Abstract
Hypervariable region 1 (HVR1) of hepatitis C virus (HCV) E2 envelope glycoprotein has been implicated in virus neutralization and persistence. We deleted HVR1 from JFH1-based HCV recombinants expressing Core/E1/E2/p7/NS2 of genotypes 1 to 6, previously found to grow efficiently in human hepatoma Huh7.5 cells. The 2aΔHVR1, 5aΔHVR1, and 6aΔHVR1 Core-NS2 recombinants retained viability in Huh7.5 cells, whereas 1aΔHVR1, 1bΔHVR1, 2bΔHVR1, 3aΔHVR1, and 4aΔHVR1 recombinants were severely attenuated. However, except for recombinant 4aΔHVR1, viruses eventually spread, and reverse genetics studies revealed adaptive envelope mutations that rescued the infectivity of 1aΔHVR1, 1bΔHVR1, 2bΔHVR1, and 3aΔHVR1 recombinants. Thus, HVR1 might have distinct functional roles for different HCV isolates. Ultracentrifugation studies showed that deletion of HVR1 did not alter HCV RNA density distribution, whereas infectious particle density changed from a range of 1.0 to 1.1 g/ml to a single peak at ∼1.1 g/ml, suggesting that HVR1 was critical for low-density HCV particle infectivity. Using chronic-phase HCV patient sera, we found three distinct neutralization profiles for the original viruses with these genotypes. In contrast, all HVR1-deleted viruses were highly sensitive with similar neutralization profiles. In vivo relevance for the role of HVR1 in protecting HCV from neutralization was demonstrated by ex vivo neutralization of 2a and 2aΔHVR1 produced in human liver chimeric mice. Due to the high density and neutralization susceptibility of HVR1-deleted viruses, we investigated whether a correlation existed between density and neutralization susceptibility for the original viruses with genotypes 1 to 6. Only the 2a virus displayed such a correlation. Our findings indicate that HVR1 of HCV shields important conserved neutralization epitopes with implications for viral persistence, immunotherapy, and vaccine development.
Approximately 180 million people are chronically infected with hepatitis C virus (HCV) with increased risk of developing liver cirrhosis and hepatocellular carcinoma (1). HCV is an enveloped positive-strand RNA virus of the Flaviviridae family. The 9.6-kb genome consists of 5′ and 3′ untranslated regions (5′ and 3′ UTRs) flanking the open reading frame (ORF) encoding a single polyprotein, which is processed into structural proteins (Core and envelope [E] glycoproteins 1 and 2), p7, and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (15). Seven HCV genotypes and multiple subtypes exist, differing at the amino acid (aa) level by ∼30% and ∼20%, respectively (15). Genotype-specific differences in response to alpha interferon-based therapy, in the risk of developing liver steatosis, and possibly in viral persistence have been reported (2, 15).
HCV immune evasion mechanisms underlying viral persistence are poorly understood, but it has been suggested that these mechanisms rely on rapid virus evolution, mediating escape from humoral and cellular adaptive immunity (9). Studies of virus neutralization were facilitated by development of HCV culture systems producing pseudoparticles (HCVpp) (5) and JFH1-based cell culture infectious viruses (HCVcc) (22, 33). Select sera from chronically infected patients were shown to contain cross-genotype-reactive neutralizing serum antibodies (18, 20, 25, 29), although their neutralization efficacy varied greatly depending on the virus genotype. The failure of these antibodies to control the virus in vivo might be linked to the emergence of escape mutants (32). However, in acutely infected HCV patients, the occurrence of neutralizing antibodies was associated with viral clearance (11, 27).
The envelope motif hypervariable region 1 (HVR1) has the highest sequence variability of the HCV genome. HVR1 was classified as the 26 or 27 N-terminal amino acids of E2 and is identifiable by cross-genotypic conserved residues (7). Variation in HVR1 is believed to arise from antibody-driven immune selection, as HVR1 contains at least one neutralization epitope (12) and does not evolve in IgG-deficient patients (21). HVR1 may act as an immunological decoy, diverting the immune system from targeting more-conserved neutralization epitopes (28). However, several studies showed that an acute-phase immune response against HVR1 was associated with viral clearance (11, 13, 36), and although HVR1-deleted genotype 1a virus was attenuated in experimentally infected chimpanzees, it adapted to produce a robust acute infection and establish persistent infection (14).
Recently, an in vitro study with a single JFH1-based recombinant Jc1, in which Core-p7 and the N-terminal part of NS2 is encoded by J6CF (35), showed that HVR1 deletion caused viral attenuation with a 10-fold decrease in infectivity. The HVR1-deleted 2a virus was found to have higher density and increased neutralization susceptibility (4). However, the study did not address in vivo relevance of these findings, and the Jc1 virus has not been shown to be infectious in vivo. Also, the focus on a single isolate raised the question of whether reported observations were representative of HCV in general. In independent studies, we compared the viability of HVR1-deleted viruses across HCV genotypes by deleting HVR1 from JFH1-based 2a recombinant J6/JFH1, in which the entire Core-NS2 is encoded by J6CF, as well as from viruses of genotypes 1 to 6 of the recently developed panel of JFH1-based viruses with genotype-specific Core-NS2 (18). This panel of HCV with and without HVR1 allowed us to perform density analysis and patient serum neutralization comparing virus with and without HVR1 across genotypes. In addition, by infecting human liver chimeric mice with 2a virus with and without HVR1, we observed similar rises in HCV RNA titers and in vitro infectivity titers of infected animal samples. These in vivo infections allowed us to verify our in vitro neutralization findings using in vivo-produced viruses with and without HVR1. Taken together, our data showed differential dependency of HVR1 and that HVR1 is probably protecting HCV from neutralization in vivo by shielding cross-genotype conserved neutralization epitopes, thereby substantiating previous reports of involvement of HVR1 in establishment of chronic infections in human patients and in chimpanzees (11, 12).
MATERIALS AND METHODS
Plasmids.
We used inter- and intragenotypic HCV recombinants with Core-NS2 of genotype 1 to 6 isolates (17, 18, 20, 22, 29) and UTRs as well as NS3-NS5B of JFH1, most of which contained cell culture-adaptive mutations (Table 1) (17, 18, 20, 29). Plasmids with HVR1 deletions and/or point mutations were made by conventional cloning techniques, and the HCV sequence of final maxipreps was confirmed in all cases as described previously (17, 18, 20, 29). For an alignment of E1 and E2 sequences of genotypes 1a to 6a, see Meunier et al. (25).
TABLE 1.
Virus stocks of JFH1-based HCV recombinants with genotype-specific Core-NS2 used for density analysis and in vitro neutralizationa
| Recombinant virus | Core-NS2 genotypeb | Adaptive mutationsc | Viral passage | Infectivity (log10 TCID50/ml)d | HCV RNA (log10 IU/ml)e | Specific infectivityf |
|---|---|---|---|---|---|---|
| H77/JFH1 | 1a | V787A, Q1247L | 2nd | 4.1 | 7.3 | 1/1,500 |
| J4/JFH1 | 1b | F886L, Q1496L | 1st | 3.7 | 7.3 | 1/4,000 |
| J6/JFH1 | 2a | None | 2nd | 5.2 | 7.6 | 1/250 |
| J6/JFH1ΔHVR1 | 2a | None | 2nd | 4.7 | 7.3 | 1/370 |
| J8/JFH1 | 2b | None | 1st | 4.4 | 7.4 | 1/1,000 |
| S52/JFH1 | 3a | I787S, S2272P | 2nd | 4.3 | 7.6 | 1/2,100 |
| S52/JFH1ΔHVR1 | 3a | A369V, I787S, S2272P | 3rd | 4.2 | 7.4 | 1/1,500 |
| ED43/JFH1 | 4a | T827A, T977S | 1st | 3.9 | 7.6 | 1/5,000 |
| SA13/JFH1 | 5a | A1021G, K1118R | 2nd | 4.3 | 7.0 | 1/500 |
| SA13/JFH1ΔHVR1 | 5a | A1021G, K1118R | 2nd | 3.8 | 7.5 | 1/5,300 |
| HK6a/JFH1 | 6a | T349S, N417T | 1st | 4.4 | 7.0 | 1/400 |
| HK6a/JFH1ΔHVR1 | 6a | T349S, N417T | 2nd | 3.5 | 7.2 | 1/4,700 |
The ORF sequences of all virus stocks were verified.
NS3-NS5B and 5′and 3′ UTRs of the recombinant viruses are of the genotype 2a isolate JFH1.
Mutations are annotated based on the H77 reference sequence (GenBank accession no. AF009606). All mutations, except for the A369V mutation for the 3aΔHVR1 recombinant, are adapting the intergenotypic recombinant viruses to cell culture (17, 18, 20, 29).
Virus titers are the averages of at least two six-replicate TCID50 determinations with a standard error of the mean (SEM) of <0.3.
HCV RNA titers are from a single determination by TaqMan RT-qPCR.
Specific infectivity was calculated as TCID50/IU. HVR1-deleted ED43/JFH1T827A,T977S was found to be nonviable. HVR1-deleted H77/JFH1V787A,Q1247L, J4/JFH1F886L,Q1496L, and J8/JFH1 could be adapted to the HVR1 deletion (Fig. 2), but the infectivity titers of adapted recombinant viruses in passages were about 103 FFUs/ml and hence too low for the generation of high-titer virus stocks.
In vitro studies in Huh7.5 cells.
Culturing, transfection, and infection of Huh7.5 cells were done as described previously (17), and cultures were evaluated every 2 or 3 days by HCV-specific immunostaining against either HCV Core or NS5A proteins (17). Infectivity titers were determined by inoculating supernatant sample dilutions on 6 × 103 cells/well plated the day before on poly-d-lysine-coated 96-well plates (Nunc). The cells were fixed and analyzed with HCV-specific NS5A immunostaining (17, 18). The effect of using undiluted samples was tested by including a minimum dilution of 1:2 in focus-forming units (FFUs) or 50% tissue culture infectious dose (TCID50) assays. Manual FFU counting was done on wells with 5 to 100 FFUs/well. In some assays, FFU counting was automated using an ImmunoSpot series 5 UV analyzer (CTL Europe GmbH) (16). In automated counting, background was defined as the mean FFU count of at least six replicates without virus and subtracted from assay values (this mean never exceeded 15 FFUs/well). The lower cutoff was set at 3 + background + 3 × standard deviation of the background, and the upper cutoff was set at 200 FFUs/well, based on the linear range of a test dilution series. Infectivity titer calculations used three independent virus dilutions, unless otherwise stated. TCID50/ml calculations used the standard Reed-Muench limiting dilution formula (28a) applied to 10-fold dilution series with six replicates. Supernatant HCV RNA titers were measured by an in-house reverse transcription (RT)-quantitative PCR (qPCR) (17). HCV ORF sequencing from culture supernatants was done by RT-nested PCR procedures, as described previously (15, 17, 18, 20, 29).
Equilibrium density gradient centrifugation.
Step gradients were made by layering 2.5 ml each of 40%, 30%, 20%, and 10% OptiPrep (iodixanol; Axis-Shield) on top of each other. The four iodixanol solutions were prepared by dilution with phosphate-buffered saline (PBS) from the 60% stock. The step gradients were left upright 24 h at 4°C for formation of semicontinuous gradients. Amicon centrifugation filters (Millipore) were used to concentrate virus stock samples (Table 1), retaining ∼250 μl. The samples were spun at 35,000 rpm (∼151,000 × relative centrifugal force [RCF]) for 18 h at 4°C using a Beckman SW-41 rotor mounted in a Beckman XL-70 ultracentrifuge. After centrifugation, ∼550-μl gradient fractions were harvested from the bottom of the tube, and 400-μl portions were weighed (model SI-114; Denver Instruments) to calculate fraction densities. The fractions were titrated for HCV infectivity and HCV RNA at 1:10 dilutions as described above.
HCV neutralization.
For neutralization studies, we used chronic-phase HCV sera of genotype 1a (H06 [29]), 4a (AA [29]), and 5a (SA3 [20]). IgG was purified from H06 and SA3 sera as described previously (20). We plated 6 × 103 Huh7.5 cells/well into poly-d-lysine-coated 96-well plates (Nunc). On the following day, 50 to 400 TCID50s of HCV was incubated for 1 h at 37°C with 2-fold dilutions of heat-inactivated (56°C for 30 min) serum or 4-fold dilutions of purified serum IgG (20) and subsequently incubated with cells for 3 h. After 48 h, cells were immunostained for NS5A (17). Neutralization was done in three replicate samples. Percent neutralization was calculated by relating FFU counts to the mean of six replicate samples incubated with virus only. Neutralization data were analyzed as variable-slope dose-response curves using GraphPad Prism 4.0. The 50% inhibitory concentration (IC50) and 90% inhibitory concentration (IC90) were approximated as the first serum dilution with greater than 50% and 90% neutralization, respectively.
Human liver chimeric mouse infections and ex vivo neutralization.
HCV infections of urokinase-type plasminogen activator, severe combined immune deficiency (uPA-SCID) mice engrafted with human hepatocytes were performed at Ghent University Hospital, with protocols approved by the hospital's Ethical Committee and Animals Ethics Committee. Animals were infected with HCV by intraperitoneal injections of 104 TCID50s. Animal care and sampling were done as described previously (23). Mouse liver repopulation by human hepatocytes was confirmed 2 days preinfection by human plasma albumin levels of >3.2 mg/ml. Sequencing HCV envelope sequences of viruses recovered from plasma or serum samples was performed as described above. However, in certain cases, random hexamers were used in reverse transcription with Superscript II (Invitrogen). HCV RNA titers were determined with Roche HCV TaqMan 48 or by in-house qPCR (17). Cell culture experiments using mouse plasma were done with 5 IU/ml heparin. To avoid cytotoxic effects of mouse plasma and serum in ex vivo neutralization experiments, a virus dose of 10 TCID50s per well was used to infect cultures in 96-well plates. Experiments were done with 24 replicate samples and scored by manual FFU counting. The remaining procedure was performed as described above. Two-tailed t tests were performed comparing infections in the presence or absence of neutralizing antibody-containing serum samples by using GraphPad Prism 4.0 with a significance threshold of P = 0.05.
RESULTS
Differential effect of HVR1 deletion on the viability of HCV strains of genotypes 1 to 6.
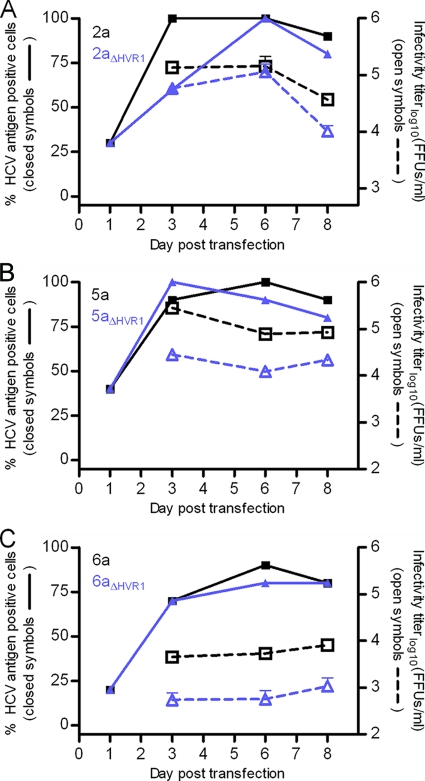
We investigated the viability of HVR1 deletion mutants of the recently developed panel of cell culture-adapted JFH1-based recombinants with Core-NS2 of genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a (17, 18, 20, 29) (Table 1, recombinants referred to by the Core-NS2 genotype). We deleted the entire HVR1 motif from the eight Core-NS2 genotype recombinants (corresponding to nucleotide [nt] 1491 to 1571 for all recombinants except for recombinant 6a for which it corresponded to nt 1494 to 1571; nucleotide and amino acid positions in this study relate to the H77 reference sequence with GenBank accession no. AF009606). After HCV RNA transfection of Huh7.5 cells, all HVR1-deleted recombinants replicated efficiently, as NS5A immunostainings of transfected cells on day 1 revealed similar percentages of brightly fluorescent cells. The 2aΔHVR1, 5aΔHVR1, and 6aΔHVR1 recombinants exhibited efficient viral spread and were only slightly attenuated with relatively high infectivity titers (Fig. 1A to C). The similar infectivities of 2a and 2aΔHVR1 viruses after transfection were further characterized by a kinetic infection experiment with three different multiplicities of infection (MOIs) showing highly similar degrees of virus spread, along with similar HCV RNA and infectivity titers of collected supernatants (data not shown). Supernatants from cell culture infections of 2nd passage 2aΔHVR1, 1st passage 5aΔHVR1, and 2nd passage 6aΔHVR1 recombinants had peak HCV RNA titers of approximately 107.5 IU/ml and peak HCV infectivity titers of 104.7, 103.8, and 103.5 TCID50s/ml, respectively (Table 1). Sequencing of the full ORFs of genomes recovered from these peak infections confirmed the HVR1 deletion and revealed no nucleotide changes. Thus, viability of the 2a, 5a, and 6a viruses lacking HVR1 did not require cell culture adaptation.
FIG. 1.
Intergenotypic JFH1-based recombinant HCV with Core-NS2 of genotypes 2a, 5a, and 6a were infection competent following HVR1-deletion. (A to C) Comparing HVR1-deleted virus to the original virus for genotypes 2a, 5a, and 6a revealed similar spread kinetics and supernatant infectivity titers following HCV RNA transfection of Huh7.5 cells. The percentage of infected cells was monitored by HCV-specific immunostaining and HCV supernatant infectivity titers were monitored by FFU assay, shown as the mean of three replicate samples with the standard deviation (SD) (error bar) (the lower cutoff was 500 FFUs/ml for genotype 2a, and it was 100 FFUs/ml for genotypes 5a and 6a). The original virus is shown in black, and the HVR1-deleted virus is shown in purple.
In contrast, the 1aΔHVR1, 1bΔHVR1, 2bΔHVR1, 3aΔHVR1, and 4aΔHVR1 recombinants were severely attenuated in Huh7.5 cells, displaying inefficient or nonexistent spread and low infectivity titers following HCV RNA transfection (Fig. 2A to E). With the exception of the 4aΔHVR1 recombinant, they all eventually spread, and we identified coding mutations by ORF sequencing of virus from culture supernatants upon spread to at least 80% of cells. We mainly observed envelope mutations and therefore focused on the effects of coding envelope mutations for the adaptation of attenuated HVR1-deleted viruses. Multiple transfections were performed, and the observed coding changes in the envelope genes are summarized in Table 2. For the 1aΔHVR1 recombinant, the number of HCV antigen-positive cells decreased following transfection and was nearly undetectable 30 days after transfection (not shown). Two additional transfections of Huh7.5 cells with the 1aΔHVR1 recombinant confirmed this attenuation. However, after 40 days, these two transfected cell cultures displayed virus spread and at least 80% of cells were HCV antigen positive on day 56, suggesting viral adaptation. ORF sequencing of HCV genomes recovered from supernatants confirmed HVR1 deletion and revealed several mutations (Table 2). In reverse genetics studies, we found that substitutions H261R (nucleotide change A1123G; E1) with Q444R (nucleotide change A1672G; E2), as well as N476D (nucleotide change A1767G; E2) with S733F (nucleotide change C2539T; E2), restored infectivity of the HVR1-deleted 1a virus (Fig. 2A). Infectivity titers of these adapted recombinants were ∼10-fold lower than those of the original 1a virus, but ORF sequencing of 1st passage viruses revealed no dominant nucleotide changes. Genotype 3aΔHVR1 developed only one coding mutation following transfection of Huh7.5 cells, A369V (nucleotide change C1447T; E1) (Table 2), which was shown in reverse genetics studies to fully restore 3aΔHVR1 infectivity (Fig. 2D). Supernatants from cell culture infection of 3rd passage 3aΔHVR1,A369V recombinant had a peak HCV RNA titer of 107.4 IU/ml and a peak HCV infectivity titer of 104.2 TCID50s/ml (Table 1). Sequencing of the complete ORFs of recovered genomes revealed no nucleotide changes. Finally, we demonstrated that the combination of S363F (nucleotide change C1429T; E1), T435P (nucleotide change A1644C; E2), and V710L (nucleotide change G2469C; E2) for the 1bΔHVR1 recombinant and Y412N (nucleotide change T1575A; E2) for the 2bΔHVR1 recombinant resulted in adaptation of these viruses and efficient virus spread (Fig. 2B and C). Sequencing of the ORFs of these viruses at different viral passages did not reveal nucleotide changes, and we consistently observed infectivity titers of about 103 FFUs/ml in viral passages. Although the difference in viability of HVR1-deleted 2a and 2b recombinants indicated that HVR1 dependence was not genotype specific, our observations showed that dependency on this region varied greatly among different HCV isolates.
FIG. 2.
Intergenotypic JFH1-based recombinant HCV with Core-NS2 of genotypes 1a, 1b, 2b, 3a, and 4a were infection impaired upon HVR1 deletion, but the infectivity of most viruses could be rescued by envelope mutations. The spread of viruses (genotypes 1a, 1b, 2b, 3a, and 4a) following HCV RNA transfection of Huh7.5 cells was moni- tored by determining the percentage of infected cells by HCV-specific immunostaining and HCV supernatant infectivity titers by FFU assay, shown as the mean of three replicates with SD (the lower cutoff was 100 FFUs/ml). The original virus is shown by black squares, HVR1-deleted virus is shown by purple triangles pointing up, and HVR1-deleted virus with one or more HVR1 deletion adaptive envelope mutations is shown by green triangles pointing down or by orange diamonds. All mutations are annotated based on the H77 reference sequence (GenBank accession no. AF009606). n.d., not detected.
TABLE 2.
Coding envelope mutations observed by full ORF sequencing of transfections and infections with HVR1-deleted HCV recombinants with Core-NS2 of genotypes 1a, 1b, 2b, and 3aa
The original HCV recombinants are described in greater detail in Table 1. The data were compiled from sequencing multiple transfection/infection ORFs (genotype 1a, 9 ORFs; genotype 1b, 3 ORFs; genotype 2b, 4 ORFs; genotype 3a, 2 ORFs).
b Amino acid change at the amino acid reference positions for HVR1-deleted JFH1 recombinants with genotype-specific Core-NS2 genes. The amino acid reference positions are for H77 reference strain (GenBank accession no. AF0096060). Only coding envelope mutations are shown. A mutation observed in more than one ORF is shown in boldface type. A mutation tested by reverse genetics and found to increase the infectivity of the HVR1-deleted recombinant virus is shown on a gray background. A small solid circle indicates that there was no change for this recombinant.
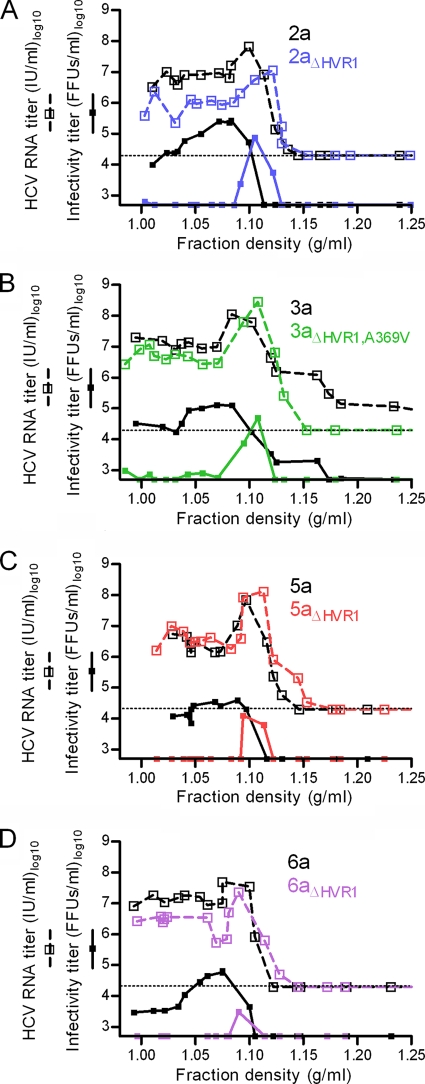
Deletion of HVR1 from HCV recombinants increased density and decreased structural heterogeneity of infectious particles irrespective of the HCV genotype.
We used virus stocks of genotypes 2a, 3a, 5a, and 6a with and without HVR1 (Table 1) to investigate changes in physicochemical properties. Performing equilibrium buoyant density centrifugation in iodixanol gradients, we found similar HCV RNA titers at densities between 1.0 and 1.15 g/ml for all genotypes both with and without HVR1 (Fig. 3A to D). However, the density of infectious HCV invariably changed from a range of 1.0 to 1.1 g/ml for the original recombinants to a single peak at ∼1.1 g/ml upon HVR1 deletion (Fig. 3A to D). This finding indicated that HVR1 deletion caused increased density and decreased heterogeneity of the infectious virus population irrespective of genotype.
FIG. 3.
HVR1 was critical for infectivity of low-density HCVcc irrespective of genotype. Equilibrium gradient density centrifugation was done in iodixanol comparing a single JFH1-based Core-NS2 genotype virus with and without HVR1 (Table 1) (genotypes 2a, 3a, 5a, and 6a). After centrifugation, ∼550-μl gradient fractions were harvested from the bottom of the tube, and 400-μl portions were weighed to calculate fraction densities. HCV RNA titration (the lower cutoff was 20,000 IU/ml and is shown as a dotted line in each graph) and infectivity titration (the lower cutoff was 500 FFUs/ml) was performed for each fraction (samples were diluted at least 1:10). Mutations are annotated based on the H77 reference sequence (GenBank accession no. AF009606).
Deletion of HVR1 resulted in greatly increased neutralization susceptibility to chronic-phase HCV serum antibodies for most HCV genotypes, resulting in similar neutralization profiles across genotypes.
To further evaluate the effect of HVR1 deletion on different HCV genotypes, we compared the susceptibility of genotypes 1a, 2a, 3a, 5a, and 6a with and without HVR1 to neutralization with chronic-phase H06 serum. This HCV chronic-phase serum is known to contain cross-reactive neutralizing antibodies with varying potency against the original recombinants of different genotypes used to generate the HVR1-deleted viruses (18, 20, 29). We observed three distinct neutralization profiles for original viruses of genotypes 1a, 2a, 3a, 5a, and 6a (Fig. 4A to C). In contrast, HVR1-deleted viruses were all highly sensitive to neutralization. The original 6a virus was efficiently neutralized at high H06 dilutions, and HVR1-deleted virus displayed only a small increase in neutralization susceptibility (Fig. 4A and Table 3). The original 1a and 5a viruses showed less steep dose-response curves, requiring a large increase in serum antibodies for a small increase in neutralization. In contrast, the HVR1-deleted viruses of both genotypes were neutralized completely at high H06 dilutions, and no difference was observed between 1aΔHVR1 viruses harboring different HVR1 deletion adaptive mutations in the envelope genes, suggesting that the relevant neutralization epitopes were unaltered (Fig. 4B and Table 3). The original 2a and 3a viruses showed a two-population dynamic with half the virus population seemingly being neutralization resistant, while the HVR1-deleted viruses were neutralized completely at high H06 dilutions (Fig. 4C and Table 3). The 50% and 90% neutralization titers of H06 against the HVR1-deleted viruses of the different genotypes were similar, with the highest titer against 6aΔHVR1 (Table 3).
FIG. 4.

HVR1 deletion from JFH1-based viruses with Core-NS2 of different genotypes resulted in increased sensitivity to neutralizing antibodies. Viruses of genotypes 1a, 2a, 3a, 5a, and 6a with HVR1 (closed symbols) and without HVR1 (open symbols) (Table 1) were neutralized by incubation for 1 h at 37°C with heat-inactivated chronic-phase patient serum H06 (genotype 1a virus-infected patient H) previously shown to contain cross-genotype-reactive neutralizing antibodies (18, 29). The number of FFUs/well was visualized following 48 h of infection by HCV-specific immunostaining. Neutralization data are shown as the means of three replicates with the standard error of the mean (SEM) (error bars) related to the mean of 6 replicates with virus only. Variable-slope dose-response curve regression was used to fit the data points. Due to the relatively low infectious titer of the 1aΔHVR1,H261R,Q444R recombinant, this virus was concentrated ∼50-fold using Amicon centrifugation columns. Mutations are annotated based on the H77 reference sequence (GenBank accession no. AF009606).
TABLE 3.
Reciprocal titers of neutralizing antibodies in chronic-phase patient sera against recombinant viruses with genotype-specific Core-NS2 of genotypes 1a, 2a, 3a, 5a, and 6a with and without HVR1a
| Virus genotype (Core-NS2) | Reciprocal serum neutralization titerb |
|||||
|---|---|---|---|---|---|---|
| 90% |
50% |
|||||
| H06 (1a) | AA (4a) | SA3 (5a) | H06 (1a) | AA (4a) | SA3 (5a) | |
| Original virus | ||||||
| 1a | <50 | • | • | 800 | • | • |
| 2a | <50 | <50 | <50 | 50-51,200c | <50 | <50 |
| 3a | <50 | • | • | 50-51,200c | • | • |
| 5a | 50 | <50 | <50 | 3,200 | 200 | 100 |
| 6a | 6,400 | • | • | 51,200 | • | • |
| Virus lacking HVR1 | ||||||
| 1aΔHVR1,H261R,Q444R | 6,400 | • | • | 51,200 | • | • |
| 1aΔHVR1,N476D,S733F | 6,400 | • | • | 25,600 | • | • |
| 2aΔHVR1 | 12,800 | 6,400 | 1,600 | 51,200 | 51,200 | 12,800 |
| 3aΔHVR1,A369V | 6,400 | • | • | 51,200 | • | • |
| 5aΔHVR1 | 6,400 | 3,200 | 1,600 | 25,600 | 25,600 | 12,800 |
| 6aΔHVR1 | 51,200 | • | • | 204,800 | • | • |
Fifty to 400 TCID50s of virus stocks with genotype-specific Core-NS2 were incubated in 3 replicates with H06, AA, or SA3 sera in 2-fold dilution series and tested in Huh7.5 cell infections as described in Materials and Methods.
Reciprocal neutralization titers are indicated as the highest dilution of H06 (genotype 1a), AA (genotype 4a), or SA3 (genotype 5a) sera leading to a reduction of FFU counts of at least 90% or 50% compared to the mean of 6 replicates of virus only. The small solid black circles indicate that no data was obtained for that virus. Mutations are annotated based on the H77 reference sequence (GenBank accession no. AF009606).
For original viruses 2a and 3a, around half the virus population was neutralized at H06 serum dilutions 50 to 51,200 (Fig. 4B).
We verified the increased neutralization susceptibility of HVR1-deleted viruses by additional neutralization experiments of genotype 2a and 5a viruses with and without HVR1 using dilutions of chronic-phase patient HCV sera AA (genotype 4a) and SA3 (genotype 5a), both of which have been shown to contain cross-genotype reactive neutralizing antibodies against HCV (20, 29). Both sera neutralized the HVR1-deleted viruses with high efficiency but had minimal effect against the corresponding original viruses (Table 3). To address the possibility of serum-specific IgG-independent effects, we attempted to neutralize 2a and 5a viruses with and without HVR1 using two HCV negative-control sera at 1:50, 1:500, 1:5,000, and 1:50,000 dilutions. We did not observe any effect of either serum against any of the tested viruses at 1:500 or higher dilutions, and we observed only a slight effect of about 50% neutralization against the two HVR1-deleted viruses at the 1:50 dilution (data not shown). Thus, the chronic-phase patient sera 50% neutralization titers for HVR1-deleted viruses of at least 1:12,800 (Table 3) were caused by HCV-specific serum antibodies. Finally, we confirmed that chronic-phase patient serum neutralization was caused specifically by serum IgG by performing neutralization of 2a, 2aΔHVR1, 5a, and 5aΔHVR1 recombinants, using IgG purified from H06 and SA3 sera (data not shown). In conclusion, our findings showed that HVR1 deletion caused viruses to become highly neutralization susceptible, resulting in similar neutralization profiles across HCV genotypes.
A relationship between virus density and neutralization susceptibility was observed only for genotype 2a.
Since we had observed increased density and increased neutralization susceptibility of HVR1-deleted viruses, we wanted to investigate whether a direct correlation between density and neutralization susceptibility existed for the original Core-NS2 recombinant viruses. Using genotype 1 to 6 virus-containing fractions from gradient centrifugations, we performed neutralization tests with four different dilutions of H06 serum (Fig. 5A to F). When comparing the neutralization susceptibility of noncentrifuged viruses (Fig. 4) with that of centrifuged viruses (Fig. 5), it seemed that the overall neutralization susceptibility was increased by the centrifugation procedure. This is in spite of the fact that iodixanol gradients have been shown to be superior for virus structure preservation in ultracentrifugation (26). Furthermore, a correlation between a higher density and higher neutralization susceptibility was observed only for the genotype 2a viruses. To investigate whether this effect on genotype 2a was reproducible, we repeated the experiment for the 2a recombinant gradient viruses using the H06, AA, and SA3 sera (Fig. 6A to C). This experiment confirmed the correlation between neutralization susceptibility and density of 2a virus particles for neutralizing antibodies from patients infected with HCV genotypes 1a (H06), 4a (AA), and 5a (SA3).
FIG. 5.
When comparing neutralization susceptibility with virus density for JFH1-based viruses with Core-NS2 of genotypes 1a to 6a, only 2a virus displayed increased neutralization at higher densities. JFH1-based viruses with Core-NS2 of genotypes 1 to 6 (virus with genotypes 1a, 2a, 3a, 4a, 5a, and 6a) from centrifugation fractions with different densities were neutralized with four dilutions of H06 serum as described in Materials and Methods. Neutralization data are shown as the means of three replicates with the SEM related to mean of 6 replicates of virus only.
FIG. 6.

Neutralization susceptibility of the JFH1-based Core-NS2 genotype 2a recombinant increased with virus density for the three neutralizing antibody-containing sera H06, AA, and SA3. The neutralization susceptibilities of ultracentrifugation fractions with different densities of original genotype 2a virus were examined using three sera, H06 (A), AA (B), and SA3 (C). Neutralization was carried out at four different serum dilutions as indicated in each graph and performed as described in Materials and Methods. Neutralization data are shown as the means of three replicates with the SEM related to the mean of 6 replicates of virus only.
In vivo-produced HVR1-deleted virus had increased neutralization susceptibility similar to observations for in vitro-produced HVR1-deleted virus.
To investigate whether the increase in neutralization susceptibility for HVR1-deleted virus was an in vitro phenomenon, we neutralized 2a and 2aΔHVR1 viruses produced in vivo in native human hepatocytes of human liver chimeric mice. We used samples from two chimeric mice originally inoculated with 104 TCID50 culture-derived 2a viruses with HVR1 (mouse B156) and without HVR1 (mouse B150R). Both animals were robustly infected, as evidenced by at least two HCV RNA titers above 106.5 IU/ml for samples taken during weeks 2 to 4. Furthermore, the samples had relatively high HCV infectivity titers in culture displaying similar specific infectivities of 1/79 and 1/63 for mouse B156 (2a infected) and mouse B150R (2aΔHVR1 infected), respectively. By sequencing the envelope genes, we found that viruses derived from the B156 and B150R mice did not have envelope mutations, and we elected to test neutralization susceptibility of in vivo-produced 2a virus on the week 3 sample from mouse B156 and of the in vivo-produced 2aΔHVR1 virus on the week 4 sample from mouse B150R.
The limited amount of mouse plasma/serum samples as well as the potential cytotoxicity necessitated using a different neutralization protocol with a single serum antibody dilution with many replicates infected at low viral doses. We initially chose to use the SA3 serum at a dilution of 1:200, which was found to have minimal effect against in vitro-produced original 2a virus and which completely neutralized the 2aΔHVR1 recombinant. In vivo-produced virus was preincubated for 1 h with either medium or a 1:200 dilution of the SA3 serum prior to infection of 24 wells with an inoculum of 10 TCID50s/well. We observed complete neutralization of the HVR1-deleted virus and no significant effect against the original 2a virus (Fig. 7A). We next investigated whether the neutralization susceptibility was similar for in vitro- and in vivo-produced viruses. We did an analogous experiment using an SA3 dilution of 1:4,000, which has been shown to neutralize in vitro-produced 2aΔHVR1 at about 70%. Here we observed similar neutralization of in vivo- and in vitro-produced HVR1-deleted 2a virus of 70 to 80% and no significant effect against the original viruses (Fig. 7B and C). Thus, we demonstrated increased neutralization susceptibility of the HVR1-deleted virus produced in vivo comparable to what was observed for the in vitro-produced virus, indicating that the protective role of HVR1 against neutralization by HCV patient sera was not an in vitro artifact.
FIG. 7.

In vivo-produced 2aΔHVR1 virus displayed greatly increased neutralization susceptibility resembling that of in vitro-produced 2aΔHVR1 virus. In vivo-produced viruses used were without dominant envelope mutations; 2a virus was from mouse B156 serum, and 2aΔHVR1 virus was from mouse B150R plasma. A virus dose of 10 TCID50s/well was used. Mouse-derived 2a and 2aΔHVR1 viruses were incubated in 24 replicates for 1 h at 37°C with either medium or a dilution of SA3 serum prior to infection of Huh7.5 cells. The number of FFUs/well was visualized following 48 h of infection by HCV-specific immunostaining. The number of FFUs/well was normalized to the virus-only infection without serum and is shown with the SEM. The asterisk for 2aΔHVR1 virus and SA3 serum diluted 1:200 indicates that no infected cells were observed. Values that were significantly different (P < 0.0001) are indicated by a bar and three asterisks. Values that were not significantly different at a significance level of P = 0.05 are indicated by a bar and ns. (A) In vivo-produced 2a and 2aΔHVR1 viruses subjected to neutralization with a 1:200 dilution of SA3 serum showed no neutralization of 2a and complete neutralization of 2aΔHVR1 as observed for in vitro-produced viruses (Table 3). (B and C) In vivo- and in vitro-produced 2aΔHVR1 subjected to neutralization with a 1:4,000 dilution of SA3 serum displayed statistically significant and similar neutralization of about 70 to 80%, whereas the original 2a viruses produced in vivo and in vitro were not neutralized.
DISCUSSION
In the present study, we found that deletion of HVR1 had a differential impact on the viability of different HCV genotype isolates, which suggests that HVR1 plays a significant role in the viral life cycle, the importance of which varies for different isolates. Comparing density distributions of infectious HCV particles and HCV RNA of the original and HVR1-deleted viruses, we found that the infectivity of low-density HCV depended on HVR1, irrespective of the virus genotype. In addition, we demonstrated that both in vitro- and in vivo-derived HVR1-deleted HCV had greatly increased neutralization susceptibility to chronic-phase patient serum antibodies compared to the original unmodified viruses.
We deleted HVR1 of the E2 glycoprotein from a recently developed panel of JFH1-based viruses with genotype 1 to 6 Core-NS2 gene sequences (18, 20, 29). Interestingly, although all HVR1-deleted viruses replicated upon transfection of Huh7.5 cells, HVR1 deletion had a differential impact on HCV viability. The deletion of HVR1 from the 2a recombinant J6/JFH1 resulted in minimal viral attenuation in vitro, whereas others reported a 10-fold decrease in infectivity when HVR1 was deleted from the closely related 2a recombinant, Jc1 (4). Jc1 is known to produce higher titers in culture than J6/JFH1. This difference is likely related to the different location of the downstream J6CF/JFH1 junction, which might affect viral replication and/or assembly and possibly impact HVR1 dependence. HVR1 deletion from genotypes 5a and 6a resulted in noticeable decreases in viral infectivity titers, but they still exhibited efficient viral spread and genetic stability. However, severely attenuated infections were observed for 1aΔHVR1, 1bΔHVR1, 2bΔHVR1, and 3aΔHVR1 recombinants; the 4aΔHVR1 recombinant was nonviable. Adaptive envelope mutations rescuing viral viability were identified for the 1aΔHVR1, 1bΔHVR1, 2bΔHVR1, and 3aΔHVR1 viruses. The high viability of the 2aΔHVR1 recombinant and the low viability of the 2bΔHVR1 recombinant indicated that the differential effect of HVR1 deletion was not genotype specific.
A comprehensive analysis of the positions of the adaptive envelope mutations is made difficult by the absence of an X-ray or nuclear magnetic resonance structure of the E1/E2 heterodimer. However, it is noteworthy that three out of the five HVR1 deletion-adapted viruses tested had mutations in the transmembrane domain of either E1 or E2 shown to be important for E1/E2 heterodimerization (8). These adaptive mutations were S733F (E2) for the 1aΔHVR1 virus, S363F (E1) for the 1bΔHVR1 virus, and A369V (E1) for the 3aΔHVR1 virus, the latter being sufficient for restoring full viability. It is also interesting that adapted 1aΔHVR1 virus had mutations in either HVR2 (E2; N476D) or HVR3 (E2; Q444R), which suggests that these regions play a role in HVR1 deletion adaptation. Furthermore, it is intriguing that the HVR2 mutation removed a glycosylation site possibly exposing parts of the virus envelope. The E1, E2, NS3, and NS5B genes have been shown to acquire coding mutations in full-length H77CΔHVR1 infections of chimpanzees (14). The Core-NS2 sequence of the 1aΔHVR1 virus used in the present study was of the H77C isolate, and we observed a need for adaptive mutations in the envelope genes in vitro. Further studies should investigate the functions of in vitro- and in vivo-derived H77C envelope mutations in response to HVR1 deletion. On the basis of these highly divergent phenotypes of HVR1-deleted HCV genotype isolates, it would appear that HVR1 plays an unknown role in the virus life cycle, the importance of which varies between isolates or subtypes. This role could relate to a differential interaction of isolates with early entry factors, such as the proposed interaction of HVR1 with scavenger receptor BI (SR-BI) (6), which should be the subject of further studies.
Using HVR1-deleted viruses of genotypes 2a, 3a, 5a, and 6a, we found that HVR1 deletion caused a shift in density of infectious HCV from a range of 1.0 to 1.1 g/ml to a single peak at ∼1.1 g/ml. Most enveloped viruses have densities of ∼1.2 g/ml (24, 34), but HCV seems to employ the very-low-density lipoprotein (VLDL) release machinery of the hepatocyte during virus assembly and is apparently released as lipo-viro particles (3). This might explain the relatively low density of virus derived from patients (3), animals (23), and cell culture (19). This result corroborates a recent finding using HVR1-deleted Jc1 (4) and indicates that infectivity of low-density HCV particles depends on HVR1 irrespective of the virus genotype.
Serum from patient H was shown to prevent H77C (genotype 1a) infection in vivo (31) and exhibited cross-genotype neutralization in vitro (20, 29). Therefore, we chose H06 serum to investigate susceptibility to neutralization of viruses with genotypes 1a, 2a, 3a, 5a, and 6a with and without HVR1. All viruses, viruses with different degrees of resistance against neutralizing antibodies, became significantly more susceptible when HVR1 was deleted. An exception to this was the original genotype 6a virus, which was highly susceptible to neutralization and which showed only a small increase in susceptibility when HVR1 was deleted. This showed that HVR1 was not the only factor determining neutralization susceptibility as corroborated by others (19, 30). It should be noted that the original 6a virus was the only recombinant with cell culture-adaptive mutations in the envelope genes (Table 1), one of which is in a known neutralization epitope (18). However, susceptibility to neutralization with H06 in vivo has been observed for the unaltered 6a virus (24a). The observation that HVR1 deletion resulted in increased neutralization susceptibility to serum antibodies could not be explained by different susceptibilities to the infection enhancing effects of serum between original and HVR1-deleted viruses. Interestingly, variation in neutralization profiles for viruses with the different genotypes disappeared upon HVR1 deletion, suggesting that HVR1 shielded cross-genotype conserved epitopes, which is corroborated by the finding that the conserved CD81 binding domains of the Jc1 virus was exposed by HVR1 deletion (4). It also suggests that sera from HCV patients with very little neutralization activity might still contain high titers of cross-genotype-reactive neutralizing antibodies, if the HVR1-dependent shielding could be overcome.
Another important difference between the different genotype viruses retaining HVR1 was that only genotype 2a displayed a correlation between higher density and increased neutralization susceptibility as described by others for a related genotype 2a virus (19). In contrast, we were not able to show such a correlation for the original viruses of genotype 1a, 3a, 4a, 5a, and 6a. Thus, our findings suggest that increased virion density does not represent the only mechanism for the increased neutralization susceptibility of HVR1-deleted viruses. The variation across genotype isolates observed in this study affirms the importance of considering the genetic heterogeneity of HCV.
The study that initially reported that the 2a recombinant J6/JFH1 was fully infectious in human liver chimeric mice did not sequence viruses derived from the plasma samples of mice (23). Importantly, we observed no apparent attenuation of HVR1-deleted virus upon infection of human liver chimeric mice with in vitro-produced 2a and 2aΔHVR1 viruses. Furthermore, we sequenced in vivo-derived viruses with and without HVR1 and found that these viruses did not depend on developing envelope mutations. Using viruses without envelope mutations, we then compared neutralization susceptibility of in vivo-produced 2a virus with and without HVR1. We found that in vivo-produced HVR1-deleted virus had greatly increased neutralization sensitivity, which is similar to what was observed for HVR1-deleted in vitro-produced virus. This indicated that one of the roles of HVR1 in vivo might in fact be related to shielding epitopes highly sensitive to neutralization. Thus, our findings might be of relevance for previous reports that HVR1 plays a role in the establishment of chronic infection (10, 11). It is possible that the viability of 2aΔHVR1 virus in human liver chimeric mice is influenced by the absence of an adaptive immune system in this model. Addressing this by experimental infections of chimpanzees would be of interest.
The present study provides important new insight into the role of the E2 HVR1 motif of HCV. This is the first demonstration of a differential importance of a specific sequence motif for different HCV isolates. Our data suggest that in the absence of HVR1, the virus exposes conserved envelope regions in vitro and in vivo, because HVR1 deletion greatly increased susceptibility to neutralization with chronic-phase HCV serum antibodies. The shift in density caused by HVR1 deletion might be a consequence of such an exposure, suggesting that it depends on a decreased association with low-density moieties, such as lipids. Although the neutralization protection of HVR1 is incomplete, it is likely an important contributor to viral persistence and might help explain why HVR1 evolution has been linked to chronic disease progression (11). Such a role for HVR1 implies a need to focus on the in vivo availability of targeted neutralization epitopes when designing antibody-mediated treatments or vaccines against HCV and suggests that overcoming this HVR1-dependent epitope shielding would render the virus more vulnerable to the host immune response. Thus, our study has important implications for the development of effective antibody-based vaccines and therapeutics.
Acknowledgments
We are grateful to Anna-Louise Sørensen, Lotte Mikkelsen, Lubna Ghanem (CO-HEP) and Lieven Verhoye (University of Ghent) for technical assistance, to Steen Ladelund for statistical advice, to Jens Ole Nielsen, Ove Andersen, and Kristian Schønning (Copenhagen University Hospital, Hvidovre, Denmark) for their support of the project, and to Robert Purcell (NIH), Harvey Alter (NIH), David Thomas (John Hopkins University School of Medicine), Michael Kew (University of the Witwatersrand), Charles Rice (Rockefeller University), and Takaji Wakita (National Institute of Infectious Diseases) for providing reagents.
This study was supported by Ph.D. stipends from the Faculty of Health Sciences, University of Copenhagen (J.P., T.B.J., and T.K.H.S.), by research grants from the Lundbeck Foundation (J.B.), The Danish Cancer Society (J.M.G. and J.B.), the Novo Nordisk Foundation (J.M.G. and J.B.), The Danish Medical Research Council (J.B.), A. P. Moeller and the Chastine Mc-Kinney Moeller Foundation (T.K.H.S., J.M.G., and J.B.), the Leo Nielsen and Karen Margrethe Nielsen Foundation (J.P. and J.B.), and Copenhagen University Hospital, Hvidovre, Denmark (T.B.J., T.K.H.S., J.M.G., and J.B.), by a Concerted Action Grant (01G00507) from Ghent University, and by the Belgian State via the Interuniversity Attraction Poles Program (P6/36-HEPRO). P.M. is a postdoctoral fellow supported by The Research Foundation-Flanders (FWO-Vlaanderen; project 31500910).
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 2.Amoroso, P., et al. 1998. Correlation between virus genotype and chronicity rate in acute hepatitis C. J. Hepatol. 28:939-944. [DOI] [PubMed] [Google Scholar]
- 3.Andre, P., et al. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankwitz, D., et al. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 84:5751-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., et al. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh, J., et al. 2000. Analysis of the hypervariable region 1 of hepatitis C virus, p. 330-335. In H. S. Margolis, M. J. Alter, T. J. Liang, and J. L. Dienstag (ed.), Viral hepatitis and liver diseases. Proceedings of the 10th International Symposium on Viral Hepatitis and Liver Disease. International Medical Press, Atlanta, GA.
- 8.Ciczora, Y., N. Callens, F. Penin, E. I. Pecheur, and J. Dubuisson. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 81:2372-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dustin, L. B., and C. M. Rice. 2007. Flying under the radar: the immunobiology of hepatitis C. Annu. Rev. Immunol. 25:71-99. [DOI] [PubMed] [Google Scholar]
- 10.Farci, P., et al. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc. Natl. Acad. Sci. U. S. A. 91:7792-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farci, P., et al. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 12.Farci, P., et al. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. U. S. A. 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, J., et al. 2004. Long-term persistence of infection in chimpanzees inoculated with an infectious hepatitis C virus clone is associated with a decrease in the viral amino acid substitution rate and low levels of heterogeneity. J. Virol. 78:9782-9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forns, X., et al. 2000. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 97:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, J. M., and J. Bukh. 2008. Cutting the Gordian knot—development and biological relevance of hepatitis C virus cell culture systems. Adv. Virus Res. 71:51-133. [DOI] [PubMed] [Google Scholar]
- 16.Gottwein, J. M., et al. 2010. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J. Virol. 84:5277-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottwein, J. M., et al. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614-1626. [DOI] [PubMed] [Google Scholar]
- 18.Gottwein, J. M., et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364-377. [DOI] [PubMed] [Google Scholar]
- 19.Grove, J., et al. 2008. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J. Virol. 82:12020-12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, T. B., et al. 2008. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J. Infect. Dis. 198:1756-1765. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, U., J. Monjardino, and H. C. Thomas. 1994. Hypervariable region of hepatitis C virus envelope glycoprotein (E2/NS1) in an agammaglobulinemic patient. Gastroenterology 106:1072-1075. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., et al. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3805-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, W. T., et al. 1991. Isolation of dengue virus with a human promonocyte cell line. Am. J. Trop. Med. Hyg. 44:494-499. [DOI] [PubMed] [Google Scholar]
- 24a.Meuleman, P., et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology, in press. [DOI] [PMC free article] [PubMed]
- 25.Meunier, J. C., et al. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. U. S. A. 102:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen, S. U., et al. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestka, J. M., et al. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. U. S. A. 104:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray, S. C., et al. 1999. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J. Virol. 73:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 29.Scheel, T. K., et al. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. U. S. A. 105:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao, W., et al. 2009. A single point mutation in E2 enhances hepatitis C virus infectivity and alters lipoprotein association of viral particles. Virology 395:67-76. [DOI] [PubMed] [Google Scholar]
- 31.Vanwolleghem, T., et al. 2008. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology 47:1846-1855. [DOI] [PubMed] [Google Scholar]
- 32.von Hahn, T., et al. 2007. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 132:667-678. [DOI] [PubMed] [Google Scholar]
- 33.Wakita, T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, C. T., and E. Barklis. 1993. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 67:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 36.Zibert, A., et al. 1997. Early antibody response against hypervariable region 1 is associated with acute self-limiting infections of hepatitis C virus. Hepatology 25:1245-1249. [DOI] [PubMed] [Google Scholar]