Abstract
Burkholderia pseudomallei, Burkholderia thailandensis, and the Burkholderia cepacia complex differ greatly in pathogenicity and epidemiology. Yet, they are occasionally misidentified by biochemical profiling, and even 16S rRNA gene sequencing may not offer adequate discrimination between certain species groups. Using the 23 B. pseudomallei, four B. thailandensis, and 16 B. cepacia complex genome sequences available, we identified gene targets specific to each of them (a Tat domain protein, a 70-kDa protein, and a 12-kDa protein for B. pseudomallei, B. thailandensis, and the B. cepacia complex, respectively), with an in-house developed algorithm. Using these targets, we designed a robust multiplex PCR assay useful for their identification and detection from soil and simulated sputum samples. For all 43 B. pseudomallei, seven B. thailandensis, and 20 B. cepacia complex (B. multivorans, n = 6; B. cenocepacia, n = 3; B. cepacia, n = 4; B. arboris, n = 2; B. contaminans, B. anthina, and B. pyrrocinia, n = 1 each; other unnamed members, n = 2) isolates, the assay produced specific products of predicted size without false positives or negatives. Of the 60 soil samples screened, 19 (31.6%) and 29 (48.3%) were positive for B. pseudomallei and the B. cepacia complex, respectively, and in four (6.7%) soil samples, the organisms were codetected. DNA sequencing confirmed that all PCR products originated from their targeted loci. This novel pan-genomic analysis approach in target selection is simple, computationally efficient, and potentially applicable to any species that harbors species-specific genes. A multiplex PCR assay for rapid and accurate identification and detection of B. pseudomallei, B. thailandensis, and the B. cepacia complex was developed and verified.
INTRODUCTION
Burkholderia pseudomallei is a highly pathogenic bacterium and has been classified as a category B bioterrorism agent by the Centers for Disease Control and Prevention (http://www.bt.cdc.gov/agent/agentlist-category.asp). A phylogenetically closely related organism, Burkholderia thailandensis, is much less virulent and causes human infection only rarely (6, 7, 49). Despite certain subtle phenotypic differences, the two species cannot be confidently distinguished by 16S rRNA gene sequencing (48), antigen agglutination tests, or API and Vitek biochemical profiling (22, 23).
The Burkholderia cepacia complex currently comprises 17 genotypic species (26). They are opportunistic pathogens implicated in chronic and occasionally serious infections of patients with cystic fibrosis and chronic granulomatous disease (3, 26, 27, 29). Although coinfection by the B. cepacia complex and B. pseudomallei in cystic fibrosis patients has been reported (31), commercial systems often fail to distinguish between the two organisms (14, 24, 43). Since the treatment of B. pseudomallei infection requires a prolonged course of antibiotics (46), this can directly affect management, as inadequate treatment is associated with a high rate of relapse (12). Moreover, misidentification is a significant health risk to the microbiological laboratory personnel (33), as laboratory-acquired melioidosis following exposure from antimicrobial susceptibility testing on a misidentified B. cepacia complex isolate has been reported (35).
Previous attempts to identify organisms in the B. pseudomallei group, i.e., B. pseudomallei, B. thailandensis, Burkholderia mallei, and Burkholderia oklahomensis (19), included sequencing of the groEL gene (48), the 16S rRNA gene (17), and various housekeeping genes by multilocus sequence typing (MLST) schemes (20, 38). Methods that do not require gene sequencing included probe-based real-time PCR and loop-mediated isothermal amplification targeting Burkholderia type III secretion system genes (10, 30, 40), single-nucleotide polymorphism (SNP) typing (42), and multiplex PCR amplification of a region flanking variable copies of a repetitive element (RE) (25). SNP-based tests, however, are susceptible to back substitution when a limited number of loci are used, and the interpretation of multiplex PCR with variable RE copies is intrinsically difficult (25). With the exceptions of the more recent use of DNA microarrays (36) and proteome profiling (44), all existing tests that differentiate among the species are based on genetic differences in homologous loci present in all species of the group, and none except gene sequencing (32) can also positively identify the B. cepacia complex. An expanded MLST scheme that can cover the entire Burkholderia genus has only recently been possible with the advent of genomics (38).
Since the first bacterial genome was published in 1995 (16), more than 1,000 prokaryotic genomes have been sequenced. As with other virulent or emerging bacterial pathogens (34, 45, 51), complete genome sequences of 23 B. pseudomallei, four B. thailandensis, and 16 B. cepacia complex isolates have been made publicly available (http://www.ncbi.nlm.nih.gov/sites/genome). In the clinical microbiology setting, however, this information has yet to be fully exploited. We hypothesize that, by use of bacterial genome data, species-specific genes can be identified and serve as targets for bacterial identification. In this study, a novel pan-genomic analysis approach using multiple genome comparison to select gene targets specific to B. pseudomallei, B. thailandensis, or the B. cepacia complex was devised. The gene targets were used to design a multiplex PCR useful for the identification of the bacteria as pure isolates. The assay was further verified using simulated sputum samples, and a pilot study to screen for the Burkholderia species in environmental samples was carried out. The potential use of this pan-genomic approach in choosing targets for identifying and detecting other clinically important bacteria is also discussed.
MATERIALS AND METHODS
Database generation and pan-genome search.
A three-step approach was devised from our previous publication on lineage-specific genes (8). First, predicted protein sequences for the sequenced genomes of B. pseudomallei, B. thailandensis, and the B. cepacia complex were downloaded from the NCBI Genome database (www.ncbi.nlm.nih.gov/sites/genome). Three pan-genome databases of protein sequences were then generated by compiling all of the sequences for B. pseudomallei, B. thailandensis, and the B. cepacia complex species using the Formatdb software in the NCBI BLAST suite (1) (Fig. 1, step 1). Using gapped BLAST (2), predicted protein sequences from the smallest genome, i.e., that with the fewest or shortest genes such that the total number of amino acid residues encoded is also the lowest, from each group were selected and used as the query set against the databases of the other two groups, since core genes common to all isolates of a species group must still be present in that genome (Fig. 1, step 2). An Expect value (E value) of 1.0 was used as a cutoff threshold, below which protein sequences were considered similar, probably not useful as discriminating targets, and eliminated. This is because, in a BLAST database search, the E value describes the expected number of hits by random chance: short and relatively dissimilar BLAST matches have high E values (i.e., a large number of short matches due to chance can be expected), and long, similar BLAST matches have low E values (i.e., few if any long stretches of sequence match arise from chance alone) (13). Thus, increasing this elimination cutoff threshold enabled the removal of more unrelated sequences and reduced the number of false positives identified as species-specific targets.
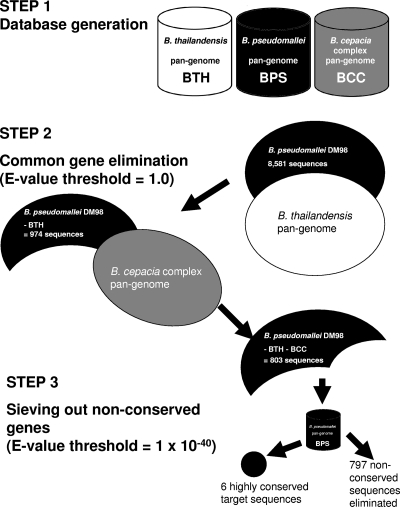
Fig. 1.
Three-step pan-genomic approach to select gene targets for bacterial identification. Step 1, generation of a pan-genome (proteome) database for each species of interest, using Formatdb software. Step 2, elimination of protein sequences common to B. pseudomallei DM98 and the pan-genomes of B. thailandensis and the B. cepacia complex. From the initial 8,581 sequences in the genome of B. pseudomallei DM98, 974 sequences were left after elimination with the B. thailandensis pan-genome. Only 803 sequences were left after further elimination with the B. cepacia complex pan-genome. Common genes were sequences that had a match with an E value less than 1.0. Step 3, sieving out nonconserved gene targets, i.e., those that fail to achieve an E value less than 1 × 10−40 in a BLASTp search. Six highly conserved targets were identified, while the remaining 797 sequences were eliminated, as they were not consistently present in the B. pseudomallei genomes.
Finally, the protein sequences with “no hits” from the search (taken as unique to the query set) were further searched against the database of the original group. An E value of 10−40 was used as an empirical cutoff below which matches were found to be highly conserved in sequence and of practical length for conventional PCR primer design. Increasing this conservation cutoff would identify a greater number of shorter and dissimilar sequences, increasing the number of false positives identified as species-specific targets (Fig. 1, step 3). To facilitate the iterative BLAST searches, an in-house Visual Basic.NET program, BLASTP Parser (available on request), was written to organize the search results and eliminate excluded sequences. The Burkholderia genomes used are shown in Table 1, and Fig. 1 illustrates the steps for identifying the B. pseudomallei-specific gene targets. The same procedures were repeated using the genomes of B. thailandensis and the B. cepacia complex.
Table 1.
List of Burkholderia genomes used in the study
| Bacterial species | Strain(s) or plasmids | GenBank accession no.a |
|---|---|---|
| B. pseudomallei | 9, 14, 91, 112, 305, 576, 668, 1655, 7894, 1106a, 1106b, 1710a, 1710b, 406e, B7210, BCC215, DM98, K96243, MSHR346, NCTC 13177, Pakistan 9, Pasteur 52237, and S13 | ABBL00000000 (w), ABBJ00000000 (w), ABBK00000000 (w), ABBP00000000 (w), AAYX00000000 (w), ACCE00000000 (w), NC_009074 (1), NC_009075 (2), AAHR00000000 (w), ABBO00000000 (w), NC_009076 (1), NC_009078 (2), AAMB00000000 (w), AAHS00000000 (w), NC_007434 (1), NC_007435 (2), AAMM00000000 (w), ABBN00000000 (w), ABBR00000000 (w), ABBI00000000 (w), NC_006350 (1), NC_006351 (2), ACOJ00000000 (w), NC_012695 (1), ABBQ00000000 (w), ACKA00000000 (w), AAHV00000000 (w), AAHW00000000 (w) |
| B. thailandensis | Bt4, E264, MSMB43, and TXDOH | ABBH00000000 (w), NC_007651 (1), NC_007650 (2), ABBM00000000 (w), ABBD00000000 (w) |
| B. cepacia complex | ||
| Burkholderia ambifaria | AMMD, IOP40-10, MC40-6, and MEX-5 | NC_008390 (1), NC_008391 (2), NC_008392 (3), NC_008385 (p), ABLC00000000 (w), NC_010551 (1), NC_010552 (2), NC_010557 (3), NC_010553 (p), ABLK00000000 (w) |
| Burkholderia cenocepacia | AU 1054, HI2424, J2315, MC0-3, and PC184 | NC_008060 (1), NC_008061 (2), NC_008062 (3), NC_008542 (1), NC_008543 (2), NC_008544 (3), NC_008545 (p), NC_011000 (1), NC_011001 (2), NC_011002 (3), NC_011003 (p), NC_010508 (1), NC_010515 (2), NC_010512 (3), AAKX00000000 (w) |
| Burkholderia cepacia | Plasmids | NC_013666 (p), NC_010099 (p) |
| Burkholderia dolosa | AUO158 | AAKY00000000 (w) |
| Burkholderia multivorans | ATCC 17616, CGD1, CGD2, and CGD2M | NC_010804 (1), NC_010084 (1), NC_010805 (2), NC_010086 (2), NC_010801 (3), NC_010087 (3), NC_010070 (p), NC_010802 (p), ACFB00000000 (w), ACFC00000000 (w), ACFD00000000 (w) |
| Burkholderia lata | 383 | NC_007510 (1), NC_007511 (2), NC_007509 (3) |
| Burkholderia vietnamiensis | G4 | NC_009256 (1), NC_009255 (2), NC_009254 (3), NC_009230 (p), NC_009227 (p), NC_009229 (p), NC_009228 (p), NC_009226 (p) |
1, chromosome 1 sequence; 2, chromosome 2 sequence; 3, chromosome 3 sequence; w, whole-genome shotgun sequence; p, plasmid sequence.
Selection of species- and complex-specific targets.
By analysis of the BLAST results, all highly conserved species- and complex-specific protein sequences were checked to determine if they were present in more than 95% of the genomes of the target organisms. This nonstringent cutoff (95% instead of 99 or 100%) was used since predicted protein sequences may be truncated at the contig borders of whole-genome shotgun sequences and fail to produce a BLAST match of sufficient length to achieve the low E value required. The sequences were then examined based on their protein sequence conservation, as indicated by the average E value achieved from the cross-genome search. The most conserved protein sequences and their respective nucleotide sequences from each group were searched against the nonredundant protein sequence (nr) and nucleotide collection (nr/nt) databases using the online BLAST suite (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to look for any homologues and sequence similarity with all other species.
Bacterial strains and identification.
All isolates were phenotypically identified by the API 20NE system (bioMérieux Vitek, Hazelwood, MO), supplemented by conventional biochemical methods. The B. pseudomallei strains included a collection of 23 clinical isolates from hospitalized patients and 20 veterinary and environmental isolates from Hong Kong. All seven B. thailandensis strains were isolated and characterized in a previous study (37). The identities of the B. pseudomallei and B. thailandensis isolates were confirmed by groEL gene sequencing (48). The B. cepacia complex panel comprised 20 strains, including 17 clinical isolates from patients in local hospitals (B. multivorans, n = 6; B. cenocepacia, n = 3; B. cepacia, n = 3; B. arboris, n = 2; B. contaminans, n = 1; other unnamed B. cepacia complex species, n = 2) and three type strains obtained from the BCCM/LMG bacterial collection (B. anthina LMG 20980, B. cepacia LMG 1222, and B. pyrrocinia LMG 14191). Species-level identification of the B. cepacia complex species was based on recA gene sequencing (28) and subsequent BLAST comparison against sequences on the MLST site (http://pubmlst.org/bcc), which is up to date for all named species in the B. cepacia complex.
Extraction of bacterial DNA for PCR amplification.
Bacterial DNA extraction was performed according to our previously published protocol (47). Briefly, 800 μl of 0.05 M NaOH was added to 200 μl of bacterial cells suspended in phosphate-buffered saline (PBS), and the mixture was incubated at 60°C for 45 min, followed by addition of 240 μl Tris-HCl (pH 7.0), achieving a final pH of 8.0. The resultant mixture was centrifuged at 13,000 × g for 5 min. Afterwards, 150 μl of the supernatant was purified using a QIAamp DNA minikit (QIAgen, Hilden, Germany) and eluted in 100 μl of nuclease-free water.
PCR amplification and DNA sequencing of the species- and complex-specific gene targets.
Specific gene targets, one each for B. pseudomallei, B. thailandensis, and the B. cepacia complex, were selected and amplified by multiplex PCR using conserved and consensus primers according to the following protocol. The PCR mixture (20 μl) contained purified bacterial DNA (1.0 μl), 1.0 M betaine monohydrate (Fluka BioChemika, Steinheim, Germany), 0.5 μM each B. pseudomallei-specific primer (LPW13372, 5′-CAAGAACGGTTTATGCG-3′, and LPW13373, 5′-GAAGTGATCCATCAAATGTC-3′), 0.5 μM each B. thailandensis-specific primer (LPW13376, 5′-CGTACAACGTCGATAGC-3′, and LPW13377, 5′-GATACCTGGACGATGTTT-3′), 1.0 μM each B. cepacia complex-specific primer (LPW13807, 5′-CCATGAACGTCGAYTAYCTYTT-3′, and LPW13808, 5′-GTCARCCGTARACGATGTC-3′) (Sigma-Aldrich, Steinheim, Germany), 2.0 μl 10× PCR buffer II, 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate (dNTP) (GeneAmp; Applied Biosystems, CA), and 1.0 U Taq polymerase (AmpliTaq Gold; Applied Biosystems, CA). Thermocycling was performed with an automated thermocycler (Veriti 96-well fast thermal cycler; Applied Biosystems, CA), with a hot start at 95°C for 10 min, 10 touchdown cycles of 95°C for 30 s, annealing for 1.5 min at temperatures decreasing from 60 to 51°C (with 1.0°C decremental steps), 72°C for 1 min, 30 cycles of 95°C for 30 s, 50°C for 1.5 min, 72°C for 1 min, and a final extension at 72°C for 10 min. Five microliters of each amplified product was electrophoresed in a 2.5% (wt/vol) agarose gel, with a molecular size marker (GeneRuler 50-bp DNA ladder; Fermentas, Ontario, Canada) in parallel. Electrophoresis in Tris-borate-EDTA buffer was performed at 100 V for 45 min. The gel was stained with ethidium bromide (0.5 μg/ml) for 25 min, rinsed, and photographed under UV light illumination.
The PCR products were gel purified using a QIAquick PCR gel extraction kit (QIAgen, Hilden, Germany). Both strands of the PCR products were sequenced with an ABI 3130xl genetic analyzer according to manufacturer's instructions (Applied Biosystems, CA), using PCR primers specific to each PCR product. The DNA sequences obtained were analyzed using a BLASTx search of the in-house Burkholderia pan-genome databases and a BLASTn search against the online nucleotide collection (nr/nt) database on NCBI to confirm the sequence identities.
Sensitivity of the assay.
Sensitivity of the multiplex PCR was assessed according to a published protocol (18), with modifications. One isolate each of B. pseudomallei, B. thailandensis, and the B. cepacia complex was grown for 16 to 18 h on horse blood agar. Single colonies were picked and suspended in PBS to reach approximately a 0.5 McFarland standard, and 10-fold dilutions were made. Viable counts were simultaneously made as previously published (9). Sputum samples known to be negative for any of the Burkholderia species tested were pooled and homogenized using 0.1% dithiothreitol in PBS (Sputasol) at room temperature for 30 min. Diluted bacterial suspensions (0.1 ml) were mixed with 0.9 ml of homogenized sputum, briefly vortexed, and centrifuged at 16,000 × g for 30 min. Eight hundred microliters of supernatant was removed from each sample, and the remaining liquid and residue were digested and purified using a QIAamp DNA minikit (QIAgen, Hilden, Germany) according to the manufacturer's protocol for body fluids and eluted in 50 μl of nuclease-free water. One microliter of purified sample was used per PCR.
PCR products of each of the targets were also cloned into a plasmid vector (pCRII-TOPO) using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Plasmids were isolated from the electrotransformed Escherichia coli strains using a Roche High Pure plasmid isolation kit (Roche Applied Science, Indianapolis, IN) and eluted in 50 μl of nuclease-free water. Dilutions of the plasmid DNA were used to assess the triplex sensitivity of the assay.
Specificity of the assay.
To test the empirical specificity of the assay, purified DNA from a panel of bacterial pathogens was used as a negative control (Table 2). The bacterial species were selected based on their phylogenetic relatedness and phenotypic similarities to the targeted Burkholderia species and the propensity of coisolation, especially in clinical samples from cystic fibrosis and bronchiectasis patients (26).
Table 2.
List of bacteria used to verify empirical specificity
| Bacterial species | Reason for inclusion (reference) | PCR result |
|---|---|---|
| Burkholderia gladioli pv. gladioli LMG 2216 | Phenotypic resemblance to the B. cepacia complex species; common cystic fibrosis pathogen in Canada and certain European countries (26) | Negative |
| Neisseria meningitidis, Neisseria gonorrhoeae, and Laribacter hongkongensis | Phylogenetic relatedness to the Burkholderia genus | Negative |
| Pseudomonas aeruginosa | Phenotypic resemblance to the B. cepacia complex species; common cystic fibrosis pathogen | Negative |
| Stenotrophomonas maltophilia, Achromobacter xylosoxidans, Chryseobacterium indologenes, Elizabethkingia meningoseptica (previously known as Chryseobacterium meningosepticum), and Ralstonia pickettii | Common opportunists associated with cystic fibrosis (26) | Negative |
Detection of B. pseudomallei, B. thailandensis, and B. cepacia complex species from soil samples using multiplex PCR.
As a pilot study, 60 soil samples were collected from sites where B. pseudomallei had been recovered (unpublished data). Each 100-g soil sample was mixed with 100 ml of purified water and incubated at 25°C overnight for soil settlement. Thirty microliters of the supernatant was inoculated into 3 ml Ashdown medium (4) and incubated at 37°C for 48 h. One milliliter of the enrichment culture was harvested, and bacterial DNA extraction was performed using a QIAamp DNA minikit (QIAgen, Hilden, Germany) according to the manufacturer's instruction. One microliter of purified DNA extract was used as the template for PCR. Agarose gel electrophoresis, PCR product purification, and DNA sequencing were also performed as described above.
Phylogenetic analysis of the species- and complex-specific sequences.
The target gene sequences were analyzed to detect genetic variation within the sequenced region. Multiple sequence alignments were performed using MUSCLE (15), followed by phylogenetic tree construction using the neighbor-joining method with 1,000 bootstrap replicates in MEGA4 (39).
Nucleotide sequence accession numbers.
The species- and complex-specific gene sequences of the 43 B. pseudomallei, seven B. thailandensis, and 20 B. cepacia complex isolates have been deposited in GenBank under accession numbers HQ166195 to HQ166261 and HQ611959 to HQ611961.
RESULTS
Specific gene targets in B. pseudomallei, B. thailandensis, and the B. cepacia complex.
The B. pseudomallei, B. thailandensis, and B. cepacia complex pan-genome protein sequence databases were based on genomes of 23, 4, and 16 strains and contained 170,866, 32,201, and 112,875 predicted protein sequences, respectively (Fig. 1). Protein sequences from 13 available plasmid sequences were also included in the database for the B. cepacia complex, as inclusion and analysis of genes from the accessory genome help avoid false positives in PCR detection. The initial query set for each group was selected based on total genome size, computed by adding the sequence lengths of the chromosomes when required. The smallest genomes selected for the groups were B. pseudomallei DM98 (8,581 predicted proteins), B. thailandensis E264 (5,633 predicted proteins), and Burkholderia dolosa AUO158 (4,822 predicted proteins). In the second step, each of the selected genomes was searched against the pan-genome protein databases of the other two groups, and only sequences with no BLAST match with an E value of less than 1.0 in the other two databases were retained. The remaining set contained 803, 203, and 60 protein sequences unique to B. pseudomallei DM98, B. thailandensis E264, and B. dolosa AUO158, respectively (Fig. 1, step 2). The final round of the pan-genome search, for targets conserved among over 95% of all genomes of each group (using an E-value cutoff of 1 × 10−40 and eliminating sequences with no match), identified six potential targets each for B. pseudomallei and B. thailandensis (Fig. 1, step 3) and five targets for the B. cepacia complex that may be useful for single-gene PCR identification (Table 3).
Table 3.
Potential gene targets to identify B. pseudomallei, B. thailandensis, and the B. cepacia complex
| Burkholderia species | GenBank accession no. | Protein description | Length (amino acids) | Note |
|---|---|---|---|---|
| B. pseudomallei | ZP_02406082 | Tat domain protein | 676 | Chosen target; corresponds to locus BPSS0658 in B. pseudomallei K96243 reference genome (http://www.ncbi.nlm.nih.gov/genomeprj/178) |
| ZP_02401787 | Conserved 31-kDa protein | 278 | Alternative target; should also detect B. pseudomallei-like B. oklahomensis | |
| ZP_02402672 | Putative lipase | 98 | Alternative target | |
| ZP_02402698 | Conserved 15-kDa protein | 135 | Alternative target | |
| ZP_02402702 | Tetratricopeptide repeat protein | 93 | Sequence similarity with Acidiphilium cryptum JF-5 | |
| ZP_02401892 | Conserved 11-kDa protein | 100 | Sequence similarity with B. mallei JHU | |
| B. thailandensis | ZP_05586505 | 70-kDa protein | 645 | Chosen target; corresponds to locus BTH_I1515 in B. thailandensis E264 genome (http://www.ncbi.nlm.nih.gov/genomeprj/10774) |
| ZP_05589956 | 16-kDa protein | 145 | Alternative target; may also detect B. pseudomallei-like B. oklahomensis | |
| ZP_05590352 | Conserved 20-kDa protein | 178 | Sequence similarity with Lutiella nitroferrum 2002 | |
| ZP_05589818 | UPF0066 family-like protein | 155 | Sequence similarity with Firmicutes and other Gram-positive organisms | |
| ZP_05587830 | Conserved 30-kDa protein | 269 | Sequence similarity with other proteobacteria | |
| ZP_05589817 | 13-kDa protein | 115 | Sequence similarity with Bacillus clausii KSM-K16 | |
| B. cepacia complex | ZP_04948143 | Conserved 12-kDa protein | 110 | Chosen target; corresponds to locus BCAM2834 in B. cenocepacia J2315 reference genome (http://www.ncbi.nlm.nih.gov/genomeprj/339) |
| ZP_04945743 | Protease subunit of ATP-dependent Clp protease | 247 | Alternative target | |
| ZP_04947479 | DUF1255 domain protein | 106 | Sequence similarity with Ralstonia eutropha H16 | |
| ZP_04947699 | DUF488 domain protein | 188 | Sequence similarity with other proteobacteria | |
| ZP_04947794 | Thiol-disulfide isomerase and thioredoxin | 248 | Sequence similarity with other proteobacteria |
Multiplex PCR identification of B. pseudomallei, B. thailandensis, and the B. cepacia complex.
By considering sequence similarity with phylogenetically related or distant species, one target was selected from the potential targets for each group. Their nucleotide sequences showed little similarity to unintended species by BLAST search. A Tat domain protein, a 70-kDa protein, and a conserved 12-kDa protein were selected for B. pseudomallei, B. thailandensis, and the B. cepacia complex, respectively (Table 3). To verify the sensitivity and specificity of the selected targets, as well as to demonstrate their application in the one-step identification of the three groups of medically important Burkholderia species, a multiplex PCR assay was designed and performed to screen our Burkholderia isolate collection. During the optimization process, the addition of betaine (as monohydrate solution) in the PCR mix enhanced sensitivity and specificity of the multiplex assay; a concentration of 1.0 M was initially used, according to Henke et al. (21), and gave good results. The use of purified DNA extract was found to be essential, as crude extract from the alkaline lysis procedure occasionally failed to yield any PCR product for some B. cepacia complex isolates tested, putatively due to the presence of PCR inhibitors. We have also found that a High Pure PCR product purification kit (Roche Diagnostics, Mannheim, Germany) may be used in place of the more specialized QIAamp DNA minikit (QIAgen, Hilden, Germany) and gave comparable results (data not shown), although the latter was used in this study because of more-comprehensive instructions on bacterial DNA purification provided by the manufacturer. For all of the 43 B. pseudomallei, seven B. thailandensis, and 20 B. cepacia complex isolates, the multiplex PCR assay produced products of predicted size, 336 bp for the B. cepacia complex, 189 bp for B. pseudomallei, and 110 bp for B. thailandensis (Fig. 2B), with no false negatives.
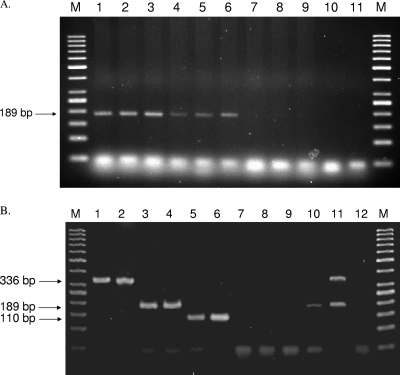
Fig. 2.
(A) DNA products from multiplex PCR identification and detection of B. pseudomallei from simulated sputum samples. Lanes M, molecular marker GeneRuler 50-bp DNA ladder; lanes 1 to 3, 100,000 CFU per 20 μl PCR mix; lanes 4 to 6, 10,000 CFU per 20 μl PCR mix; lanes 7 to 9, 1,000 CFU per 20 μl PCR mix; lane 10, Burkholderia gladioli LMG 2216; lane 11, Achromobacter xylosoxidans PW1766. CFU counts per PCR mix were estimated from the known amount of bacteria added to sputum samples, assuming no loss during the subsequent sedimentation, digestion, and DNA purification steps. (B) DNA products from multiplex PCR identification and detection of B. pseudomallei, B. thailandensis, and the B. cepacia complex. Lanes M, molecular marker GeneRuler 50-bp DNA ladder; lane 1, B. cepacia complex isolate BCEP01; lane 2, B. cepacia complex isolate BCEP17; lane 3, B. pseudomallei clinical isolate B1; lane 4, B. pseudomallei veterinary isolate BC183; lane 5, B. thailandensis isolate Bt1; lane 6, B. thailandensis isolate Bt7; lane 7, Chromobacterium violaceum; lane 8, Neisseria gonorrhoeae; lane 9, PCR-grade water used as a negative control; lane 10, soil sample 0706-01 (screened positive for B. pseudomallei by multiplex PCR); lane 11, soil sample 1005-09 (screened positive for B. pseudomallei and B. cepacia complex species by multiplex PCR); lane 12, soil sample 0706-04 (screened negative for B. pseudomallei, B. thailandensis, and B. cepacia complex species by multiplex PCR).
Specificity of the multiplex PCR.
There were no false positives for the whole panel of negative controls tested (Table 2). DNA sequencing further confirmed that PCR products amplified from the positive samples all originated from their targeted loci.
Sensitivity of the multiplex PCR.
The viable counts of the B. pseudomallei, B. thailandensis, and B. cepacia complex bacterial suspensions were 1.3 × 108, 0.6 × 108, and 1.0 × 108 CFU/ml, respectively. After adjusting for the differences in viable count values, the sensitivities of the multiplex PCR were estimated to be 1,000 CFU per 20 μl PCR mix for B. thailandensis and 10,000 CFU for B. pseudomallei and the B. cepacia complex in simulated sputum samples (Fig. 2A). The limit of triplex detection (triple positive) was determined to be less than 1,000 copies per 20 μl PCR mix by use of purified plasmid DNA in nuclease-free water.
Detection of B. pseudomallei, B. thailandensis, and B. cepacia complex species from soil samples.
Of the 60 soil samples screened by multiplex PCR, 19 (31.6%) were positive for B. pseudomallei and 29 (48.3%) were positive for the B. cepacia complex. In four (6.7%) of the samples, B. pseudomallei and the B. cepacia complex were codetected (Fig. 2B). None of the samples was positive for B. thailandensis. DNA sequencing confirmed that PCR products originated from the targeted loci.
Phylogenetic analysis of the species- and complex-specific sequences.
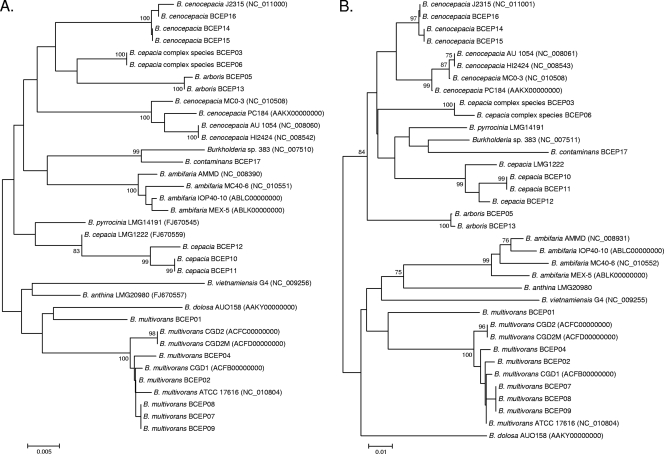
One polymorphic site was noted in the 43 sequences from the B. pseudomallei isolates. There were two sequence types. For the seven B. thailandensis isolates, the sequences were identical. Phylogenetic tree construction using species-specific sequences was therefore not attempted for the two species. Substantial sequence variation exists in the sequences obtained from B. cepacia complex isolates. In the 295 nucleotide positions analyzed, 193 (65.4%) were conserved, and 102 (34.6%) were variable. This contrasted with the recA gene, a housekeeping gene, in which 844 (84.7%) and 152 (15.2%) of the 996 analyzed nucleotide sites were conserved and variable, respectively. The phylogenetic tree constructed using the B. cepacia complex-specific gene was similar to the recA gene tree. Both trees offered good species-level discrimination (Fig. 3).
Fig. 3.
The recA (A) and B. cepacia complex-specific gene (B) phylogenetic trees were inferred using the neighbor-joining method. The bootstrap percentage support values from 1,000 replicates are shown when ≥75%. The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the LogDet (Tamura-Kumar) method and are reported as number of base substitutions per site. The differences in the composition bias among sequences were considered in evolutionary comparisons. The analysis involved 36 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 996 (A) or 295 (B) positions in the final data set.
DISCUSSION
We report a novel pan-genomic approach in target selection for multiplex PCR identification and detection of B. pseudomallei, B. thailandensis, and the B. cepacia complex. As a proof-of-concept study, we identified species- and complex-specific gene targets which were subsequently validated using a panel of 70 isolates, simulated sputum samples, and environmental soil samples. The B. cepacia complex-specific gene sequenced in the current study also showed a degree of genetic polymorphism useful as an alternative form of species discrimination. We propose and advocate this in silico-in vitro approach: generation of lineage-specific targets by extensive in silico analysis and validation and in vitro confirmation using laboratory isolates and application via a technology routinely accessible to the clinical laboratory, i.e., multiplex PCR. The effectiveness of this approach is evident.
This approach is simple, computationally efficient, and applicable to any species that harbors species-specific genes. As in the case of B. pseudomallei, starting from the initial list of 8,581 predicted protein-coding genes in the genome of B. pseudomallei DM98 (Fig. 1, step 1), the in silico search eliminated 7,778 genes (90.6%) that were also found in the chromosome and plasmid sequences of B. thailandensis and B. cepacia complex species, with only 803 genes (9.35%) remaining (Fig. 1, step 2). After the nonconserved gene targets, i.e., those that are present in the genome of B. pseudomallei DM98 but not other B. pseudomallei genomes, were sieved out, only six gene targets (0.06%) were left for manual examination (Fig. 1, step 3). We are aware of alternative approaches, such as pairwise BLASTp (50) or Markov clustering (11), used for the identification of lineage-specific genes, and yet the computational power demanded by such methods appears prohibitive; the “Benchmark Data set” of 100 proteomes used by the authors of OrthoMCL, a Markov clustering-based algorithm, took “3 days to run all-v-all BLAST on a 500 cpu compute cluster” and “16 h for the orthomclPairs processing to find pairs” (http://orthomcl.org/common/downloads/software/v2.0/UserGuide.txt). There is also no guarantee that the computationally intensive alternatives require less manual input. In contrast, the streamlined process reported here required less than 1 h on an ordinary personal computer (Intel 1.8-GHz CPU, 1GB RAM) for our data set and produced a manageable number of targets for manual selection. This speed is primarily attributed to the early elimination of a large number of orthologous and similar targets (Fig. 1, step 2), while the selection of a small genome as the starting query set also reduced the search time (data not shown). Of note, quick reduction of query set size in the one-way BLASTp procedure using our approach has an especially marked effect when the target and nontarget organisms are genetically closely related, as it allows more orthologues or significantly similar sequences to be eliminated. This is clearly desirable, as single-gene identification by PCR is also more useful in such cases. Moreover, by using a pan-genomic analysis approach and including accessory genome components by using multiple genomes and plasmid sequences, specificity of the targets is enhanced and the additional cost is nonetheless minimal. In contrast, while in vitro experiments by two-dimensional PAGE (44) or subtractive hybridization (5) can generate differential profiles that may be useful for identifying bacteria, usually only few isolates are used and genome database searches are still inevitable. An entirely in silico screening step using publicly available genome data and free software programs is arguably more efficient and economical.
The in vitro verification of the targets using actual isolates is essential to the current approach. As the selected targets were cross-validated and screened using a panel of positives (i.e., our Burkholderia isolate collection) and realistic negatives (i.e., phylogenetically related or diagnostically challenging bacteria), robustness of the assay was increased, and the theoretical rates of false positives and negatives lowered. It is clear that while sophisticated algorithms and fast computers have greatly facilitated our design of diagnostic tests, verification with real-world samples remains essential. As demonstrated by this study, the multiplex PCR assay was successful in detecting and identifying the Burkholderia species from simulated sputum samples and did not yield any false positives with the important cystic fibrosis pathogens tested, including Pseudomonas aeruginosa and Stenotrophomonas maltophilia, among others (Table 2). Moreover, from our pilot study using environmental soil samples, no false positives were found by DNA sequencing, and this highlights a potential use of our approach, i.e., as an alternative to culture-based techniques or even metagenomic studies by shotgun sequencing, in elucidating the prevalence of multiple species in microbial communities. Although we are aware that our gene targets may require further validation before they are accepted into practice, we also note that the current approach has allowed the detection of species not represented by the published genome sequences. In the case of the B. cepacia complex, while the analyzed genomes spanned only 6 of the 17 known species, the complex-specific gene target could also detect the B. cepacia, B. arboris, B. contaminans, B. anthina, and B. pyrrocinia strains in our collection, which had no published genome available.
Despite all of these advantages, the approach illustrated by this proof-of-concept study has its limitations. The presence of a positive PCR signal, or even a confirmed sequencing result, does not signify the presence of a viable organism. Unlike widely accepted gene targets, such as 16S rRNA genes, the specificity and species sensitivity of each of the targets have to be validated according to the species being investigated. The reader should be cautioned that the probing of nonhousekeeping genes is, in general, more susceptible to gene loss and horizontal gene transfer, although the increased level of sequence (or even copy number) variation may point to applications in typing. Availability of multiple genome sequences for the target group is a prerequisite, since a single genome is unlikely to capture the genetic variability of a species or taxonomic group. An adaptation for such was made in this study: during the pan-genome search, the B. cepacia complex genomes were not analyzed at the species level, as at the time of the study, only five genomes were available even for the most richly sampled species of Burkholderia cenocepacia, but 17 genomes were available for 16 strains of the B. cepacia complex as a whole (Burkholderia multivorans ATCC 17616 was sequenced by two groups independently). Given the genetic variability of the large B. cepacia complex, it may also be prudent to validate the complex-specific gene target in species not represented in our study, e.g., Burkholderia stabilis and Burkholderia diffusa.
Currently, this approach is already feasible for clinically important bacteria with abundant genome data, such as P. aeruginosa, Staphylococcus aureus, and Staphylococcus epidermidis, but requires additional genome data for single-genome species, such as other coagulase-negative staphylococci (41) and alpha-hemolytic streptococci (47). With the commencement of the human microbiome project (http://nihroadmap.nih.gov/hmp/) and various initiatives to sequence and resequence commensal and pathogen genomes, it is expected that the sequence data required for the described identification approach will be made available in the very near future, enabling the low-cost identification and discrimination of all clinically relevant species. Targets identified by this approach, by virtue of their lineage specificity, may also be useful for the design of real-time PCR and in situ hybridization assays.
ACKNOWLEDGMENTS
This work was partly supported by the Committee of Research and Conference Grants, The University of Hong Kong, and the Croucher Foundation.
We thank N. J. White for providing us the B. thailandensis strains, and we thank Herman Tse, Hoi-Wah Tsoi, Shirly Curreem, and Alan Tsang for inspiring discussions.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aris R. M., Routh J. C., LiPuma J. J., Heath D. G., Gilligan P. H. 2001. Lung transplantation for cystic fibrosis patients with Burkholderia cepacia complex. Survival linked to genomovar type. Am. J. Respir. Crit. Care Med. 164:2102–2106 [DOI] [PubMed] [Google Scholar]
- 4. Ashdown L. R. 1979. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology 11:293–297 [DOI] [PubMed] [Google Scholar]
- 5. Bernier S. P., Sokol P. A. 2005. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia. J. Bacteriol. 187:5278–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brett P. J., DeShazer D., Woods D. E. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48(Pt. 1):317–320 [DOI] [PubMed] [Google Scholar]
- 7. Brett P. J., Deshazer D., Woods D. E. 1997. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol. Infect. 118:137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai J. J., Woo P. C., Lau S. K., Smith D. K., Yuen K. Y. 2006. Accelerated evolutionary rate may be responsible for the emergence of lineage-specific genes in Ascomycota. J. Mol. Evol. 63:1–11 [DOI] [PubMed] [Google Scholar]
- 9. Chan C. M., et al. 1996. Single-tube nested PCR in the diagnosis of tuberculosis. J. Clin. Pathol. 49:290–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chantratita N., et al. 2008. Loop-mediated isothermal amplification method targeting the TTS1 gene cluster for detection of Burkholderia pseudomallei and diagnosis of melioidosis. J. Clin. Microbiol. 46:568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen F., Mackey A. J., Stoeckert C. J., Jr., Roos D. S. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34:D363–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng A. C., Currie B. J. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins J. F., Coulson A. F., Lyall A. 1988. The significance of protein sequence similarities. Comput. Appl. Biosci. 4:67–71 [DOI] [PubMed] [Google Scholar]
- 14. Deepak R. N., Crawley B., Phang E. 2008. Burkholderia pseudomallei identification: a comparison between the API 20NE and VITEK2GN systems. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1):S42–S44 [DOI] [PubMed] [Google Scholar]
- 15. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fleischmann R. D., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512 [DOI] [PubMed] [Google Scholar]
- 17. Gee J. E., et al. 2003. Use of 16S rRNA gene sequencing for rapid identification and differentiation of Burkholderia pseudomallei and B. mallei. J. Clin. Microbiol. 41:4647–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gillespie S. H., Ullman C., Smith M. D., Emery V. 1994. Detection of Streptococcus pneumoniae in sputum samples by PCR. J. Clin. Microbiol. 32:1308–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glass M. B., Steigerwalt A. G., Jordan J. G., Wilkins P. P., Gee J. E. 2006. Burkholderia oklahomensis sp. nov., a Burkholderia pseudomallei-like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int. J. Syst. Evol. Microbiol. 56:2171–2176 [DOI] [PubMed] [Google Scholar]
- 20. Godoy D., et al. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henke W., Herdel K., Jung K., Schnorr D., Loening S. A. 1997. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 25:3957–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inglis T. J., Chiang D., Lee G. S., Chor-Kiang L. 1998. Potential misidentification of Burkholderia pseudomallei by API 20NE. Pathology 30:62–64 [DOI] [PubMed] [Google Scholar]
- 23. Inglis T. J., Merritt A., Chidlow G., Aravena-Roman M., Harnett G. 2005. Comparison of diagnostic laboratory methods for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 43:2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kiratisin P., Santanirand P., Chantratita N., Kaewdaeng S. 2007. Accuracy of commercial systems for identification of Burkholderia pseudomallei versus Burkholderia cepacia. Diagn. Microbiol. Infect. Dis. 59:277–281 [DOI] [PubMed] [Google Scholar]
- 25. Lee M. A., Wang D., Yap E. H. 2005. Detection and differentiation of Burkholderia pseudomallei, Burkholderia mallei and Burkholderia thailandensis by multiplex PCR. FEMS Immunol. Med. Microbiol. 43:413–417 [DOI] [PubMed] [Google Scholar]
- 26. Lipuma J. J. 2010. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 23:299–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lipuma J. J., et al. 1994. Inapparent transmission of Pseudomonas (Burkholderia) cepacia among patients with cystic fibrosis. Pediatr. Infect. Dis. J. 13:716–719 [DOI] [PubMed] [Google Scholar]
- 28. Mahenthiralingam E., et al. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller M. B., Gilligan P. H. 2003. Laboratory aspects of management of chronic pulmonary infections in patients with cystic fibrosis. J. Clin. Microbiol. 41:4009–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Novak R. T., et al. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J. Clin. Microbiol. 44:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Carroll M. R., et al. 2003. Burkholderia pseudomallei: another emerging pathogen in cystic fibrosis. Thorax 58:1087–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Payne G. W., et al. 2005. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 71:3917–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peacock S. J., et al. 2008. Management of accidental laboratory exposure to Burkholderia pseudomallei and B. mallei. Emerg. Infect. Dis. 14:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Read T. D., et al. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86 [DOI] [PubMed] [Google Scholar]
- 35. Schlech W. F., III, et al. 1981. Laboratory-acquired infection with Pseudomonas pseudomallei (melioidosis). N. Engl. J. Med. 305:1133–1135 [DOI] [PubMed] [Google Scholar]
- 36. Schmoock G., et al. 2009. DNA microarray-based detection and identification of Burkholderia mallei, Burkholderia pseudomallei and Burkholderia spp. Mol. Cell. Probes 23:178–187 [DOI] [PubMed] [Google Scholar]
- 37. Smith M. D., Angus B. J., Wuthiekanun V., White N. J. 1997. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect. Immun. 65:4319–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spilker T., et al. 2009. Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 47:2607–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 40. Thibault F. M., Valade E., Vidal D. R. 2004. Identification and discrimination of Burkholderia pseudomallei, B. mallei, and B. thailandensis by real-time PCR targeting type III secretion system genes. J. Clin. Microbiol. 42:5871–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tse H., et al. 2010. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J. Bacteriol. 192:1471–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. U'Ren J. M., et al. 2005. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 43:5771–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weissert C., Dollenmaier G., Rafeiner P., Riehm J., Schultze D. 2009. Burkholderia pseudomallei misidentified by automated system. Emerg. Infect. Dis. 15:1799–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wongtrakoongate P., Mongkoldhumrongkul N., Chaijan S., Kamchonwongpaisan S., Tungpradabkul S. 2007. Comparative proteomic profiles and the potential markers between Burkholderia pseudomallei and Burkholderia thailandensis. Mol. Cell. Probes 21:81–91 [DOI] [PubMed] [Google Scholar]
- 45. Woo P. C., et al. 2009. The complete genome and proteome of Laribacter hongkongensis reveal potential mechanisms for adaptations to different temperatures and habitats. PLoS Genet. 5:e1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woo P. C., et al. 2003. Seronegative bacteremic melioidosis caused by Burkholderia pseudomallei with ambiguous biochemical profile: clinical importance of accurate identification by 16S rRNA gene and groEL gene sequencing. J. Clin. Microbiol. 41:3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Woo P. C., et al. 2002. Streptococcus sinensis sp. nov., a novel species isolated from a patient with infective endocarditis. J. Clin. Microbiol. 40:805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Woo P. C., Woo G. K., Lau S. K., Wong S. S., Yuen K. 2002. Single gene target bacterial identification. groEL gene sequencing for discriminating clinical isolates of Burkholderia pseudomallei and Burkholderia thailandensis. Diagn. Microbiol. Infect. Dis. 44:143–149 [DOI] [PubMed] [Google Scholar]
- 49. Woods D. E. 1999. Species versus biotype status. J. Clin. Microbiol. 37:3786–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang X., Jawdy S., Tschaplinski T. J., Tuskan G. A. 2009. Genome-wide identification of lineage-specific genes in Arabidopsis, Oryza and Populus. Genomics 93:473–480 [DOI] [PubMed] [Google Scholar]
- 51. Yuen K. Y., et al. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 39:4227–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]