Abstract
Type I interferon protects cells from virus infection through the induction of a group of genes collectively named interferon-stimulated genes (ISGs). In this study, we utilized short hairpin RNA (shRNA) to deplete ISGs in SupT1 cells in order to identify ISGs that suppress the production of human immunodeficiency virus type 1 (HIV-1). Among the ISG candidates thus identified were interferon-induced transmembrane (IFITM) proteins, including IFITM1, IFITM2, and IFITM3, that potently inhibit HIV-1 replication at least partially through interfering with virus entry. Further mutagenesis analysis shows that the intracellular region, rather than the N- and C-terminal extracellular domains, is essential for the antiviral activity of IFITM1. Altogether, these data suggest that the IFITM proteins serve as important components of the innate immune system to restrict HIV-1 infection.
Mammalian cells produce type I interferon in response to virus infection (reviewed in references 8 and 25). Viral proteins and viral nucleic acids are detected by pathogen recognition receptors (PRRs) as pathogen-associated molecular patterns (PAMPs). These PRRs are either membrane associated, such as Toll-like receptor 3 (TLR3) and TLR7 on endosomes, or cytosolic, such as retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), nucleotide-binding oligomerization domain-containing 2 (NOD2), and absent in melanoma 2 (AIM2). These recognition events activate signaling transduction pathways that trigger production of type I interferon and other cytokines. Type I interferon then augments the expression of hundreds of genes, named interferon-stimulated genes (ISGs), that subvert virus replication by a variety of mechanisms (reviewed in references 50 and 53).
Human immunodeficiency virus type 1 (HIV-1) infection also triggers type I interferon production (17). Plasmacytoid dendritic cells, the major interferon producers, release large quantities of interferon, partially through recognition of HIV-1 RNA by TLR7 (23). Although the clinical benefit of engaging interferon in treating HIV-1 infection is uncertain due to the controversial results of clinical trials (16, 31, 51, 60), HIV-1 replication in cultured cells is clearly inhibited by interferon (4, 19, 22, 24, 48, 49, 68). This inhibition must have exerted selection pressure on HIV-1 in vivo in light of the multiple measures that the virus has evolved to evade interferon restriction. For example, HIV-1 infection causes a gradual decrease in the number of plasmacytoid dendritic cells, as well as a reduced capacity of these cells to produce interferon (13, 35, 62). In addition, HIV-1 infection causes depletion of interferon regulatory factor 3 (IRF-3), which plays a key role in Toll-like receptor- and RIG-I-mediated innate immune signaling (12, 45). Equally important, HIV-1 encodes proteins that antagonize individual ISGs. Examples are Vpu (viral protein u), which counteracts BST-2 (bone marrow stromal cell antigen 2, also known as tetherin/CD317/HM1.24) that otherwise blocks virus release from the cell surface (44, 64), and Vif (viral infectivity factor), which antagonizes APOBEC3G (apolipoprotein B mRNA editing enzyme 3G), a cytidine deaminase that causes hypermutation of HIV-1 cDNA (57).
Interferon impedes multiple steps of HIV-1 replication in both the early and late phases (1, 11, 18-22, 24, 28-30, 40, 49, 58, 59, 67, 68). These inhibition activities are the functions of distinct ISGs. For example, PKR (protein kinase R) inhibits HIV-1 production by suppressing protein translation (43). RNase L degrades HIV-1 RNA, thus restricting HIV-1 replication (36). TRIM22 (tripartite motif protein 22) disrupts Gag assembly (3). BST-2 blocks the release of HIV-1 particles from the cell surface (44, 64). ISG15 impedes HIV-1 virus production by causing ISGylation of viral Gag protein and cellular factors, such as CHMP5 (chromatin-modifying protein 5) (46, 47). APOBEC3G restricts HIV-1 replication by causing hypermutation of viral cDNA during reverse transcription (37, 57). TRIM5α from Old World monkeys targets viral core following HIV-1 entry and destroys the viral reverse transcription complex (37, 63). Although APOBEC3G and TRIM5α are constitutively expressed, their levels are elevated by interferon, which potentiates their antiviral function (10, 54). Recent studies by Brass et al. identified the interferon-induced transmembrane (IFITM) proteins as potent inhibitors of influenza A H1N1 virus, West Nile virus, and dengue virus in their genome-wide RNA interference (RNAi) screen (6). In this study, we further show that IFITM proteins also inhibit HIV-1 replication.
MATERIALS AND METHODS
Plasmid DNA, viruses, and antibodies.
The cDNA clones of IFITM1, IFITM2, and IFITM3 were kindly provided by Ju-Tao Guo (26). The cDNA sequences of these ifitm genes were amplified by PCR using primer pairs IFITM1-S/IFITM1-A, IFITM2-S/IFITM2-A, and IFITM3-S/IFITM3-A (the primer sequences are provided in Table S1 in the supplemental material). The PCR products were digested with restriction enzymes BamHI and EcoRI and inserted into the pRetroX-Tight-Pur retroviral vector (Clontech) to create DNA constructs Tet-IFITM1, Tet-IFITM2, and Tet-IFITM3. A Flag tag was attached to the N terminus of each IFITM. IFITM1 mutants were generated by PCR using primers that are listed in Table S1 in the supplemental material. The infectious HIV-1 proviral DNA clone BH10 was obtained from the NIH AIDS Research and Reference Reagent Program. The NLEY1-IRES (internal ribosome entry site) and the NLEY1-ES-IRES DNA constructs were kindly provided by David Levy (33) and the pCMV-BlaM-Vpr plasmid DNA by Warner Greene (9). HIV-1 stocks were generated by transfecting 293T cells with the proviral DNA clone BH10, NLEY1-IRES, or NLEY1-ES-IRES. Note that both the BH10 and NLEY1-IRES viruses are infectious, whereas the NLEY1-ES-IRES virus lacks HIV-1 envelope protein and needs to be pseudotyped for entry. When necessary, the vesicular stomatitis virus (VSV) glycoprotein (G) was used to pseudotype HIV-1 particles. The amounts of viruses in the stock were determined by measuring the activity of viral reverse transcriptase or the level of viral CA(p24) antigen.
Anti-Flag and anti-tubulin antibodies were purchased from Sigma; anti-HIV-1 p24 antibody from ID Lab Inc.; anti-IFITM1, anti-IFITM2, and anti-IFITM3 antibodies from Proteintech Group; and phycoerythrin (PE)-conjugated anti-human CD4 antibody from BD Biosciences. α2b interferon (IFN-α2b) and G418 were purchased from Invitrogen and puromycin and doxycycline from Sigma.
Microarrays.
SupT1 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% glutamine. Following treatment with IFN-α2b (1,000 U/ml) for 8 or 16 h, total cellular RNA was extracted using the Trizol reagent (Invitrogen). RNA samples were submitted to McGill University and Génome Québec Innovation Centre for microarray analysis using the Illumina expression BeadChip.
Screening for anti-HIV-1 ISGs.
Mission short hairpin RNA (shRNA) transduction particles were purchased from Sigma to knock down ISGs in SupT1 cells. Each ISG was targeted on average by 3 to 5 shRNA clones. A control shRNA that had the scrambled RNA target sequence was also purchased from Sigma. HIV-1 stock was prepared by transfecting 293T cells with a subtype B HIV-1 DNA named BH10, together with a plasmid expressing the glycoprotein (G) of VSV. VSV G protein was used to enhance the infection efficiency of wild-type HIV-1 particles, and the VSV G-containing wild-type HIV-1 particles were used only in screening experiments. The level of viruses was titrated by infecting TZM-bl indicator cells (65).
First, stable SupT1 cell lines were created to express each individual shRNA. This was achieved by infecting 1 × 106 SupT1 cells with 50 μl of the Mission shRNA transduction particles supplemented with Polybrene (5 μg/ml) in a 24-well plate. After 48 h, puromycin (2 μg/ml) was added to select for stably transduced cells. Cells (106) were then treated with IFN-α2b (500 U/ml) for 16 h before infection with HIV-1. After the inocula were removed, the cells were kept in culture for a further 24-hour period before the levels of infectious HIV-1 in the supernatants were measured by infecting the TZM-bl cells. An ISG was considered a candidate if more than two of its shRNA clones alleviated IFN-α2b-posed restriction by >2-fold. The candidate ISGs were then tested in a second round of HIV-1 infection for validation.
Creating doxycycline-inducible IFITM cell lines.
Retroviral particles were prepared by transfecting 1 μg of each of the Tet-IFITM1, Tet-IFITM2, and Tet-IFITM3 plasmid DNAs into the packaging cells GP2-293 (Clontech), together with 0.1 μg of the VSV G DNA. These viruses were used to infect SupT1 cells, together with viruses expressing the rtTA activator (Clontech). Stably transduced cell lines were selected in RPMI 1640 medium supplemented with puromycin (2 μg/ml), G418 (1 mg/ml), and tetracycline-free serum (Clontech). To check the expression of the IFITM proteins, cell lines were exposed to doxycycline (500 ng/ml) for 16 h before the cell lysates were collected and examined by Western blotting with an anti-Flag antibody (Sigma). Doxycycline-inducible IFITM MT-2 and 293 cell lines were created following the same protocol.
HIV-1 virion fusion assay.
The HIV-1 virion fusion assay was performed as described previously (9). HIV-1 particles containing β-lactamase-Vpr (BlaM-Vpr) chimera proteins were produced by transfecting 293T cells with 3 μg of BH10 DNA and 1 μg of pCMV-BlaM-Vpr DNA using Lipofectamine 2000 (Invitrogen).The virus particles thus generated bore HIV-1 envelope proteins gp120/gp41. After 48 h, the culture supernatants were first clarified by passing them through a 0.22-μm filter and then concentrated by ultracentrifugation at 100,000 × g for 1 h at 4°C. The pelleted viruses were suspended in complete Dulbecco's modified Eagle's medium (DMEM), aliquoted, and stored at −80°C.
SupT1 cell lines were exposed to doxycycline (500 ng/ml) for 16 h before spinoculation with HIV-1 particles containing BlaM-Vpr. After a 2-hour incubation at 37°C, the cells were washed with CO2-independent medium (Invitrogen) and loaded with CCF2/AM substrate (Invitrogen) by incubating the cells with 100 μl of loading solution for 1 h at room temperature in the dark. The loading solution was prepared by mixing 2 μl of CCF2/AM (1 nM) with 8 μl of 0.1% acetic acid containing 100 mg/ml Pluronic-F127surfactant (solution B, provided by Invitrogen with the CCF2/AM loading kit) and 1 ml of CO2-independent medium. Finally, the cells were washed with 200 μl of development medium, and the BlaM reaction was developed in 200 μl development medium for 16 h at room temperature in the dark. The development medium was prepared by mixing 10 μl of probenecid (250 mM) with 1 ml CO2-independent medium and 100 μl tetracycline-free FBS. After being washed once with cold 1× phosphate-buffered saline (containing 2% FBS), the cells were fixed with 1% paraformaldehyde. The levels of CCF2/AM and its cleaved products were measured by flow cytometry.
siRNA knockdown of IFITM proteins in TZM-bl cells.
Short interfering RNA (siRNA) oligonucleotides were purchased from Ambion to target IFITM1 (siRNA ID, s16192 [GGUCCACCGUGAUCAACAU]), IFITM2 (siRNA IDs, s20771 [CCACGUACUCUAUCUUCCA] and s230492 [GCCCUUGACCUGUAUUCCA]), and IFITM3 (siRNA ID, s195035 [CCCACGUACUCCAACUUCC]). TZM-bl cells were seeded in 24-well plates 1 day before siRNA transfection was performed with Lipofectamine 2000 (Invitrogen). Two siRNA mixtures were used to simultaneously deplete IFITM1, IFITM2, and IFITM3; these mixtures were s16192/s20771/s195035 and s16192/s230492/s195035. Ten nanomolar each siRNA was used in transfection. After two sequential siRNA transfections, the cells were exposed to the wild-type HIV-1 named BH10 for 2 h before the viruses were washed off. Luciferase activity in TZM-bl cells was measured 40 h later. The levels of IFITM proteins were assessed by Western blotting using anti-IFITM1, anti-IFITM2, and anti-IFITM3 antibodies, respectively.
Virus infection assays.
IFITM SupT1 cells (106 cells for each line) were treated with doxycycline (500 ng/ml) for 16 h before being infected with HIV-1 named NLEY1-IRES equivalent to 100 ng CA(p24). The NLEY1-IRES virus bears HIV-1 envelope protein, is infectious, and expresses the yellow fluorescent protein (YFP) (33). The inoculated viruses were washed off after an overnight incubation. After 24 h, the cells were washed with cold 1× phosphate-buffered saline and fixed with 1% paraformaldehyde (in 1× phosphate-buffered saline). YFP-positive cells were scored by flow cytometry. The amounts of viruses in the supernatants were determined by infecting TZM-bl indicator cells.
For spreading infection, 2 × 106 IFITM SupT1 cells were induced with doxycycline (500 ng/ml) for 16 h before being infected with wild-type HIV-1 (BH10) equivalent to 20 ng CA(p24). Viral replication was monitored by measuring the levels of viral reverse transcriptase activity in supernatants over different time intervals.
Measuring HIV-1 reverse transcription products.
SupT1 cells (2 × 106) were exposed to the wild-type BH10 viruses [equivalent to 100 ng CA(p24)] for 1 h at 37°C before the virus inocula were washed off. As a control, nevirapine (1 μM) was used to block reverse transcription. Cells were collected 16 h after infection, and total DNA was extracted with the DNeasy tissue kit (Qiagen). Real-time PCR was performed with equal amounts of DNA from different samples using the following primer pairs: 5′-TTAGACCAGATCTGAGCCTGGGAG-3′/5′-GGGTCTGAGGGATCTCTAGTTACC-3′ to amplify the early viral DNA and 5′-TGTGTGCCCGTCTGTTGTG-3′/5′-GAGTCCTGCGTCGAGA-3′ to amplify the late viral DNA (34). The reactions were performed with the LightCycler FastStart DNA Master SYBR green 1 system (Roche) in accordance with the manufacturer's instructions. The PCR conditions were 95°C for 10 s, 62°C for 5 s, and 72°C for 7 s. In order to measure the viral DNA that is integrated into cellular chromosomal DNA, the first round of PCR was performed with primers 5′-GCCTCCCAAAGTGCTGGGATTACAG-3′/5′-GTTCCTGCTATGTCAC TTCC-3′, which bind to Alu and HIV-1 Gag DNA sequences (Alu-gag PCR). The reactions were performed at 94°C for 1 min to denature DNA templates, followed by 12 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 3.3 min. The amplified DNA products were quantified in real-time PCR as described above with primers 5′-TTAAGCCTCAATAAAGCTTGCC-3′/5′-GTTCGGGCGCCACTGCTAGA-3′, which amplify the HIV-1 long terminal repeat (LTR) region (73).
Immunofluorescence microscopy.
293 cells were seeded into slide chambers prior to transfection with vector DNA expressing Flag-tagged IFITM1, IFITM2, and IFITM3. Forty hours after transfection, the cells were rinsed with serum-free DMEM and incubated with Alexa Fluor 555-conjugated transferrin (5 μg/ml in serum-free DMEM; Invitrogen) for 10 min before fixation in 4% paraformaldehyde (in 1× phosphate-buffered saline) for 10 min at room temperature. After permeabilization with 0.1% Triton X-100 for 10 min at room temperature and incubation in a blocking solution (2% FBS) for 2 h, the cells were stained with rabbit anti-Flag primary antibody (1:500 dilution; 2 h at room temperature) and Alexa Fluor 488-conjugated secondary anti-rabbit antibody (1:2,000 dilution; 40 min at room temperature). Images were taken using the Zeiss Pascal laser scanning confocal microscope.
RESULTS
Knockdown of IFITM1 increases HIV-1 production in the presence of IFN-α2b.
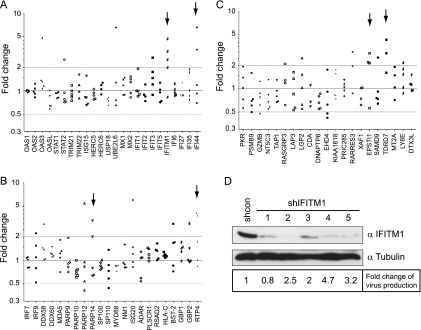
With an aim to identify ISGs that have anti-HIV-1 activity, we first tested the effect of IFN-α2b on HIV-1 production in several human cell lines, as well as in cord blood mononuclear cells (CBMCs), in order to find a cell line that was suitable for the screening assay. The results showed a greater than 100-fold decrease in HIV-1 production in both SupT1 cells and CBMCs following IFN-α2b treatment, as opposed to a less pronounced HIV-1 reduction in Jurkat, U937, and 293T cells (see Fig. S1 in the supplemental material). We then proceeded with SupT1 cells and performed microarray analysis to determine the genes that were upregulated by IFN-α2b. A total of 94 genes showed a more than 2-fold increase in their mRNA expression (see Table S2 in the supplemental material). In order to determine which of these ISGs inhibit HIV-1 production, we used the Mission shRNA products from Sigma to knock down each individual ISG. Sixty-seven out of these 94 ISGs were covered by shRNA clones that were available at Sigma (see Table S2 in the supplemental material). We first created stably transduced shRNA SupT1 cell lines, including a scrambled shRNA cell line as the control, and then treated these cells with IFN-α2b for 16 h before infection with wild-type HIV-1 (BH10). Production of infectious HIV-1 particles was monitored by infecting the TZM-bl indicator cells (65). An ISG was selected as an anti-HIV-1 candidate if at least two of its shRNA clones increased virus production by >2-fold over the control cell line in the presence of IFN-α2b. Six such ISGs were identified: IFITM1, IFI44, PARP14, RTP4, EPSTI1, and TDRD7 (Fig. 1A to C). Further experiments showed that IFITM1 was substantially downregulated by each of the five shRNA clones and that four out of these five shRNAs increased HIV-1 production by >2-fold (Fig. 1D). It is notable that interferon-mediated inhibition of HIV-1 production was not fully alleviated by knocking down any of the 67 ISGs. It is therefore possible that the ISG with the strongest inhibition activity was not covered in the 67 ISGs tested in this study or that multiple ISGs were needed to strongly suppress HIV-1 infection.
FIG. 1.
(A to C) Effect of ISG knockdown on HIV-1 production in the presence of IFN-α2b. Each ISG was targeted with 3 to 5 shRNA clones. The shRNA-transduced SupT1 cells were treated with IFN-α2b for 16 h before being infected with a subtype B HIV-1 (BH10). Production of infectious HIV-1 particles was determined by infecting TZM-bl indicator cells. The fold change for each shRNA was calculated by dividing the amount of infectious HIV-1 made from the shRNA SupT1 cells by that from the control shRNA SupT1 cells. The results are summarized in three graphs. The candidate genes are highlighted by arrows. (D) Western blots of SupT1 cells that were stably transduced with either the control shRNA vector or the shIFITM1 clones. Cells were treated with IFN-α2b (500 U/ml) for 20 h prior to Western blotting. The fold change in HIV-1 production for each shIFITM1 clone is shown. The results shown are the averages of two independent infection experiments.
IFITM proteins diminish HIV-1 replication in SupT1 cells.
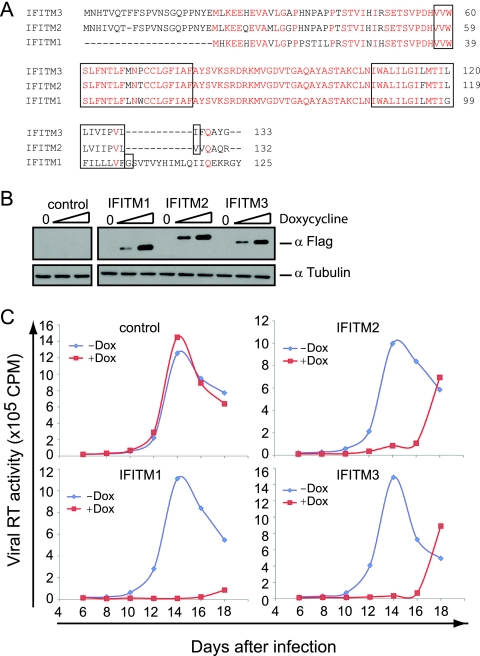
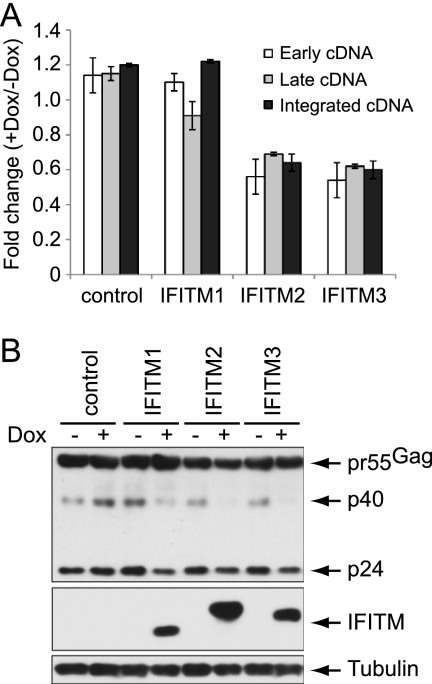
In order to assess the anti-HIV-1 activity of an individual ISG more specifically, we utilized the Tet-On expression system and generated SupT1 stable cell lines that were able to express a single ISG in response to tetracycline (or doxycycline). With this approach, we found that doxycycline-induced IFITM1 strongly inhibited HIV-1 replication in SupT1 cells (Fig. 2A, B, and C), which is consistent with the increased HIV-1 production seen with shRNA knockdown of IFITM1 (Fig. 1D). IFITM1 has three homologs in the human genome, i.e., IFITM2, IFITM3, and IFITM5 (38, 42). Unlike IFITM1, -2, and -3, IFITM5 is solely expressed in osteoblasts and is involved in bone mineralization (42); thus, it was not further investigated in this study. When the effects of IFITM2 and IFITM3 on HIV-1 replication were examined using the Tet-On system, we found that IFITM2 and IFITM3 also markedly suppressed HIV-1 infection, albeit to a lesser extent than IFITM1 (Fig. 2C). Further experiments showed that the induced expression of IFITM1, IFITM2, or IFITM3 did not affect cell proliferation, the cell cycle, or cell surface expression of CD4 (see Fig. S2 in the supplemental material), supporting a likely direct inhibition of HIV-1 replication by IFITM1, IFITM2, and IFITM3.
FIG. 2.
IFITM proteins inhibit HIV-1 replication in SupT1 cells. (A) Illustration of the domain structures of IFITM proteins. The transmembrane domains are highlighted in boxes. A Flag tag was attached to the N termini of IFITM proteins. The conserved amino acid residues are highlighted in red letters. (B) Doxycycline-induced expression of IFITM proteins in SupT1 cells. Stably transduced SupT1 cells were exposed to different amounts of doxycycline (0, 50, and 500 ng/ml) for 16 h before the cell lysates were harvested and subjected to Western blotting using anti-Flag antibodies to detect the expression of IFITM proteins. Control represents a cell line that was stably transduced with the empty retroviral vector. (C) HIV-1 replication in IFITM-expressing SupT1 cells. The cells were treated with doxycycline (Dox) (500 ng/ml) for 16 h before they were exposed to wild-type HIV-1. Virus production over different time intervals was monitored by measuring viral reverse transcriptase activity in the culture supernatants. The results shown represent four independent infection experiments.
IFITM2 and IFITM3 impede HIV-1 entry.
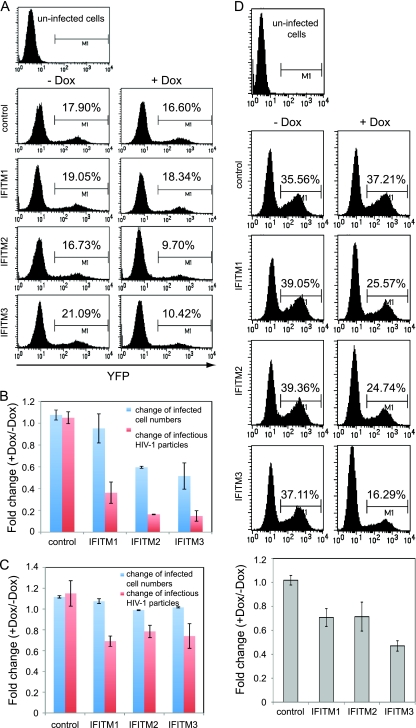
We next asked at what step(s) HIV-1 replication was affected by IFITM proteins. To answer this question, we used in the subsequent one-cycle infection assays an infectious HIV-1 reporter virus named NLEY1-IRES, which expresses YFP (33). Cells were first treated with doxycycline to induce IFITM expression and then infected with the NLEY1-IRES virus for 40 h. The number of YFP-positive cells was scored by flow cytometry (Fig. 3A), and the amounts of infectious viruses in supernatants were determined by infecting the TZM-bl indicator cells (Fig. 3B). The results showed that IFITM2 and IFITM3 reduced the number of infected cells by 40 to 50% and decreased the amounts of infectious viruses by 4- to 5-fold; interestingly, IFITM1 exerted a less pronounced inhibition effect (Fig. 3A and B). Similar observations were made in MT-2 cells (see Fig. S3 in the supplemental material). When SupT1 cells were infected by the NLEY1-IRES virus prior to the addition of doxycycline, neither the number of infected cells nor the amounts of infectious HIV-1 particles were evidently affected by any of the induced IFITM proteins (Fig. 3C). These results indicate that IFITM proteins likely target an early step in HIV-1 replication. In support of this notion, HIV-1 production was not affected by ectopic expression of IFITM proteins in 293 cells that were transfected with HIV-1 DNA clone BH10 (data not shown). We also measured the effect of IFITM proteins on infection by HIV-1 NLEY1-ES-IRES that was pseudotyped with VSV G protein. The results revealed that IFITM3 reduced virus infection by approximately 55% whereas IFITM1 and IFITM2 led to 30% reduction (Fig. 3D). The more dramatic impact of IFITM3 on VSV G protein-mediated infection is consistent with a recent report showing that IFITM3 inhibits VSV infection by targeting distinct steps of the virus life cycle (66).
FIG. 3.
IFITM proteins inhibit an early step of HIV-1 replication. (A) IFITM2 and IFITM3 diminished the number of HIV-1-infected cells. Following doxycycline treatment (0 and 500 ng/ml) for 16 h, SupT1 cells were infected with the NLEY1-IRES virus, which expresses YFP. Forty hours after infection, the number of YFP-positive cells was scored by flow cytometry. (B) IFITM proteins suppressed production of infectious HIV-1 particles. The amounts of infectious virus particles were determined by infecting TZM-bl indicator cells. The fold change was calculated by dividing the values from doxycycline-treated cells (+Dox) by the values from untreated cells (−Dox). The results shown are the averages of three independent infection experiments shown in panel A. The error bars indicate standard deviations. (C) SupT1 cells were infected with the NLEY1-IRES virus for 2 h before exposure to doxycycline (500 ng/ml) treatment. The effects of IFITM proteins on the number of infected cells and the amounts of infectious HIV-1 are summarized in the bar graph. (D) The IFITM SupT1 cell lines were first treated with Dox (500 ng/ml) for 16 h and then infected with NLEY1-ES-IRES viruses that were pseudotyped with VSV G protein. The number of YFP-positive cells was scored 40 h after infection. The results of three independent infections are summarized in the bar graph.
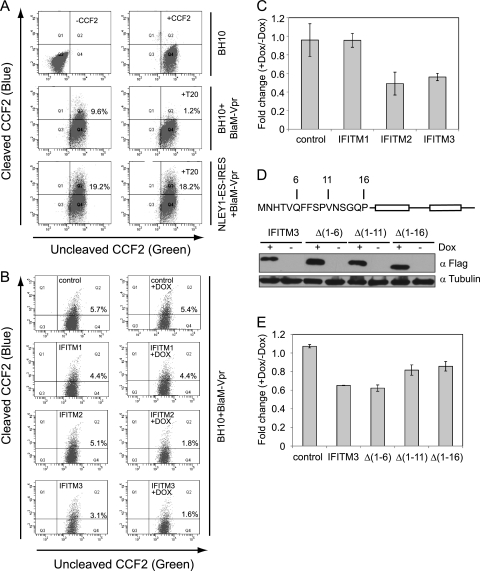
We then performed a HIV-1 virion fusion assay to test whether IFITM proteins affect virus entry. The fusion virus particles were prepared by transfecting 293T cells with an infectious HIV-1 proviral DNA clone, BH10, together with BlaM-Vpr DNA. The HIV-1 entry inhibitor T20 (1 μg/ml) effectively blocked the entry of BH10/BlaM-Vpr viruses but did not affect entry mediated by VSV G protein (Fig. 4A). When the BH10/BlaM-Vpr viruses were used to infect the IFITM cell lines, IFITM2 and IFITM3 diminished HIV-1 entry by 2- to 3-fold, whereas IFITM1 did not exert any such effect (Fig. 4B and C). Since IFITM1 differs from IFITM2 and IFITM3 mainly by the lack of an amino acid stretch in the N-terminal region, we speculated that the elongated N-terminal region may allow IFITM2 and IFITM3 to affect virus entry. To test this, we created three IFITM3 mutants named Δ(1-6), Δ(1-11), and Δ(1-16) that lacked various lengths of the N-terminal sequences (Fig. 4D). The results of HIV-1 virion fusion assays showed that the Δ(1-6) mutant and the wild-type IFITM3 diminished virus entry by about 40% as opposed to a 20% reduction caused by the Δ(1-11) and the Δ(1-16) mutants (Fig. 4E), suggesting a role of the 7-FFSPV-11 peptide in restricting virus entry. Taken together, these data suggest that IFITM2 and IFITM3 interfere with HIV-1 entry, which represents one mechanism underlying their anti-HIV-1 activities.
FIG. 4.
Effect of IFITM proteins on HIV-1 entry. (A) T20 blocks the infection of SupT1 cells by the BH10/BlaM-Vpr viruses. SupT1 cells were infected with the BH10/BlaM-Vpr or the NLEY1-ES-IRES/VSV G/BlaM-Vpr viruses in the absence or presence of the entry inhibitor T20 (1 μg/ml) for 2 h before cells were harvested to detect BlaM activity. The numbers of cells with successful HIV-1 entries are scored in windows Q2. (B and C) IFITM SupT1 cell lines were treated with Dox (500 ng/ml) for 16 h before being infected with BH10/BlaM-Vpr viruses. The results of three independent HIV-1/BlaM-Vpr fusion experiments are summarized in the bar graph. The error bars indicate standard deviations. (D) Illustration of IFITM3 mutations. The N-terminal sequences of IFITM3 were deleted to create mutants Δ(1-6), Δ(1-11), and Δ(1-16). Their expression in the Dox-inducible SupT1 cell lines was examined in Western blots. (E) The Δ(1-11) and Δ(1-16) mutants are less effective in inhibiting HIV-1 entry than the wild-type IFITM3. The results of three independent HIV-1 fusion experiments are shown in the bar graph.
IFITM2 and IFITM3 associate with endocytosis.
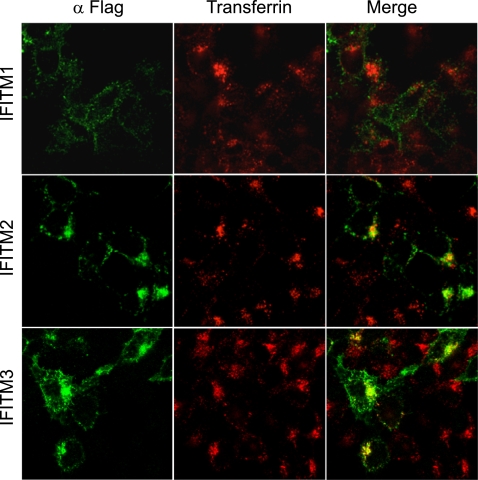
Virus entry involves binding to the receptor molecules on cell surfaces and subsequent endocytosis of virus particles (39). Recent studies suggest that HIV-1 entry also involves the endocytosis process (41). Different viruses escape the endosome pathway at different stages. Some viruses, such as influenza virus, travel to the late endosomes to accomplish membrane fusion. We envision that in order to elicit a direct effect on virus entry, IFITM proteins may need to associate with the endocytosis pathway. To test this, we fed cells with Alexa Fluor 555-conjugated transferrin and determined whether IFITM proteins colocalized with the endocytosed transferrin. Although IFITM1 exhibited barely any colocalization with transferrin, a strong association was observed between the endocytosed transferrin and IFITM2 or IFITM3 (Fig. 5). This result suggests that IFITM2 and IFITM3, but not IFITM1, associate with the endocytosis pathway and may exert an effect on the fate of the endocytosed cargos, such as virus particles.
FIG. 5.
Association of IFITM2 and IFITM3 with endocytosis. 293 cells were transiently transfected with vector DNA that expressed Flag-tagged IFITM1, IFITM2, or IFITM3. Forty hours after transfection, the cells were fed with Alexa Fluor 555-conjugated transferrin (red) at 37°C for 10 min before being fixed with 4% paraformaldehyde (in 1× phosphate-buffered saline). IFITM proteins (green) were detected by immunostaining them with anti-Flag antibody.
IFITM1 suppresses HIV-1 Gag production.
IFITM1 does not affect HIV-1 entry yet efficiently inhibits virus replication in SupT1 cells. In order to gain insights into this restriction activity of IFITM1, we first measured the effect of IFITM1 on HIV-1 reverse transcription by real-time PCR. Viral DNA was not detected by PCR in SupT1 cells that were infected by HIV-1 in the presence of a reverse transcriptase inhibitor named nevirapine (1 μM) (data not shown), which validates the idea that the amplified DNA products from HIV-1-infected cells in the absence of nevirapine treatment are reverse transcribed viral DNA. Consistent with the 40% to 50% decrease in virus entry caused by IFITM2 and IFITM3, the early and the late HIV-1 cDNA products diminished by similar magnitudes (Fig. 6A). No effect of IFITM1 was observed on the production of HIV-1 cDNA (Fig. 6A). We next performed Alu-gag PCR to assess the level of HIV-1 DNA that was integrated into cellular chromosomal DNA (73). The results did not reveal any effect of IFITM1 on HIV-1 DNA integration (Fig. 6A). Interestingly, when HIV-1 Gag protein and its processing products, including p40 and p24, were examined in Western blots, a significant reduction was detected in SupT1 cells that expressed any of the three IFITM proteins (Fig. 6B), suggesting that IFITM1, as well as IFITM2 and IFITM3, adversely affect viral Gag expression, which must have led to diminished virus production.
FIG. 6.
Effects of IFITM proteins on HIV-1 cDNA synthesis and viral Gag expression. (A) IFITM-inducible SupT1 cells were infected with HIV-1 (BH10). Real-time PCR was performed using primers that amplify either the early or the late viral cDNA products. The levels of integrated viral DNA were determined by Alu-gag PCR. The results shown are the averages of three independent infections. The error bars indicate standard deviations. (B) Western blots of HIV-1 Gag protein expression in IFITM-inducible SupT1 cells that were infected with HIV-1 (BH10). The results shown represent three independent infection experiments.
The C-terminal sequence of IFITM1 regulates its antiviral activity.
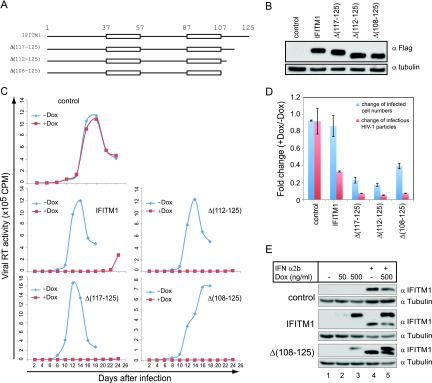
It is interesting that IFITM1 does not affect HIV-1 entry but inhibits HIV-1 replication in SupT1 cells more severely than IFITM2 and IFITM3. In order to understand the antiviral activity of IFITM1, we performed mutagenesis studies to determine which domains of IFITM1 are critical for inhibiting HIV-1 replication. IFITM1 can be divided into five domains interspersed with its two transmembrane domains (Fig. 7A). We first deleted the C-terminal sequence of IFITM1 and generated three mutants named Δ(117-125), Δ(112-125), and Δ(108-125) (Fig. 7A and B). The results of spreading-infection assays showed that these three mutants inhibited HIV-1 replication in SupT1 cells even more effectively than did the wild-type IFITM1 (Fig. 7C). Consistent with this, these mutants reduced the number of infected cells and the amounts of infectious virus particles to a greater degree than the wild-type IFITM1 (Fig. 7D). In order to exclude the possibility that the increased antiviral activities of these IFITM1 mutants were a result of their artificially high expression, we compared the doxycycline-induced expression of one mutant, Δ(108-125), to that of the IFN-α2b-induced endogenous IFITM1. Comparable levels in Δ(108-125) and endogenous IFITM1 were shown by the results of Western blotting (Fig. 7E). These data suggest not only that the C-terminal sequence of IFITM1 is dispensable for the antiviral activity of IFITM1, but that its presence may impair the antiviral function of the protein.
FIG. 7.
The anti-HIV-1 activity of IFITM1 does not depend on its C-terminal sequence. (A) Illustration of IFITM1 deletion mutants. Sequences from the IFITM1 C terminus were deleted. (B) Doxycycline-induced expression of IFITM1 mutants in SupT1 cells. (C) HIV-1 replication in SupT1 cells that express IFITM1 mutants. SupT1 cells were cultured in media with or without doxycycline (500 ng/ml). The results shown represent three independent spreading-infection experiments. (D) Infection of SupT1 cells with the NLEY1-IRES virus. For details, refer to the legend to Fig. 3B. (E) Comparison of doxycycline-induced expression of IFITM1 and Δ(108-125) to that of IFN-α2b-induced endogenous IFITM1. Cell lines were exposed to doxycycline (500 ng/ml), IFN-α2b (500 U/ml), or both for 16 h before the levels of IFITM1 and Δ(108-125) were assessed in Western blots using anti-IFITM1 antibody. Tubulin was probed as the internal control.
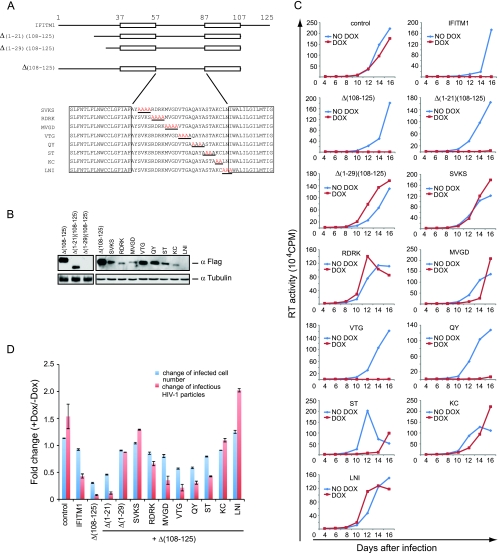
We next assessed the role of the N-terminal sequence of IFITM1 in inhibiting HIV-1 replication. Two mutants were accordingly created by deleting either 21 or 29 amino acids from the N terminus in the context of the Δ(108-125) deletion that not only inhibited HIV-1 replication more strongly than wild-type IFITM1, but also severely diminished the number of infected cells in the one-cycle infection assay. Despites its low expression level, the Δ(1-21)/Δ(108-125) mutant inhibited HIV-1 replication as efficiently as the wild-type IFITM1 and the Δ(108-125) mutant (Fig. 8). HIV-1 replication was not affected by the Δ(1-29)/Δ(108-125) mutant due to the loss of its expression (Fig. 8). These results suggest that at least the first 21 amino acids of the N-terminal region are not required for IFITM1 to inhibit HIV-1 replication.
FIG. 8.
Roles of the N-terminal sequence and the loop sequence of IFITM1 in suppressing HIV-1 infection. (A) Illustration of IFITM1 mutants. All mutants lack the C-terminal sequence from amino acids 108 to 125. Red letters indicate the mutated amino acids. The mutated peptide sequence for each location is underlined. The transmembrane domains are shown in boxes. (B) Expression of the IFITM1 mutants in stably transduced SupT1 cell lines upon treatment with doxycycline (500 ng/ml). (C) Replication of HIV-1 in SupT1 cell lines that express IFITM1 mutants. The results shown represent three independent spreading-infection experiments. (D) Effects of IFITM1 mutants on HIV-1 infection in one-cycle infection assays. For details, see the legend to Fig. 3B.
The intracellular region of IFITM1 was also subjected to mutagenesis analysis by changing every 4 amino acids to alanines. Eight mutants were created in this way (Fig. 8A). The results of Western blots showed that these mutants were not equally well expressed (Fig. 8B). Nevertheless, the VTG and QY mutants suppressed HIV-1 replication as effectively as the wild-type IFITM1 and the Δ(108-125) mutant (Fig. 8C and D). MVGK and ST partially repressed HIV-1 replication, whereas the SVKS, RDRK, KC, and LNI mutants did not exert an evident inhibitory effect (Fig. 8C and D). However, given the low expression levels of these six mutants, the data are insufficient to conclude that there is a direct involvement of the mutated amino acids in inhibiting HIV-1 infection. Nonetheless, the results do suggest that the amino acids altered in the VTG and QY mutants do not have a role in the IFITM1 anti-HIV-1 function.
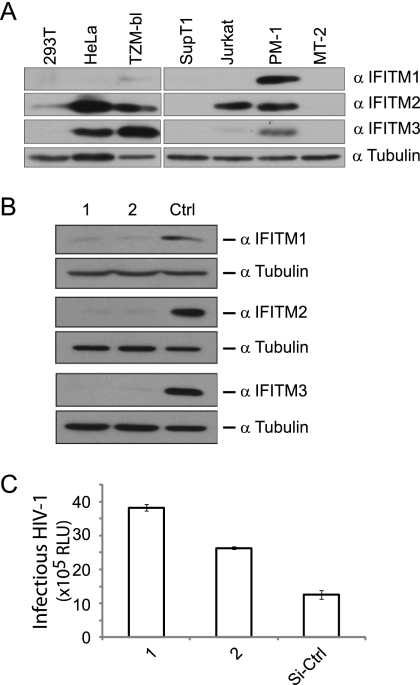
Knockdown of IFITM proteins increases susceptibility of TZM-bl cells to HIV-1 infection.
IFITM proteins were recently shown to inhibit infection by influenza A virus, West Nile virus, and dengue virus (6, 27). However, an inhibition of HIV-1 infection was not measured in the HeLa-CD4+ cells when IFITM3 was knocked down with siRNA oligonucleotides (6). Consistent with this, we also failed to observe an increase in HIV-1 infection when depleting each of the three IFITM proteins in the TZM-bl cells (data not shown). However, we subsequently noted that TZM-bl, which is a HeLa-derived HIV-1 indicator cell line (65), expresses considerable amounts of IFITM2 and IFITM3 proteins (Fig. 9A). Since both IFITM2 and IFITM3 inhibited HIV-1 entry, we decided to deplete all three IFITM proteins by transfecting the TZM-bl cells with siRNA oligonucleotide mixtures that target all three ifitm genes (Fig. 9B). Indeed, this treatment led to a 2- to 3-fold increase in HIV-1 infection (Fig. 9C), which suggests that the endogenous IFITM proteins play a role in restricting HIV-1 infection.
FIG. 9.
Knockdown of IFITM1, IFITM2, and IFITM3 increases HIV-1 infection of TZM-bl cells. (A) Expression of endogenous IFITM1, IFITM2, and IFITM3 in different cell lines. Western blots were probed with anti-IFITM1, -IFITM2, or -IFITM3 antibody. (B) Western blots to assess the knockdown of IFITM1, IFITM2, and IFITM3 in TZM-bl cells that were transfected with mixtures of siRNA oligonucleotides. (C) Following transfection with siRNA oligonucleotides, TZM-bl cells were exposed to HIV-1 infection. The levels of luciferase activity in the TZM-bl cell lysates were measured 40 h after infection. The values (relative luciferase units [RLU]) represent the susceptibility of TZM-bl cells to HIV-1 infection under different siRNA treatments. The results shown are the averages of three independent experiments.
DISCUSSION
There are four human ifitm genes, i.e., ifitm1, ifitm2, ifitm3, and ifitm5 (38, 42). IFITM5 is expressed in osteoblasts and plays a role in bone mineralization (42). IFITM1, IFITM2, and IFITM3 are ubiquitously expressed, with IFITM1 and IFITM3 responding to interferon stimulation due to the presence of a functional interferon stimulation response element (ISRE) in their promoters (38). IFITM proteins have been reported to play a role in a number of cell functions, such as oncogenesis (7, 15), cell adhesion (14), and immune cell signaling (5). High levels of IFITM proteins have also been observed in certain types of tumors, such as colorectal tumors and astrocytoma (2, 56). Nonetheless, these proteins do not seem to play a vital role in embryogenesis, since mice that are knocked out for all ifitm genes appear to develop normally (32). Encoded by interferon-stimulated genes, the antiviral functions of IFITM proteins have not been well appreciated until the recent discovery of their strong inhibition activities against influenza A virus, West Nile virus, dengue virus, and vesicular stomatitis virus (6, 27, 66). Studies also show that palmitoylation of the cysteine residues is required for the antiviral function of IFITM3 (72). Our current study demonstrates that IFITM proteins also inhibit HIV-1 infection, thus expanding the spectrum of viruses under the control of this small ISG family.
One mechanism behind the restriction activity of IFITM proteins involves impeding virus entry. Studies by Brass and colleagues have shown that murine leukemia virus (MLV) particles that were pseudotyped with envelope proteins from influenza A virus, West Nile virus, and dengue virus, but not from alphaviruses, were sensitive to restriction by IFITM proteins (6). One implication of this observation is that IFITM proteins inhibit the entry step that is mediated by the envelope proteins of certain viruses. This function of IFITM is now supported by the results of an assay that directly measures HIV-1 entry (Fig. 4).
Brass and colleagues depleted the endogenous IFITM3 protein in a CD4+ HeLa cell line and did not observe an effect on infection by an HIV-1 IIIb strain (6). This is most likely because HeLa cells express all three IFITM proteins, and each IFITM protein is a potent inhibitor of HIV-1 infection. Indeed, when we performed similar experiments in TZM-bl cells, which are HeLa-derived CD4-expressing cells, an increase in HIV-1 infection was observed only when all three IFITM proteins were simultaneously depleted by siRNA oligonucleotides (Fig. 9).
IFITM proteins are membrane associated, a feature that is considered necessary for directly interfering with virus entry (6). The IFITM protein family is characterized by two transmembrane domains and a highly conserved intracellular region. The N and C termini project away from the plasma membrane toward the extracellular environment (6, 61, 66). Interestingly, this topological presentation inversely mirrors that of BST-2, which inhibits the release of a variety of enveloped viruses from the cell surface (reviewed in reference 55). Although the molecular mechanism behind the IFITM restriction action remains to be elucidated, it seems unlikely that it directly targets viral envelope proteins or viral receptors, given the different viruses that IFITM proteins inhibit. Since virus entry often proceeds through endocytosis (reviewed in reference 39), it is possible that IFITM proteins are involved in endocytosis, such as by detecting and eliminating virus invaders before infection is established. In support of this speculation, it has been shown that IFITM1 is associated with the CD81-antibody complexes when they are endocytosed by cells (61). Our results further demonstrate an association of IFITM2 and IFITM3 with endocytosed transferrin (Fig. 5).
Among the viruses tested thus far, arenaviruses are resistant to restriction by IFITM proteins (6). This suggests that arenavirus envelope proteins may have a mechanism to evade IFITM restriction. Entry of arenaviruses, such as Lassa virus (LASV), lymphocytic choriomeningitis virus (LCMV), and machupo virus (MACH), begins with endocytosis at the plasma membrane and ends at the late endosome with low-pH-triggered membrane fusion (reviewed in reference 52). It is noted that LASV and LCMV exploit alpha-dystroglycan (alpha-DG) as the receptor and enter cells via an endocytosis process that is independent of clatherin, dynamin, and caveolin, whereas MACH uses transferrin receptor 1 to enter cells via clatherin-mediated endocytosis. In addition, the glycoproteins (GPs) of arenaviruses contain a stable signal peptide (SSP) of unusual length that becomes an integral component of the mature GP complex (including GP1, GP2, and SSP) (69-71). This SSP component may have a role in communication between the cytoplasmic domain (CTD) and the fusion-active ectodomain, which represents a unique feature of arenavirus GP-mediated entry. Further studies should determine which of these features or other unknown properties of arenavirus GPs allow these viruses to be resistant to IFITM proteins.
Targeting virus entry may not be the only mechanism by which IFITM proteins inhibit HIV-1 replication. For example, the 2- to 3-fold decrease in entry as a result of IFITM2 or IFITM3 expression does not seem to fully account for the over 5-fold decrease in HIV-1 production as measured in the one-cycle virus infection assay (Fig. 3B). It is possible that the results of the virion fusion assay may have underestimated the impact of IFITM proteins on virus entry in the context of a natural HIV-1 infection, during which cells are exposed to a dose of viruses much lower than the large quantity of concentrated HIV-1 particles that were used for the virion fusion assay. It is also notable that IFITM1 does not affect HIV-1 entry yet strongly inhibits HIV-1 replication. The latter observation adds to the argument that IFITM proteins may affect more than one step of HIV-1 replication.
The effect of interferon on HIV-1 entry was recently assessed by Goujon and Malim in a number of human immune cell lines (20). Their results revealed a moderate inhibitory effect in CD4+ T cells, which is, to some extent, consistent with the 2- to 3-fold reduction in HIV-1 entry seen with IFITM2 or IFITM3 in our study (Fig. 4). Given the constitutive expression of IFITM proteins in different cell lines (Fig. 9A), the exact impact of IFITM proteins on HIV-1 entry awaits further experimentation using IFITM knockout cells.
In summary, our results demonstrate that IFITM proteins restrict HIV-1 infection. Given the evidence that other viruses, including influenza A virus, West Nile virus, and dengue virus, are inhibited by IFITM proteins, this small ISG family emerges as an integral component of the interferon-mediated innate immune system that helps to defend against important human viral pathogens.
Supplementary Material
Acknowledgments
We thank Warner Greene, David Levy, and Ju-Tao Guo for providing valuable reagents and Vicky Cheng for critically reading the manuscript.
This work was supported by funding from the Canadian Institutes of Health Research and the Canadian Foundation for AIDS Research.
Footnotes
Published ahead of print on 22 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agy, M. B., R. L. Acker, C. H. Sherbert, and M. G. Katze. 1995. Interferon treatment inhibits virus replication in HIV-1- and SIV-infected CD4+ T-cell lines by distinct mechanisms: evidence for decreased stability and aberrant processing of HIV-1 proteins. Virology 214:379-386. [DOI] [PubMed] [Google Scholar]
- 2.Andreu, P., et al. 2006. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 66:1949-1955. [DOI] [PubMed] [Google Scholar]
- 3.Barr, S. D., J. R. Smiley, and F. D. Bushman. 2008. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 4:e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bednarik, D. P., J. D. Mosca, N. B. Raj, and P. M. Pitha. 1989. Inhibition of human immunodeficiency virus (HIV) replication by HIV-trans-activated alpha 2-interferon. Proc. Natl. Acad. Sci. U. S. A. 86:4958-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradbury, L. E., G. S. Kansas, S. Levy, R. L. Evans, and T. F. Tedder. 1992. The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. J. Immunol. 149:2841-2850. [PubMed] [Google Scholar]
- 6.Brass, A. L., et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brem, R., K. Oraszlan-Szovik, S. Foser, B. Bohrmann, and U. Certa. 2003. Inhibition of proliferation by 1-8U in interferon-alpha-responsive and non-responsive cell lines. Cell. Mol. Life Sci. 60:1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan, K., and A. G. Bowie. 2010. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 13:503-507. [DOI] [PubMed] [Google Scholar]
- 9.Cavrois, M., J. Neidleman, M. Bigos, and W. C. Greene. 2004. Fluorescence resonance energy transfer-based HIV-1 virion fusion assay. Methods Mol. Biol. 263:333-344. [DOI] [PubMed] [Google Scholar]
- 10.Chen, K., et al. 2006. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 80:7645-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dianzani, F., et al. 1998. Effects of IFN alpha on late stages of HIV-1 replication cycle. Biochimie 80:745-754. [DOI] [PubMed] [Google Scholar]
- 12.Doehle, B. P., F. Hladik, J. P. McNevin, M. J. McElrath, and M. Gale, Jr. 2009. Human immunodeficiency virus type 1 mediates global disruption of innate antiviral signaling and immune defenses within infected cells. J. Virol. 83:10395-10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaghy, H., et al. 2001. Loss of blood CD11c(+) myeloid and CD11c(-) plasmacytoid dendritic cells in patients with HIV-1 infection correlates with HIV-1 RNA virus load. Blood 98:2574-2576. [DOI] [PubMed] [Google Scholar]
- 14.Evans, S. S., R. P. Collea, J. A. Leasure, and D. B. Lee. 1993. IFN-alpha induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J. Immunol. 150:736-747. [PubMed] [Google Scholar]
- 15.Evans, S. S., D. B. Lee, T. Han, T. B. Tomasi, and R. L. Evans. 1990. Monoclonal antibody to the interferon-inducible protein Leu-13 triggers aggregation and inhibits proliferation of leukemic B cells. Blood 76:2583-2593. [PubMed] [Google Scholar]
- 16.Fernandez-Cruz, E., et al. 1995. Zidovudine plus interferon-alpha versus zidovudine alone in HIV-infected symptomatic or asymptomatic persons with CD4+ cell counts > 150 x 10(6)/L: results of the Zidon trial. Zidon Study Group. AIDS 9:1025-1035. [PubMed] [Google Scholar]
- 17.Francis, M. L., and M. S. Meltzer. 1993. Induction of IFN-alpha by HIV-1 in monocyte-enriched PBMC requires gp120-CD4 interaction but not virus replication. J. Immunol. 151:2208-2216. [PubMed] [Google Scholar]
- 18.Gendelman, H. E., et al. 1990. Restriction of HIV replication in infected T cells and monocytes by interferon-alpha. AIDS Res. Hum. Retroviruses 6:1045-1049. [DOI] [PubMed] [Google Scholar]
- 19.Gendelman, H. E., et al. 1990. Regulation of HIV replication in infected monocytes by IFN-alpha. Mechanisms for viral restriction. J. Immunol. 145:2669-2676. [PubMed] [Google Scholar]
- 20.Goujon, C., and M. H. Malim. 2010. Characterization of the interferon-{alpha} induced post-entry block to HIV-1 infection in primary human macrophages and T cells. J. Virol. 84:9254-9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen, B. D., et al. 1992. Loss of infectivity by progeny virus from alpha interferon-treated human immunodeficiency virus type 1-infected T cells is associated with defective assembly of envelope gp120. J. Virol. 66:7543-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartshorn, K. L., D. Neumeyer, M. W. Vogt, R. T. Schooley, and M. S. Hirsch. 1987. Activity of interferons alpha, beta, and gamma against human immunodeficiency virus replication in vitro. AIDS Res. Hum. Retroviruses 3:125-133. [DOI] [PubMed] [Google Scholar]
- 23.Heil, F., et al. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303:1526-1529. [DOI] [PubMed] [Google Scholar]
- 24.Ho, D. D., et al. 1985. Recombinant human interferon alfa-A suppresses HTLV-III replication in vitro. Lancet i:602-604. [DOI] [PubMed] [Google Scholar]
- 25.Ishii, K. J., S. Koyama, A. Nakagawa, C. Coban, and S. Akira. 2008. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe 3:352-363. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, D., et al. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, D., et al. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infection. J. Virol. 84:8332-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornbluth, R. S., P. S. Oh, J. R. Munis, P. H. Cleveland, and D. D. Richman. 1989. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J. Exp. Med. 169:1137-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornbluth, R. S., P. S. Oh, J. R. Munis, P. H. Cleveland, and D. D. Richman. 1990. The role of interferons in the control of HIV replication in macrophages. Clin. Immunol. Immunopathol. 54:200-219. [DOI] [PubMed] [Google Scholar]
- 30.Korth, M. J., M. D. Taylor, and M. G. Katze. 1998. Interferon inhibits the replication of HIV-1, SIV, and SHIV chimeric viruses by distinct mechanisms. Virology. 247:265-273. [DOI] [PubMed] [Google Scholar]
- 31.Lane, H. C., et al. 1990. Interferon-alpha in patients with asymptomatic human immunodeficiency virus (HIV) infection. A randomized, placebo-controlled trial. Ann. Intern. Med. 112:805-811. [DOI] [PubMed] [Google Scholar]
- 32.Lange, U. C., et al. 2008. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol. Cell. Biol. 28:4688-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. U. S. A. 101:4204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 282:32065-32074. [DOI] [PubMed] [Google Scholar]
- 35.Lichtner, M., et al. 2006. Circulating dendritic cells and interferon-alpha production in patients with tuberculosis: correlation with clinical outcome and treatment response. Clin. Exp. Immunol. 143:329-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maitra, R. K., and R. H. Silverman. 1998. Regulation of human immunodeficiency virus replication by 2′,5′-oligoadenylate-dependent RNase L. J. Virol. 72:1146-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malim, M. H., and M. Emerman. 2008. HIV-1 accessory proteins-ensuring viral survival in a hostile environment. Cell Host Microbe 3:388-398. [DOI] [PubMed] [Google Scholar]
- 38.Martensen, P. M., and J. Justesen. 2004. Small ISGs coming forward. J. Interferon Cytokine Res. 24:1-19. [DOI] [PubMed] [Google Scholar]
- 39.Mercer, J., M. Schelhaas, and A. Helenius. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803-833. [DOI] [PubMed] [Google Scholar]
- 40.Michaelis, B., and J. A. Levy. 1989. HIV replication can be blocked by recombinant human interferon beta. AIDS 3:27-31. [PubMed] [Google Scholar]
- 41.Miyauchi, K., Y. Kim, O. Latinovic, V. Morozov, and G. B. Melikyan. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moffatt, P., et al. 2008. Bril: a novel bone-specific modulator of mineralization. J. Bone Miner. Res. 23:1497-1508. [DOI] [PubMed] [Google Scholar]
- 43.Nagai, K., et al. 1997. Induction of CD4 expression and human immunodeficiency virus type 1 replication by mutants of the interferon-inducible protein kinase PKR. J. Virol. 71:1718-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 45.Okumura, A., et al. 2008. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology 373:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okumura, A., G. Lu, I. Pitha-Rowe, and P. M. Pitha. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U. S. A. 103:1440-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pincetic, A., Z. Kuang, E. J. Seo, and J. Leis. 2010. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J. Virol. 84:4725-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pitha, P. M. 1994. Multiple effects of interferon on the replication of human immunodeficiency virus type 1. Antiviral Res. 24:205-219. [DOI] [PubMed] [Google Scholar]
- 49.Poli, G., J. M. Orenstein, A. Kinter, T. M. Folks, and A. S. Fauci. 1989. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science 244:575-577. [DOI] [PubMed] [Google Scholar]
- 50.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1-47. [DOI] [PubMed] [Google Scholar]
- 51.Rivero, J., M. Limonta, A. Aguilera, M. Fraga, and P. Lopez Saura. 1994. Use of recombinant interferon-alpha in human immunodeficiency virus (HIV)-infected individuals. Biotherapy 8:23-31. [DOI] [PubMed] [Google Scholar]
- 52.Rojek, J. M., and S. Kunz. 2008. Cell entry by human pathogenic arenaviruses. Cell Microbiol. 10:828-835. [DOI] [PubMed] [Google Scholar]
- 53.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakuma, R., A. A. Mael, and Y. Ikeda. 2007. Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J. Virol. 81:10201-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauter, D., A. Specht, and F. Kirchhoff. 2010. Tetherin: holding on and letting go. Cell 141:392-398. [DOI] [PubMed] [Google Scholar]
- 56.Seyfried, N. T., et al. 2008. Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: a membrane proteomics approach. Cancer Lett. 263:243-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 58.Shirazi, Y., and P. M. Pitha. 1992. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 66:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirazi, Y., and P. M. Pitha. 1993. Interferon alpha-mediated inhibition of human immunodeficiency virus type 1 provirus synthesis in T-cells. Virology 193:303-312. [DOI] [PubMed] [Google Scholar]
- 60.Skillman, D. R., et al. 1996. Phase I trial of interferon alfa-n3 in early-stage human immunodeficiency virus type 1 disease: evidence for drug safety, tolerance, and antiviral activity. J. Infect. Dis. 173:1107-1114. [DOI] [PubMed] [Google Scholar]
- 61.Smith, R. A., J. Young, J. J. Weis, and J. H. Weis. 2006. Expression of the mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun. 7:113-121. [DOI] [PubMed] [Google Scholar]
- 62.Soumelis, V., et al. 2001. Depletion of circulating natural type 1 interferon-producing cells in HIV-infected AIDS patients. Blood 98:906-912. [DOI] [PubMed] [Google Scholar]
- 63.Stremlau, M., et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 64.Van Damme, N., et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei, X., et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weidner, J. M., et al. 2010. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84:12646-12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada, O., N. Hattori, T. Kurimura, M. Kita, and T. Kishida. 1988. Inhibition of growth of HIV by human natural interferon in vitro. AIDS Res. Hum. Retroviruses 4:287-294. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto, J. K., F. Barre-Sinoussi, V. Bolton, N. C. Pedersen, and M. B. Gardner. 1986. Human alpha- and beta-interferon but not gamma- suppress the in vitro replication of LAV, HTLV-III, and ARV-2. J. Interferon Res. 6:143-152. [DOI] [PubMed] [Google Scholar]
- 69.York, J., and J. H. Nunberg. 2007. A novel zinc-binding domain is essential for formation of the functional Junin virus envelope glycoprotein complex. J. Virol. 81:13385-13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.York, J., and J. H. Nunberg. 2007. Distinct requirements for signal peptidase processing and function in the stable signal peptide subunit of the Junin virus envelope glycoprotein. Virology 359:72-81. [DOI] [PubMed] [Google Scholar]
- 71.York, J., and J. H. Nunberg. 2006. Role of the stable signal peptide of Junin arenavirus envelope glycoprotein in pH-dependent membrane fusion. J. Virol. 80:7775-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yount, J. S., et al. 2010. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6:610-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu, J. J., et al. 2008. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 379:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.