Emerg. Infect. Dis. 12:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cama V. A., et al. 2007. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 196:684–691 [DOI] [PubMed] [Google Scholar]
- 6. Chalmers R. M., Elwin K., Hadfield S. J., Robinson G. The epidemiology of sporadic human cryptosporidiosis caused by Cryptosporidium cuniculus in the United Kingdom, 2007 and 2008. Emerg. Infect. Dis., in press doi:10.3201/eid1703.100410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chalmers R. M., Pollock K. G. J. 2010. Cryptosporidium in Scotland 2008: reference laboratory data. HPS Wkly. Rep. 44:18–20 [Google Scholar]
- 8. Chalmers R. M., Elwin K., Thomas A. L., Guy E. C., Mason B. 2009. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 14(2):6–14 [DOI] [PubMed] [Google Scholar]
- 9. Chalmers R. M., et al. 2010. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in northwest Wales. J. Water Health 8:311–325 [DOI] [PubMed] [Google Scholar]
- 10. Chalmers R. M., et al. 2009. Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerg. Infect. Dis. 15:829–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chalmers R. M., Smith R., Elwin K., Clifton-Hadley F. A., Giles M. 2010. Epidemiology of anthroponotic and zoonotic human cryptosporidiosis in England and Wales, 2004–2006. Epidemiol. Infect. 12:1–13 [DOI] [PubMed] [Google Scholar]
- 12. Coetzee N., Edeghere O., Orendi J., Chalmers R., Morgan L. 2008. A swimming pool-associated outbreak of cryptosporidiosis in Staffordshire, England, October to December 2007. Euro Surveill. 13:14–16 [PubMed] [Google Scholar]
- 13. Di Giovanni G. D., LeChevallier M. W. 2005. Quantitative-PCR assessment of Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 71:1495–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elwin K., Chalmers R. M., Roberts R., Guy E. C., Casemore D. P. 2001. Modification of a rapid method for the identification of gene-specific polymorphisms in Cryptosporidium parvum and its application to clinical and epidemiological investigations. Appl. Environ. Microbiol. 67:5581–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fayer R., Santin M. 2009. Cryptosporidium xiaoi n. sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries). Vet. Parasitol. 164:192–200 [DOI] [PubMed] [Google Scholar]
- 16. Gatei W., et al. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7:197–205 [DOI] [PubMed] [Google Scholar]
- 17. Haque R., et al. 2007. Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am. J. Trop. Med. Hyg. 76:713–717 [PubMed] [Google Scholar]
- 18. Higgins J. A., et al. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47:323–337 [DOI] [PubMed] [Google Scholar]
- 19. Jiang J., Alderisio K. A., Xiao L. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71:4446–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jothikumar N., da Silva A. J., Moura I., Qvarnstrom Y., Hill V. R. 2008. Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J. Med. Microbiol. 57:1099–1105 [DOI] [PubMed] [Google Scholar]
- 21. Khan S. M., et al. 2010. Molecular characterization and assessment of zoonotic transmission of Cryptosporidium from dairy cattle in West Bengal, India. Vet. Parasitol. 171:41–47 [DOI] [PubMed] [Google Scholar]
- 22. Limor J. R., Lal A. A., Xiao L. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mallon M., et al. 2003. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J. Mol. Evol. 56:407–417 [DOI] [PubMed] [Google Scholar]
- 24. Mallon M. E., MacLeod A., Wastling J. M., Smith H., Tait A. 2003. Multilocus genotyping of Cryptosporidium parvum type 2: population genetics and sub-structuring. Infect. Genet. Evol. 3:207–218 [DOI] [PubMed] [Google Scholar]
- 25. McLauchlin J., Amar C., Pedraza-Diaz S., Nichols G. L. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morgan U. M., Constantine C. C., Forbes D. A., Thompson R. C. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825–830 [PubMed] [Google Scholar]
- 27. Nagamani K., et al. 2007. Molecular characterisation of Cryptosporidium: an emerging parasite. Indian J. Med. Microbiol. 25:133–136 [DOI] [PubMed] [Google Scholar]
- 28. Patel S., Pedraza-Diaz S., McLauchlin J., Casemore D. P. 1998. Molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Outbreak Control Team South and West Devon 1995, Incident Management Team and Further Epidemiological and Microbiological Studies Subgroup North Thames 1997. Commun. Dis. Public Health 1:231–233 [PubMed] [Google Scholar]
- 29. Peng M. M., et al. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reed C., Sturbaum G. D., Hoover P. J., Sterling C. R. 2002. Cryptosporidium parvum mixed genotypes detected by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 68:427–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson G., et al. 2010. Re-description of Cryptosporidium cuniculus (Apicomplexa: Cryptosporidiidae): morphology, biology and phylogeny. Int. J. Parasitol. 40:1539–1548 [DOI] [PubMed] [Google Scholar]
- 32. Rochelle P. A., et al. 1996. Development of a rapid detection procedure for Cryptosporidium, using in vitro cell culture combined with PCR. J. Eukaryot. Microbiol. 43:72S. [DOI] [PubMed] [Google Scholar]
- 33. Spano F., Putignani L., McLauchlin J., Casemore D. P., Crisanti A. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209–217 [DOI] [PubMed] [Google Scholar]
- 34. Sulaiman I. M., et al. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43:2805–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sulaiman I. M., Lal A. A., Xiao L. 2002. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 88:388–394 [DOI] [PubMed] [Google Scholar]
- 36. Tanriverdi S., Arslan M. O., Akiyoshi D. E., Tzipori S., Widmer G. 2003. Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol. Biochem. Parasitol. 130:13–22 [DOI] [PubMed] [Google Scholar]
- 37. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verweij J. J., et al. 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J. Clin. Microbiol. 42:1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whiley D. M., Sloots T. P. 2006. Sequence variation can affect the performance of minor groove binder TaqMan probes in viral diagnostic assays. J. Clin. Virol. 35:81–83 [DOI] [PubMed] [Google Scholar]
- 40. Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89 [DOI] [PubMed] [Google Scholar]
- 41. Xiao L., Ryan U. 2008. Molecular epidemiology, p. 119–163 In Fayer R., Xiao L. (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 42. Yao Y., Nellaker C., Karlsson H. 2006. Evaluation of minor groove binding probe and Taqman probe PCR assays: influence of mismatches and template complexity on quantification. Mol. Cell Probes 20:311–316 [DOI] [PubMed] [Google Scholar]