Abstract
Epstein-Barr virus (EBV) undergoes latent and lytic replication cycles, and its reactivation from latency to lytic replication is initiated by expression of the two viral immediate-early transactivators, Zta and Rta. In vitro, reactivation of EBV can be induced by anti-immunoglobulin, tetradecanoyl phorbol acetate, and histone deacetylase inhibitor (HDACi). We have discovered that protein kinase C delta (PKCδ) is required specifically for EBV reactivation by HDACi. Overexpression of PKCδ is sufficient to induce the activity of the Zta promoter (Zp) but not of the Rta promoter (Rp). Deletion analysis revealed that the ZID element of Zp is important for PKCδ activation. Moreover, the Sp1 putative sequence on ZID is essential for PKCδ-induced Zp activity, and the physiological binding of Sp1 on ZID has been confirmed. After HDACi treatment, activated PKCδ can phosphorylate Sp1 at serine residues and might result in dissociation of the HDAC2 repressor from ZID. HDACi-mediated HDAC2-Sp1 dissociation can be inhibited by the PKCδ inhibitor, Rotterlin. Furthermore, overexpression of HDAC2 can suppress the HDACi-induced Zp activity. Consequently, we hypothesize that HDACi induces PKCδ activation, causing phosphorylation of Sp1, and that the interplay between PKCδ and Sp1 results in the release of HDAC2 repressor from Zp and initiation of Zta expression.
Epstein-Barr virus (EBV), a human oncogenic virus, infects two types of human cells predominantly, B lymphocytes and epithelial cells, and EBV infection also is associated with an array of malignancies derived from these two cell types, including Burkitt's lymphoma, Hodgkin's lymphoma, nasopharyngeal carcinoma, and gastric carcinoma (49). Being a human gammaherpesvirus, EBV undergoes two cycles: latency and lytic replication. Although most latent products are important for its ability to immortalize cells, and the virus is present in a latent form in most EBV-associated cancer tissues, serological studies revealed that elevated antibody titers against some lytic antigens, or increased viral DNA load in serum, are risk factors for tumor development (8, 33). In addition, experiments using the SCID mouse model suggested that Zta, an EBV lytic transactivator, is crucial for tumor formation (43). In vitro, experimental approaches also revealed that EBV lytic products may contribute to the pathogenesis of EBV-associated diseases, via upregulation of cytokines, cell growth factors or antiapoptotic proteins (5, 48, 50). Thus, the reactivation of EBV not only leads to the production of viral progeny but also facilitates viral pathogenesis.
In vitro, EBV persists predominantly in a latent form in host cells. However, lytic replication can be elicited conditionally by many different chemicals and physical stimuli or by ectopic transfection with two transactivators, Zta or Rta (49). These inducing agents provide the best available models for the study of essential factors involved in the switch of EBV from latency to lytic replication. Tetradecanoyl phorbol acetate (TPA) and anti-Ig are two well-studied inducing agents for EBV in a B-cell model (19, 45). Mechanistically, both inducers reactivate Zta expression to elicit viral replication via the protein kinase C (PKC) pathway (14, 15, 21). However, most studies have been performed in a B-cell model, and little is known about the mechanism of EBV reactivation in epithelial cells due to the lack of an appropriate culture model. We have established several EBV-harboring epithelial cell lines using a cell-to-cell coculture approach (6). Of note, we found that both sodium butyrate (SB) and trichostatin A (TSA), two histone deacetyltransferase inhibitors, are efficient inducers of EBV reactivation in an epithelial cell model. Furthermore, PKCδ was shown to be responsible for this specific induction (31).
The PKC family are serine/threonine kinases and are classified into three different isoforms, conventional (PKCα, β1, β2, and γ), novel (PKCδ, ɛ, θ, and η) and atypical (PKCξ, ι/λ, and μ) on the basis of their structure and the substrates required for activation, such as by diacylglycerol (DAG) and calcium (41). In our previous studies, we found that the PKCδ specific inhibitor, Rottlerin, alone is sufficient to block TSA-induced EBV reactivation (31). Moreover, the activation of PKCδ can be indicated by phosphorylation at threonine 505 residues, cleavage into catalytic fragments, and protein translocation to the nucleus, and all have been observed after TSA treatment (31). Our data indicate that TSA can induce PKCδ activation, which is an important mediator of EBV lytic cycle progression. It remains elusive how PKCδ kinase is involved in EBV reactivation after TSA induction.
It is well documented that the EBV lytic cycle is initiated by the activation of two promoters, the BZLF1 promoter (Zp) and the BRLF1 promoter (Rp), directing the expression of the immediate-early genes Zta and Rta, which in turn activate the downstream lytic genes individually or coordinately (4). Normally, Zp and Rp are inactive in latently infected cells, while the lytic cycle inducing agents, which disrupt viral latency, can turn on both promoters (3, 14, 19). In particular, several cis elements on Zp and Rp, containing cellular transcription factor binding sites, are defined and have been shown to regulate Zp and Rp activity. This raises the possibility that PKCδ may recruit, directly or indirectly, particular transcription factors to Zp or Rp for transcriptional activation after TSA treatment. Rp contains binding sites for cellular transcription factors, including NF1, Sp1, YY1, and Zif268 (22, 58-60). The −221 to +12 region of Zp harbors the positive cis elements for transactivation and is responsive to lytic cycle-inducing agents (46). According to sequence characteristics, this region can be classified into three distinct domains. The ZI (ZIA, ZIB, ZIC, and ZID) domain is A-T rich and is bound by the transcription factors, specificity protein 1 (Sp1) and Sp3, and myocyte enhancer factor 2D (MEF2D) (35, 36). ZII shares sequence homology with AP-1 and contains binding sites for CREB, ATF-1, and ATF-2 (17, 37, 51). ZIII (ZIIIA and ZIIIB) is the autoregulation domain of Zp, the Zta responsive element (ZRE) binding site, which can bind Zta itself (18). In the presence of inducers of EBV reactivation, ZI and ZII are activated, and a trace amount of Zta is expressed. After that, Zta can bind to its own ZRE on ZIII and thus induce the abundant expression of Zta, which can then elicit the lytic cascade (46).
Using an EBV-harboring NPC cell line as a model system, we sought to reveal further the molecular mechanism whereby PKCδ elicited the activation of the EBV immediate-early gene. The data from the present study demonstrated that overexpression of PKCδ is sufficient to induce Zp activation, rather than Rp. The response element of PKCδ-mediated Zp activation is located in the ZID element (−99 to −80 region), in which the Sp1 binding site plays an essential role for this activation. The regulation of Zp activation after TSA treatment is through PKCδ-dependent Sp1 phosphorylation, which influences the interaction between Sp1 and HDAC2 repressor, releases HDAC2 from Zp, and activates Zta expression.
MATERIALS AND METHODS
Cell culture.
NPC-TW01 is an EBV-negative NPC cell line which has lost its original infecting EBV during passage (32). Two EBV-positive NPC cell lines, NPC-TW01-NA (NA) and NPC-TW04-4A (4A), and an EBV-positive keratinocyte cell line, RHEK-RA (RA), were generated by reinfection with a recombinant Akata-EBV strain and selection by neomycin (Amresco, Solon, OH) (4, 6). NPC-TW01, NA, and RA cells were cultured with Dulbecco modified Eagle medium (HyClone, South Logan, UT) containing 1 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 8% fetal calf serum (HyClone). 4A cells were cultured with complete RPMI (HyClone) medium containing 1 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 8% fetal calf serum.
Induction of viral lytic cycle and inhibitor treatment.
To induce EBV reactivation in NA cells, 1.25 μM TSA (Calbiochem, La Jolla, CA) was added to medium when cells were grown to 80 to 90% confluence and incubated for 24 h. Rottlerin can inhibit PKCδ activity effectively. The treatment with Rottlerin (Calbiochem) was optimized at a concentration of 5 μM and incubation for 1 h. Mithramycin (Calbiochem) is an inhibitor that inhibits Sp1 binding to the GC-rich consensus promoter sequence. The treatment was carried out at concentrations of 50 nM and 500 nM, followed by incubation for 4 h. Then, the cells were treated with 1.25 μM TSA for 24 h and harvested for the reporter assay or Western blot.
Plasmids construction.
The plasmids of pEGFP-N1-derived wild-type PKCδ (pEFGP-WT-PKCδ), C-terminal catalytic domain-deleted mutant (pEGFP-CD-PKCδ), N-terminal-deleted mutant of PKCδ (pEGFP-CF-PKCδ), and vector control EGFP-N1 (Clontech, Inc., Mountain View, CA) were kindly provided by Hong-Chen Chen (Department of Life Science and the Graduate Institute of Biomedical Sciences, National Chung Hsing University, Taichung, Taiwan). The Flag-HDAC2 (pCDNA3-Flag-HDAC2) and Flag-HDAC3 (pCEP4f-Flag-HDAC3) expression plasmids were a gift from Li-Jung Juan (Genomics Research Center, Academia Sinica, Taipei, Taiwan) (27). The luciferase reporters of Zp and Rp inserted into the pGL2-Basic vector (Promega, Madison, WI) have been described in our previous studies (11). The 5′-serial-deleted mutants of Zp regions and ZID element mutant constructs (ZID m1 to m6) were generated by means of PCR-based site-directed mutagenesis.
RNA interference.
NA cells were transfected with plasmids containing specific oligonucleotides for luciferase (shLuc) or Sp1 (shSp1) acquired from National RNAi Core Facility (Academia Sinica, Taiwan). Transfected cells were reseeded each day and then transfected with siRNA three times.
Luciferase reporter assay.
NPC-TW01 cells were seeded at a density of 2 × 105 cells/well in a 12-well plate, and then plasmid DNA transfection was performed by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Reporter plasmids were transfected transiently with the effectors, as well as a Renilla luciferase expression plasmid (pRL-TK, internal control), into NPC-TW01 cells. After 48 h, the cells were harvested and subjected to a luciferase assay with a Dual-Glo assay kit (Promega). Activation was calculated by normalizing firefly luciferase activity to Renilla luciferase. By comparison with the control experiment, for which the activity was set to 1, the relative promoter activities are indicated as n-fold induction over the activity of the control. Each experiment was performed in duplicate and repeated three times.
Whole-cell lysate and nuclear protein preparation.
All of the transfected or induced cells were harvested at the time points indicated, washed once with phosphate-buffered saline (PBS) solution, and resuspended in lysis buffer (3% sodium dodecyl sulfate [SDS], 2 M urea, 2% 2-mercaptoethanol). Nuclear extracts were prepared as follows: 107 cells in a 10-cm dish were gently washed with PBS, and covered with 1 ml of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.04% NP-40, complete protease inhibitor cocktail [Roche, Mannheim, Germany], 50 mM NaF, and 1 mM Na3VaO4). After incubation for 10 min at room temperature, the cells were harvested by scraping, the cell lysates were centrifuged at 13,000 rpm (Eppendorf, 5415C) at 4°C for 5 min, and the supernatants (cytosolic fraction) were recovered. The obtained precipitates (nuclear fraction) were resuspended in buffer B (20 mM HEPES [pH 7.9], 0.4 M NaCl, 1 mM EDTA, 10% glycerol, complete protease inhibitor cocktail, 50 mM NaF, and 1 mM Na3VaO4). The protein concentration was determined by the Bradford method using Bio-Rad protein assay reagents (Bio-Rad, Hercules, CA). Cytosolic and nuclear fractions were revealed by using antibodies against tubulin (cytosolic fraction) and lamin C (nuclear fraction).
EMSA.
Oligonucleotides of wild-type ZID or ZID mutants were annealed and end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). The binding reaction was carried out in a total volume of 20 μl containing 8 μg of nuclear extracts, 62.5 nM γ-32P-labeled probes, 20 mM Tris-HCl, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 2 μg of poly(dI-dC). For the antibody supershift of the electrophoretic mobility shift assay (EMSA), 2 μg of anti-Sp1 (clone PEP2; Santa Cruz, CA) or anti-Sp3 (clone D-20; Santa Cruz) antibodies were added to the reactions. All reactions were incubated at room temperature for 30 min and then electrophoresed in 5% native polyacrylamide gels with Tris-borate buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA). The gels were dried and exposed to X-ray film.
Immunoblotting.
The extracted protein lysates were mixed with SDS-PAGE sample buffer (100 mM Tris [pH 6.8], 4% SDS, 0.2% bromophenol blue, and 20% glycerol), resolved by SDS-PAGE, and transferred onto Hybond-C membranes. The membranes were blocked in washing buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 0.2% Tween 20) containing 5% skim milk at room temperature for 1 h. Then, the membranes were incubated at 4°C overnight with primary antibodies against green fluorescent protein (GFP) (clone JL-8; Clontech), Zta (clones 1B4 and 4F10), PKCδ (clone C-20; Santa Cruz), Sp1 (clone PEP2; Santa Cruz), phosphothreonine (p-Thr; clone PTR-8; Sigma, St. Louis, MO), phosphoserine (p-Ser; clone PSR-5; Sigma, St. Louis, MO), and Flag (clone M2; Sigma). Antibodies against α-tubulin (clone CP06; Calbiochem) or β-actin (clone AC-15; Sigma) or glyceraldehyde 3-phosphate dehydrogenase (clone 6C5; Biodesign, Saco, ME) were used as normalization controls, when appropriate. After washing three times for 10 min with washing buffer, the blots were incubated with peroxidase-conjugated anti-mouse or anti-rabbit IgG secondary antibody (Jackson Immunoresearch Laboratories, West Grove, PA) at room temperature for 1 h. The luminescence signals were revealed by using a Western Lightning Chemiluminescence Reagent Plus kit (Perkin-Elmer Life Sciences, Inc., Waltham, MA) prior to exposure to X-ray films.
ChIP assay.
A chromatin immunoprecipitation assay (ChIP) assay was performed as described previously (4). Briefly, NA cells were fixed after treatment, cross-linked by 1% formaldehyde, and harvested by using nuclear lysis buffer (50 mM Tris [pH 8.0], 10 mM EDTA, and 1% SDS with complete protease inhibitors). The nuclear extracts were sonicated to yield soluble chromatin extracts (500- to 1,000-bp DNA fragments). The soluble chromatin extracts were diluted with 10 volumes of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, 1 mM dithiothreitol, and 50 μg of salmon sperm DNA), were precleared by using 50% protein A-Sepharose (GE Healthcare, Piscataway, NJ) for 1 h at 4°C, and then incubated overnight at 4°C with 4 μg of anti-Sp1, anti-PKCδ, or anti-HDAC2 (clone H-54; Santa Cruz) antibodies. Preimmune normal rabbit IgG was used as a negative control. The immunoprecipitated complexes were sequentially washed and then eluted by buffer containing 1% SDS, 0.25 M NaCl, and 0.1 M NaHCO3 to de-cross-link the interaction of DNA and protein at 65°C overnight. Finally, the immunoprecipitated DNA was eluted and analyzed by PCR with the primers ChIP-Zp+12 (5′-GCAAGGTGCAATGTTTAGTGAG, forward) and ChIP-Zp−221 (5′-CCATGCATATTTCAACTGGGC, reverse).
DAPA.
DNA affinity protein binding assay (DAPA) was performed as described previously (11). A 5′-biotin end-labeled double-stranded probe harboring the sequence of ZID element of Zp (ZID WT) and an Sp1 site mutated probe (ZID m2) with the same sequence except that the important binding site of Sp1 in Zp was changed from 5′-CACACC to 5′-ATAACC, synthesized by Pollster Biotech, Inc. (Taipei, Taiwan). Nuclear extracts (150 μg) were incubated with 0.4 μg of biotin-labeled probe in a binding buffer [20 mM Tris-HCl, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 2 μg of poly(dI-dC)]. At the same time, streptavidin MagneSphere paramagnetic beads (Promega) were preblocked in binding buffer containing 4 μg of bovine serum albumin/μl with gentle shaking for 1 h at 4°C. Thereafter, 100 μg of paramagnetic beads was incubated in a DNA-protein mixture reaction for another 1 h at 4°C. The beads were then spun down and washed three times in the binding buffer containing 250 mM NaCl and 0.25% Triton. Finally, the precipitated protein complexes captured by the beads were analyzed by immunoblotting.
IP assay.
The nuclear extracts were precleared with 100 μl of 20% protein A-Sepharose (GE Healthcare) in immunoprecipitation (IP) binding buffer (1% NP-40, 50 mM Tris-HCl, 150 mM NaCl, and 2 mM EDTA with complete protease inhibitors) for 1 h at 4°C. The reactions were mixed with 2 μg of specific antibodies against Sp1, PKCδ, HDAC2, or preimmune normal rabbit IgG at 4°C overnight. The immunocomplexes were then incubated with 100 μl of 20% protein A-Sepharose at 4°C for 2 h and washed for three times with IP binding buffer at 4°C. The Sepharose-coupled immunocomplexes were then dissolved in SDS sample buffer and detected by immunoblotting.
RESULTS
The ZID element of ZP is required for PKCδ-mediated Zp induction.
Zta gene expression governs the initiation of EBV reactivation and is driven by two promoters, Zp and Rp (38). Zp and Rp are normally inactive in latently infected cells, but these promoters can be activated in the presence of viral lytic inducing agents.
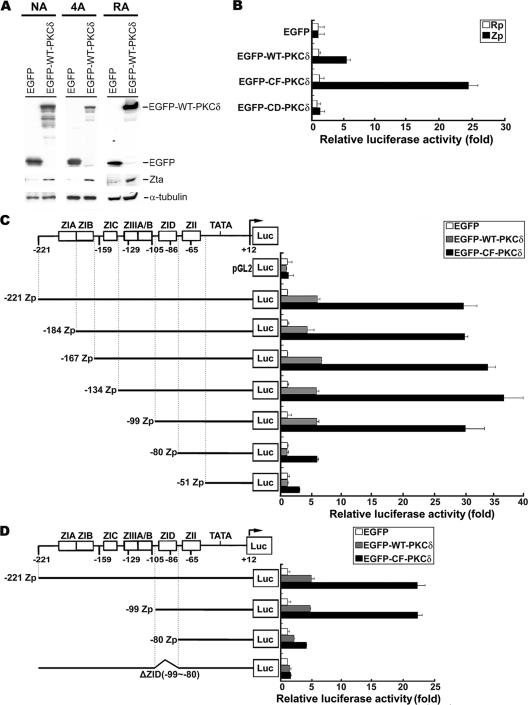
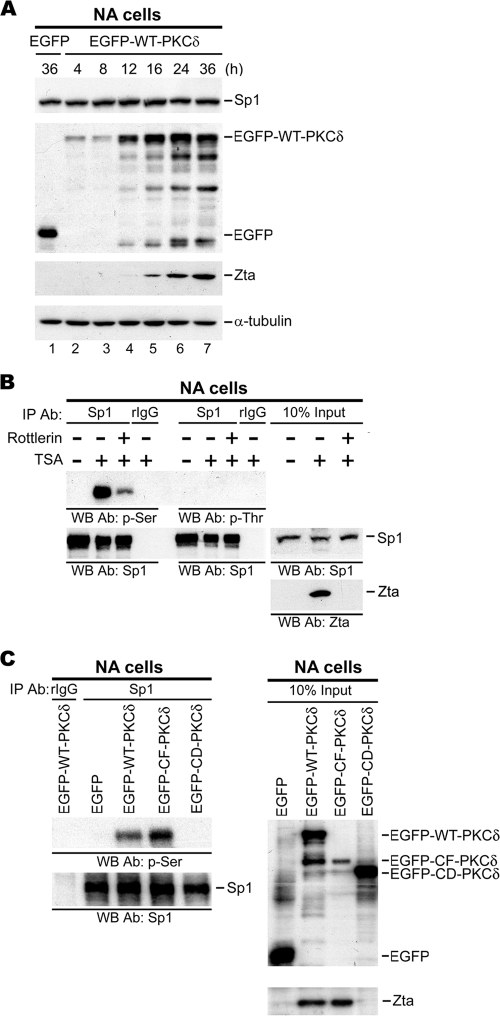
In our previous study, we demonstrated that PKCδ is responsible for HDACi-triggered reactivation of EBV (31). In Fig. 1 A, Zta expression was induced in three tested EBV-carrying cell lines: NA, 4A, and RA transfected with WT-PKCδ (4, 6). We sought to determine whether PKCδ alone is sufficient to induce Zp or Rp activation. In a luciferase reporter assay, overexpression of EGFP-derived wild-type PKCδ (EGFP-WT-PKCδ) led to increased activity of Zp, but not Rp, that was ∼6-fold greater than that of the control vector (EGFP). In addition, expression of a constitutively active form of PKCδ (EGFP-CF-PKCδ) increased Zp activity ∼27-fold; however, the kinase-inactive form of PKCδ (EGFP-DN-PKCδ) failed to turn on Zp activity (Fig. 1B). These data indicate that PKCδ is sufficient to upregulate activity of Zp and that this upregulation is dependent on kinase activity.
FIG. 1.
Identification of ZID element required for PKCδ-induced Zp activation. (A) The NA, 4A, and RA cells were transfected with EGFP and EGFP-WT-PKCδ plasmids. Cell lysates were harvested, and immunoblot analysis was performed with the indicated antibodies. (B) Analysis of Zp and Rp activity in response to PKCδ induction. NPC-TW01 cells were cotransfected with either Zp (▪) or Rp (□) reporter plasmid, together with EGFP-derived PKCδ expression plasmids (EGFP, EGFP-WT-PKCδ, EGFP-CF-PKCδ, or EGFP-CD-PKCδ) and Renilla luciferase plasmid (pRL-TK). The “relative luciferase activity (fold)” is defined as a normalization of luciferase activity to Renilla activity, followed by standardization with that of the control vector EGFP. (C) Schematic diagram of the −221 to +21 Zp that drives the luciferase gene in the promoter plasmid (left panel). Analysis of activities of Zp deletion mutants in response to PKCδ induction (right panel). NPC-TW01 cells were cotransfected with either of these deletion mutant constructs or vector control (pGL2) in combination with EGFP-derived PKCδ expression plasmids (EGFP, EGFP-WT-PKCδ, or EGFP-CF-PKCδ) and Renilla luciferase plasmid (pRL-TK). The relative luciferase activities were calculated as described above. (D) Analysis of the importance of ZID element in Zp activation mediated by PKCδ. This experiment was performed as described above.
Next, we mapped the response element of Zp required for PKCδ induction. A series of 5′-deletion constructs was generated over the −221 to +12 region of Zp and cotransfected with PKCδ expression plasmids into NPC-TW01 cells for a reporter assay. Serial deletion from positions −221 to −99 did not affect Zp activity (Fig. 1C). However, further deletion from −99 to −80 caused a significant decrease in Zp activity (Fig. 1C), suggesting that the regulatory element for PKCδ activation is within the −99 to −80 region of Zp. It has been reported that this region contains the ZID element, one of the positive cis-active elements of Zp (18). To provide further evidence that the ZID element of Zp is required for PKCδ induction of Zp, a mutant reporter construct of Zp was generated with a ZID internal deletion (ΔZID), and the construct was still unable to be activated by PKCδ (Fig. 1D). This observation suggests that the ZID element is necessary for PKCδ-mediated activation of Zp.
The Sp1 binding site residing in ZID element is important for PKCδ induction.
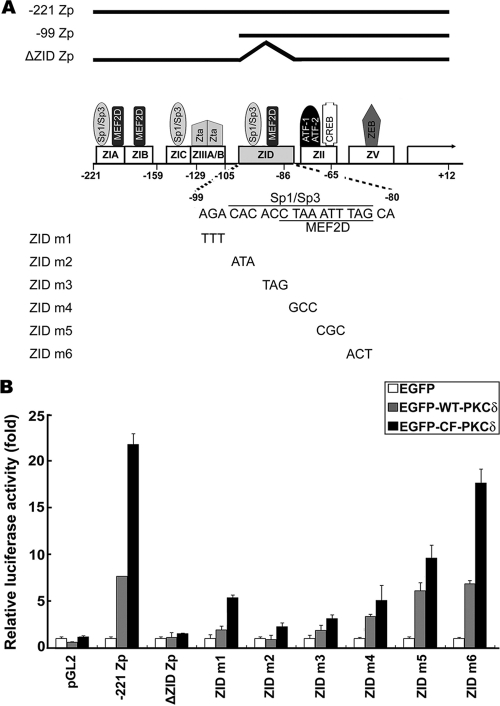
According to nucleotide sequence analysis, the ZID domain contains several cellular transcription factor binding sites, including Sp1, Sp3, and MEF2D, as illustrated in Fig. 2 A (30, 32, 35, 36, 46, 54). To determine which transcription factor binding site in ZID is involved in PKCδ-mediated activation of Zp, we introduced a series of 3-bp mutations into the ZID element sequence (from −99 to −80) by PCR-based site-directed mutagenesis (Fig. 2A). A reporter assay was conducted on NPC-TW01 cells transfected with wild-type Zp (−221Zp), ΔZID Zp, and six ZID mutants (ZID m1, m2, m3, m4, m5, and m6). The data in Fig. 2B show that the luciferase activities were reduced markedly in ZID m1 to m4 mutants compared to wild-type Zp. Of note, the luciferase activities of ZID m2 and m3, which are mutated in the GC-rich region with homology to the Sp1/Sp3 consensus binding site, were decreased to a level similar to that of the ΔZID mutant. On the other hand, the promoter activities of ZID m5 and m6, both of which are mutated in the MEF2D binding site, did not differ significantly from the wild type (Fig. 2B). Taken together, these results suggest that the Sp1/Sp3 binding motif on the ZID element is crucial for PKCδ-mediated transactivation of Zp.
FIG. 2.
Importance of Sp1/Sp3 binding site on ZID for PKCδ induction of Zp. (A) Schematic diagram of defined domain structure and transcription factor binding sites on Zp (−221 to +12). The sequence mutation of ZID reporter mutants is also indicated at the bottom. (B) Analysis of luciferase activities of ZID reporter mutants upon PKCδ induction. NPC-TW01 cells were cotransfected with ZID mutants, −221 Zp, ΔZID, or vector control (pGL2) in combination with EGFP-derived PKCδ expression plasmids (EGFP, EGFP-WT-PKCδ, or EGFP-CF-PKCδ) and Renilla luciferase plasmid (pRL-TK). After 48 h of transfection, the luciferase and Renilla activities were measured and calculated by normalizing the luciferase activity to the Renilla activity and then standardized with that of control vector EGFP. Each experiment was carried out in duplicate and error bars are shown.
Sp1 but not Sp3 is bound by the ZID element of Zp.
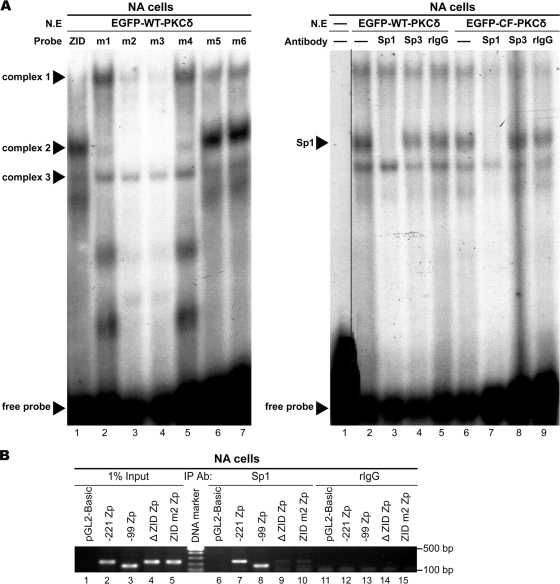
An EMSA was carried out to examine the possibility of interaction between the ZID element and transcription factor Sp1 or Sp3. Nuclear extracts from NA cells transiently expressing PKCδ were probed with a γ-32P-labeled ZID element. The specific DNA-protein complexes were detected in the reactions containing the ZID, the m5 and m6 probe, and partially in m1 and m4, but not in those of m2 and m3 (Fig. 3 A, left panel). These results are correlated well with the reporter assay (Fig. 2B). Furthermore, the DNA-complex 2 on ZID was abolished in the presence of antibody specific to Sp1 (Fig. 3A, right panel, lanes 3 and 7), while antibody against Sp3 had no effect on any complex formation (Fig. 3A, right panel, lanes 4 and 8). The results were seen in both the wild-type (EGFP-WT-PKCδ, lanes 2 to 5) and the constitutively active (EGFP-CF-PKCδ, lanes 6 to 9) forms of PKCδ.
FIG. 3.
Binding of Sp1 to Zp ZID. (A) Dissection of interaction between ZID and Sp1 or Sp3. EMSA is performed with nuclear extracts (N.E) isolated from NA cells transfected with EGFP-WT-PKCδ for 24 h. Oligonucleotides of ZID WT (lane 1) or ZID mutants (lanes 2 to 7) were labeled with γ-32P and incubated with the nuclear extracts. The specific complexes 1 to 3 and free probe are indicated (left panel). Furthermore, γ-32P-labeled ZID element probe containing the putative Sp1/Sp3 binding site was incubated without nuclear extracts (lane 1) or with nuclear extracts prepared from NA cells transfected with EGFP-WT-PKCδ (lanes 2 to 5) or EGFP-CF-PKCδ (lanes 6 to 9) for 24 h. The reaction was carried out in absence or presence of 4 μg of antibodies against Sp1, Sp3 or irrelevant rabbit IgG control (right panel). (B) Confirmation of Sp1 binding to ZID element. The nuclear extracts were prepared from EBV-positive NA cells transfected with the indicated Zp constructs (−221 Zp, −99Zp, ΔZID Zp, and ZID m2 Zp) or control vector (pGL2). These nuclear extracts were sonicated to yield soluble chromatin extracts (500- to 1,000-bp DNA fragments). Subsequently, 200-μg chromatin extracts were immunoprecipitated by using 2 μg of anti-Sp1 antibody or control rabbit IgG (rIgG) and subjected to PCR analysis for detection of Zp DNA (−221 to +12 region). Total crude chromatin extracts were used as input controls.
We next carried out a ChIP assay to confirm the interaction between endogenous Sp1 and ZID in vivo. Several mutant constructs, including −221 Zp, −99 Zp, ΔZID Zp, and ZID m2 Zp were subcloned into the pGL2-Basic vector and then transfected into NA cells. After treatment with TSA for 24 h, the cells were subjected to ChIP analysis. As shown in Fig. 3B, Sp1 was coprecipitated with chromatin containing −221 Zp and the DNA-retained ZID element (−99 Zp), but not the DNA with the ZID deletion (ΔZID Zp) and ZID mutated at the Sp1 site (ZID m2). These observations indicated that Sp1 can bind to the ZID element of Zp.
The DNA-binding ability of Sp1 is essential for activating Zp.
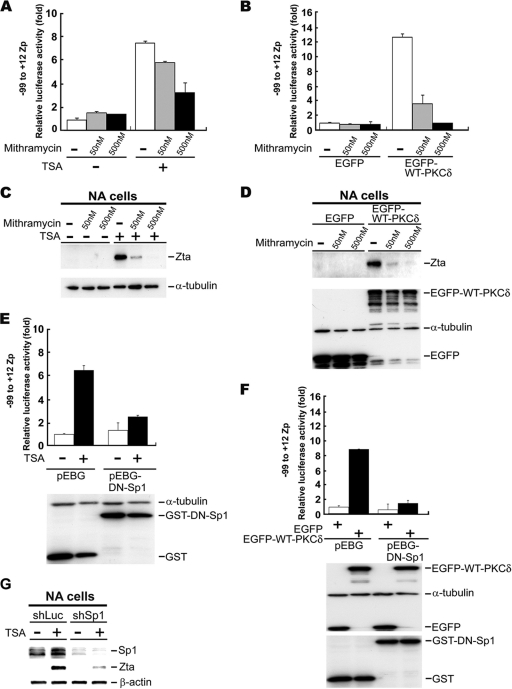
Sp1 is an important cellular transcription factor and regulates a subset of gene expression via direct or indirect binding to promoters (47, 52). Sp1 can activate or repress gene expression (47, 52). Therefore, we sought to explore the mechanism through which it regulates Zp activity in PKCδ-mediated TSA induction. First, we tested whether the DNA-binding activity of Sp1 is required for TSA-induced Zp activity. Mithramycin, a functional inhibitor of Sp1 that prevents it from binding to GC-rich promoter regions, was used in the study. The luciferase activity of the ZID-mini promoter (containing the −99 to +12 region of Zp) decreased in the presence of mithramycin, in a dose-dependent manner, after TSA treatment (Fig. 4 A). The induction of Zp activity by PKCδ was blocked in the presence of mithramycin (Fig. 4B). To investigate further the effect of this Sp1 inhibitor in PKCδ-induced EBV lytic cycle progression, NA cells were transfected with the PKCδ expression plasmid and then treated with mithramycin. Clearly, the Zta expression induced by TSA or PKCδ was inhibited in the presence of mithramycin (Fig. 4C and D).
FIG. 4.
DNA binding ability of Sp1 is essential for Zp activation. (A and B) Blockage of mithramycin on TSA or PKCδ-induced −99 to +12 Zp activity. NPC-TW01 cells transfected with −99 to +12 Zp reporter plasmid and Renilla luciferase plasmid (pRL-TK) were treated with mithramycin at 4 h posttransfection. After 1 h, the cells were incubated with (+) or without (−) TSA (A) for another 24 h or were transfected with PKCδ expression plasmid (B). Then, the luciferase activities were measured and normalized to Renilla activity. Relative promoter activities are indicated as n-fold inductions over the activity of nontreatment (A) or control vector EGFP (B). (C and D) Blockage of mithramycin on TSA-or PKCδ-induced Zta expression. EBV-harboring NA cells were used in the assay, and experiments were performed as described above. Cells were harvested at 24 h posttreatment for immunoblot analysis with the indicated antibodies. (E and F) Blockage of DN-Sp1 on TSA- or PKCδ-induced −99 to +12 Zp activation. The procedure was performed as described above except that the mithramycin treatment was replaced by transfection with DN-Sp1 expression plasmid (pEBG-DN-Sp1) or control plasmid (pEGB). (G) NA cells were transfected with plasmids containing specific oligonucleotides targeting luciferase (shLuc) or Sp1 (shSp1) and then treated with TSA. Cell lysates were harvested, and immunoblot analysis was performed with the indicated antibodies.
To confirm that the DNA-binding activity of Sp1 is crucial for activation of Zp, NPC-TW01 cells were transfected with the ZID-mini promoter and a dominant-negative Sp1 expression plasmid (pEBG-DN-Sp1) or control plasmid pEGB. As shown in Fig. 4E and F, the reporter assay showed that the induction of Zp activity by TSA treatment or PKCδ overexpression was abolished by the expression of dominant-negative Sp1 (DN-Sp1), which lacks the transactivation domain (1, 42). Consistently, a decrease in Zta expression was observed in Sp1 knockdown NA cells (Fig. 4G). These results also indicate that the Sp1 is important for Zta expression after TSA and PKCδ induction.
TSA-triggered serine phosphorylation of Sp1 is by activated PKCδ.
The results presented above indicate that interaction between Sp1 and the ZID element on Zp is required for PKCδ-mediated expression of Zta (Fig. 4). This raises the question of how PKCδ stimulates Sp1-dependent transcription of Zta following TSA treatment. We were curious as to whether PKCδ-induced expression of Zta resulted from an alteration of the amount of Sp1 protein, which has a major effect on the transcriptional activity of Sp1. The results from Western blotting indicated that the level of Sp1 protein did not differ significantly between PKCδ and control vector transfectants, although the level of Zta protein increased in a time-dependent manner after WT-PKCδ transfection (Fig. 5 A). Alternatively, it has been reported that the transcriptional activity of Sp1 also could be influenced by posttranslation modification (47, 52). In order to elucidate the influence of PKCδ on Sp1, we determined whether there is any alteration of Sp1 phosphorylation status after TSA treatment. To achieve this, nuclear extracts from NA cells with or without treatment with TSA and Rottlerin, a PKCδ inhibitor, were immunoprecipitated by Sp1-specific antibody. The immunocomplexes were then analyzed for the phosphorylation status of the Sp1 protein using antibodies against phosphoserine and phosphothreonine. Apparently, TSA treatment caused Sp1 phosphorylation at serine residues, but not threonine residues (Fig. 5B). Moreover, TSA-induced Sp1 phosphorylation was dependent on the kinase activity of PKCδ, because the phosphorylation level was reduced in the presence of Rottlerin (Fig. 5B). The specific role of PKCδ in Sp1 serine phosphorylation was confirmed by overexpression of different mutant forms of PKCδ. Of note, the results from IP-Western blotting showed that only plasmids containing the PKCδ kinase domain, such as WT-PKCδ and CF-PKCδ, could enhance Sp1 phosphorylation markedly and not kinase-inactive PKCδ (CD-PKCδ) or control (Fig. 5C). This led to the conclusion that PKCδ modulates Sp1 phosphorylation during the induction of Zta expression by TSA.
FIG. 5.
Requirement of PKCδ kinase activity in TSA-induced Sp1 serine phosphorylation. (A) Effect of PKCδ on Sp1 expression level. NA cells transfected with EGFP-WT-PKCδ expression plasmid or control EGFP are harvested at the times indicated. Western blotting was conducted to assess the expression of proteins indicated. (B) Effect of TSA and Rottlerin on the phosphorylation of Sp1. NA cells were pretreated with or without Rottlerin (5 μΜ) for 1 h, followed by incubation with TSA (+) or not (−) for 24 h. The respective nuclear extracts were immunoprecipitated with anti-Sp1 antibody or control rabbit IgG (rIgG) and analyzed by Western blotting with antibodies against p-Ser, p-Thr, and Sp1. (C) Effect of PKCδ kinase activity on phosphorylation status of Sp1. Nuclear extracts from NA cells transfected with control EGFP or EGFP-derived PKCδ expression plasmids (EGFP-WT-PKCδ, EGFP-CF-PKCδ, or EGFP-CD-PKCδ) were immunoprecipitated with anti-Sp1 antibody or control rIgG. The immunocomplexes were revealed by Western blotting (left panel). Total nuclear extracts were used as input controls (right panel).
Activated PKCδ is recruited to the ZID element of Zp through association with Sp1.
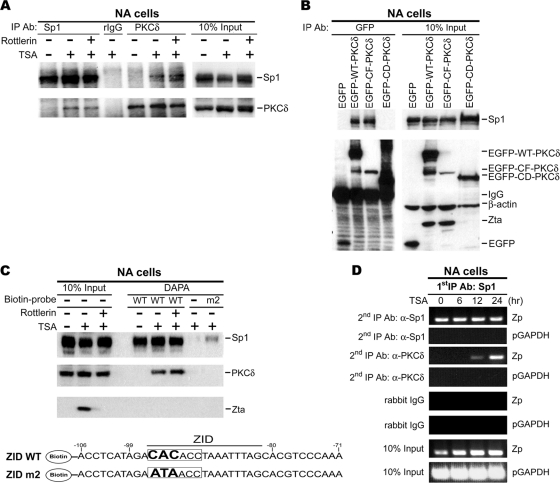
Our previous study demonstrated that TSA treatment not only induces PKCδ kinase activity but also provokes its translocation to the nucleus (31). We wondered whether Sp1 could be a nuclear partner of PKCδ. After TSA treatment with or without Rottlerin, PKCδ was detectable in the Sp1-immunoprecipitate; Sp1 could also be detected in the PKCδ immunocomplex in a reciprocal experiment, while the interaction was not affected by pretreatment with Rottlerin (Fig. 6 A). Interaction between PKCδ and Sp1 was not detected when cells were not treated with TSA (Fig. 6A). Furthermore, the domain required for PKCδ to associate with Sp1 was defined by overexpression of PKCδ domain mutants. The data from IP-Western blotting revealed that WT-PKCδ and CF-PKCδ, but not CD-PKCδ, could interact with Sp1, suggesting that PKCδ associated with Sp1 via its C-terminal kinase domain (Fig. 6B).
FIG. 6.
Recruitment of PKCδ by Sp1 onto the ZID region of Zp after TSA treatment. (A) Demonstration of interaction between PKCδ and Sp1 after TSA treatment. NA cells were stimulated with 1.25 μΜ TSA (+) or not (−) for 24 h in the presence or absence of Rottlerin. Nuclear extracts were immunoprecipitated with antibody against Sp1 or PKCδ and then analyzed by immunoblotting. (B) Examination of the PKCδ domain crucial for Sp1 interaction. NA cells were transfected with EGFP-WT-PKCδ, EGFP-CF-PKCδ, or EGFP-CD-PKCδ expression plasmid or its vector control (EGFP) and harvested at 24 h posttransfection. Nuclear extracts were subjected to immunoprecipitation with anti-GFP antibody, followed by immunoblot analysis. (C) Examination of the capacity of PKCδ binding to Sp1 binding site of ZID element upon TSA treatment. Nuclear extracts were prepared from NA cells that were pretreated with or without Rottlerin for 1 h before incubation with TSA (+) or mock treatment (−) for 24 h. DAPA was performed with the biotin end-labeled oligonucleotides containing the sequence of ZID element from −106 to −71 of Zp (ZID WT) and the Sp1 binding site mutant (ZID m2). The DNA sequences of these probes are shown. Proteins captured by biotin probe were analyzed by Western blotting. A reaction containing the nuclear extracts and streptavidin magnetic beads but without probe (−) was prepared in parallel to rule out nonspecific binding to the beads. Total nuclear extracts were used as the input controls. (D) Examination of a potential interaction between PKCδ and Sp1 on Zp. A re-ChIP assay was carried out on the chromatin extracts from NA cells, following TSA treatment and immunoprecipitated by anti-Sp1 antibody. These Sp1 immunoprecipitants were reimmunoprecipitated with antibody against PKCδ or control rabbit IgG. Then, DNA was extracted and subjected to Zp-targeted PCR with primers (−221 to +12). The detection of GAPDH promoter (pGAPDH, from nucleotide −93 to +60) serves as a negative control.
On the basis of the data presented above showing that the ZID element contains a binding site for Sp1 and that Sp1 physically interacts with PKCδ following TSA treatment (Fig. 3 and 6A), we postulated that PKCδ might be recruited to Zp through its interaction with Sp1. To test this hypothesis, a DAPA was performed. Two biotin-labeled probes, containing the sequence of wild-type ZID (ZID WT) or ZID mutated in the Sp1 binding site (ZID m2), were used for the experiment. Consistent with the observation above, Sp1 was constantly associated with ZID probe, but the association of PKCδ with ZID was seen only in the presence of TSA. However, the interaction of PKCδ and ZID was still detectable after pretreatment with Rottlerin (Fig. 6C). In addition, the results from the experiment with the ZID m2 probe suggested that, whenever the binding of Sp1 was barely detectable, the binding of PKCδ was undetectable (Fig. 6C). Together, PKCδ could interact with ZID-bound Sp1 following TSA induction, and this interaction may facilitate the binding of PKCδ to the ZID element of Zp, suggesting that Sp1 is the essential mediator of PKCδ association with the ZID element.
A re-ChIP assay was carried out to confirm the interaction between Sp1 and PKCδ on Zp in vivo. For the re-ChIP, chromatin extracts were first immunoprecipitated with anti-Sp1 antibody, and then the Sp1-immunocomplexes were reacted with anti-Sp1 or anti-PKCδ antibody for a secondary immunoprecipitation. Figure 6D shows that the Zp DNA associated first with Sp1 could be recovered in the secondary anti-PKCδ immunoprecipitates, suggesting that PKCδ could bind to Zp through direct interaction with Sp1 to regulate Zp activation.
HDAC2 is disassociated from ZID after TSA treatment.
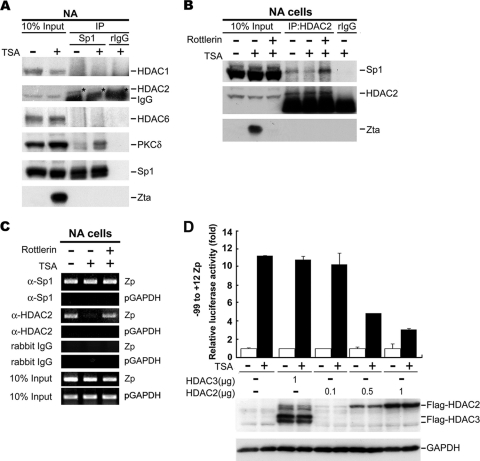
In view of a variety of regulatory mechanisms of Sp1 for different cellular genes (52), we speculated that posttranslational modification of Sp1, resulting in alteration of the DNA-bound complex, is probably involved in our case. This modification may include the ability to bind to the promoter or a change in the protein-protein interaction between Sp1 and its coactivators or corepressors (52). Based on the fact that the Sp1 DNA-binding level on Zp did not change (Fig. 6C and D) and that the serine phosphorylation of Sp1 was enhanced during TSA treatment, we focused our study on searching for candidate protein(s) that associated dynamically with Sp1 after TSA treatment. It has been reported that corepressors such as HDAC1, HDAC2, and HDAC6 may be recruited by Sp1 to repress gene expression (7, 9, 25, 34). On the other hand, coactivators such as p300 and CBP also could be recruited by Sp1 to the promoters to transactivate gene expression (20, 28). We sought to determine which corepressors or coactivators interact with Sp1. The results from IP-Western blotting showed that only HDAC2 was present in Sp1 immunoprecipitates and the level of association was decreased after TSA treatment (Fig. 7 A). The other coactivators, such as p300 and CBP, were not detectable in the Sp1-immune complexes (data not shown).
FIG. 7.
Interaction of Sp1 with HDAC2 during TSA treatment. (A) Effect of TSA on the interaction between Sp1 and transcription corepressors (left panel) or coactivators (right panel). NA cells treated with or without TSA for 24 h were lysed and immunoprecipitated with anti-Sp1 antibody or control rIgG. The immunocomplexes were analyzed by Western blotting to detect the proteins indicated. Total nuclear extracts were used as input control. HDAC2 is marked by asterisks (*). (B) Effect of TSA and Rottlerin on binding of Sp1 and HDAC2. NA cells were pretreated with or without Rottlerin for 1 h before exposure to 24 h of TSA (+) or mock treatment (−). Nuclear extracts were immunoprecipitated with anti-HDAC2 antibody or control rIgG, followed by Western blotting. (C) Effects of TSA and Rottlerin on the binding of endogenous HDAC2 and Zp. NA cells were pretreated with Rottlerin and then incubated with or without TSA for 24 h. The nuclear extracts were subjected to ChIP assay. Soluble chromatin extracts were immunoprecipitated by anti-Sp1, anti-HDAC2 antibodies or rIgG. Finally, the ChIP DNA were detected by PCR with Zp primers (−221 to +12). The detection of GAPDH promoter (pGAPDH, from nucleotide −93 to +60) serves as a negative control. (D) Effect of HDAC2 and HDAC3 on TSA-mediated Zp activation. Zp reporter construct (−99 to +12), Flag-HDAC3, or Flag-HDAC2 expression plasmids were cotransfected into NPC-TW01 cells. After 4 h, the cells were subjected to TSA treatment for 48 h, cell lysates were harvested for luciferase reporter assay (upper panel), and the remaining lysates were analyzed by Western blotting and probed with the indicated antibodies (lower panel).
HDAC2 release from the ZID element is dependent on PKCδ kinase activity.
To determine whether the interaction between HDAC2 and Sp1 is PKCδ kinase dependent, NA cells are preincubated with or without Rottlerin. As shown in Fig. 7B, HDAC2 could interact physically with Sp1 in NA cells under normal conditions and the interaction declined following TSA treatment. It is of interest that the reduction of Sp1-HDAC2 interaction induced by TSA was rescued in the presence of Rottlerin (Fig. 7B). These observations indicate that the reduction of Sp1/HDAC2 interaction induced by TSA is dependent on the kinase activity of PKCδ.
Next, we sought to determine whether HDAC2 and Sp1 are associated with Zp in vivo. Chromatin extracts were immunoprecipitated with anti-Sp1 or anti-HDAC2 antibodies, and the protein bound DNA was detected by PCR using primers encompassing the region −221 to +12 of Zp. Consistent with these data, the association of Zp and Sp1 was observed continuously (Fig. 7C, upper panel). In the absence of TSA treatment, EBV remained in a latent state and HDAC2 was bound to Zp (Fig. 7C, middle panel). During viral lytic progression induced by TSA, the amount of HDAC2 binding to Zp decreased (Fig. 7C, middle panel). The TSA-mediated reduction of HDAC2/Zp association was prevented by the Rottlerin treatment (Fig. 7C, middle panel). These results implied that TSA induced the release of bound HDAC2 from the Sp1 complex on Zp, and the disassociation of this complex was dependent on PKCδ kinase activity. As reported previously, association of HDAC2 and Sp1 on the promoter repressed its transcriptional activity (25, 53, 57). Accordingly, the specificity of HDAC2 inhibition of TSA-mediated Zp activation was examined. As shown in Fig. 7D, Zp activity could be activated up to 11-fold by treatment with TSA. Of note, this elevated Zp activity could be repressed upon HDAC2 overexpression in a dose-dependent manner (Fig. 7D). However, the overexpression of HDAC3 did not affect the promoter activity, even with TSA treatment (Fig. 7D). Viewed in this light, HDAC2 can be considered to be the specific repressor on Zp during EBV latency in epithelial cells.
DISCUSSION
Transcription in eukaryotes is regulated in part by dynamic changes in the structure of chromatin. Histone acetylation typically is involved in this process and is carried out by two classes of enzymes: histone acetyltransferases (HATs) and histone deacetylases (HDACs) (40). Generally, there is a positive correlation between the level of histone acetylation and transcriptional activity. During the last decade, a number of HDAC inhibitors (HDACi) have been shown to impair the function of HDAC and to induce epigenetic modifications, resulting in the alteration of gene expression, cell proliferation, differentiation, or apoptosis (40). Among the inhibitors of HDAC activity, trichostatin A (TSA), originally identified as an antifungal agent, is one of the most efficient (40). In addition to influencing the expression of cellular genes, such as p21, cyclooxygenase-1 (COX-1), and cyclin D3 (29, 56), TSA exerts a great effect on viral gene expression. It is one of the compounds used commonly to trigger the EBV switch from latency to the lytic cycle, although the mechanism of action is unclear.
According to our and other studies, the human tumor virus, EBV, modulates cellular signaling, biological activities, and proliferation potently in order to maintain its life cycle and produce its progeny. After infection, EBV may maintain itself in epithelial cells in a latent form and can be activated whenever the cellular environment is suitable for its replication. During reactivation, EBV takes advantage of the cellular transcriptional machinery to transcribe its gene products, especially the key lytic switch transactivator, Zta. Structurally, Zp, the Zta promoter, contains several transcriptional factor binding sites. Under certain circumstances, such as chemical induction or cellular stress, essential transcriptional factors are recruited onto Zp and stimulate the expression of Zta (46). The virus then utilizes Zta to activate the sequential expression of lytic proteins on the one hand and, on the other, Zta may accelerate the cellular activity to ensure the cellular machinery is available for viral replication. This may drive the host cells toward a dysfunctional state and may contribute to the pathogenesis of EBV-associated diseases. Therefore, the disruption of Zp silencing is the key step to induction of the EBV lytic cycle and the onset of human disease.
In the present study, we demonstrated that the activity of Zp is repressed by HDAC2 during viral latency. Treatment with HDACi leads to the phosphorylation of PKCδ and the phosphorylated PKCδ is then translocated from the cytosol to the nucleus. PKCδ then interacts with ZID-bound Sp1, leading to the recruitment of PKCδ onto the ZID element of Zp, and successively phosphorylates Sp1. This may cause the release of HDAC2 from Zp, thereby activating the promoter.
In the EBV-carrying B-cell model, the major response elements for TPA and anti-Ig treatment are located in the ZIA, ZIB, ZIC, and ZII elements of Zp (46). In the present study, we first showed that the ZID element of Zp is required for PKCδ-elicited EBV reactivation (Fig. 1). Indirectly, Carol et al. showed that mutations in the ZID site strongly reduce Zp activity after induction by anti-Ig in Akata cells (39). Therefore, both studies show clearly that the ZID element may play the essential role for Zp activity induced by different stimuli and in different cell models.
Furthermore, it was of interest to demonstrate that Sp1 is the critical mediator of PKCδ-triggered EBV reactivation. In fact, there is one clue from another study to the binding of Sp1 on ZID. Liu et al. reported that the ZIA domain can be bound by the MEF2D transcription factor and Sp1/Sp3 complexes (35, 36); notably, this binding can be abolished by the presence of the ZID element, suggesting that ZID sequences can compete against the formation of complexes of these transcriptional factors with ZIA (2). Therefore, it is suggested that ZID, as well as ZIA, may contain a binding site for MEF2D and Sp1/Sp3 complexes. Our ZID element mutation analysis showed that the binding site for Sp1/Sp3, but not MEF2D, is essential for PKCδ to stimulate Zp activity (Fig. 2B). Moreover, our data from EMSA, DAPA, and ChIP (Fig. 3 and 6) confirmed that Sp1 binds to the ZID element. The binding of MEF2D to ZID also has been demonstrated in anti-immunoglobulin-treated Akata cells (39).
Sp1 belongs to the zinc finger-containing Sp transcription factor family, which can bind to GC-rich motifs in promoters (44). Structurally, Sp1 contains three domains: the activation domain, the DNA-binding domain, and the protein-protein interacting domain (44). Functionally, Sp1 can regulate an array of biological activities via positive activation or negative repression of gene expression. Sp1 transcriptional activity can be regulated not only quantitatively, through protein expression, but also qualitatively by posttranslational modification, such as phosphorylation, acetylation, sumoylation, ubiquitylation, etc. (47). Of these modifications, phosphorylation has been studied most extensively (10, 47). There are 61 putative phosphorylation sites on the Sp1 protein, with most being serine residues (47). One outcome of phosphorylation affects the recruitment of Sp1 with its interacting proteins and, subsequently, its transcriptional activity. It has been reported that infection with many viruses may alter the phosphorylation status of Sp1 and change its transcription activity. For instance, the HIV Tat protein can promote Sp1 phosphorylation and thus may enhance the activity of the long terminal repeat (LTR) (12).
According to other studies, Sp1 may be involved in cellular gene expression via the PKC signaling pathway. For example, Sp1 sites in the cyclin D3 promoter are required for its activation by the HDAC inhibitor, and this activation occurs via a PKCδ-mediated pathway (29). The data from the present study show that the DNA-binding ability of Sp1 is important for Zp activation and Zta expression following TSA treatment or overexpression PKCδ (Fig. 4). Quantitatively, we did not detect any alteration in the Sp1 protein level (Fig. 5A) or binding to the promoter (Fig. 6C and D) during EBV reactivation, suggesting that other mechanisms may be involved. Posttranslational modifications of Sp1 are known to have a significant impact on Sp1 site-regulated gene expression (47, 52). On the basis of the data presented here, TSA-stimulated phosphorylation of Sp1 appears to require PKCδ activity, and this phosphorylation is essential for EBV reactivation (Fig. 5B). However, we are still investigating which sites of Sp1 are phosphorylated by PKCδ. In fact, we demonstrated interaction between PKCδ and Sp1, suggesting that Sp1 may be target for phosphorylation by PKCδ after TSA treatment (Fig. 6A). Moreover, we provide by re-ChIP the first evidence that PKCδ binds on Zp mediated by association with Sp1 (Fig. 6D).
Several studies have shown interactions of Sp1 with a variety of proteins, including p107, class I HDACs (HDAC1, HDAC2, and HDAC6), p300, and CBP (20, 24, 25, 28, 34, 53, 61). Modulation of the interactions between Sp1 and these partner proteins has been shown to be crucial for the regulation of Sp1-dependent gene expression. In our system, IP-screening analysis showed that only HDAC2 is associated with Sp1, and this interaction can be disrupted after TSA induction (Fig. 7A). Furthermore, overexpression of HDAC2 can inhibit TSA-mediated Zp activation specifically (Fig. 7D). It seems that chromatin structure may contribute to the maintenance of viral latency by suppression of lytic cycle gene expression. A similar finding was demonstrated in the case of HIV, in that Sp1 and c-Myc recruit HDAC1 to its LTR promoter to maintain the latency of HIV-1 (26). For Kaposi's sarcoma-associated herpesvirus (KSHV), the Sp1 binding site and transcriptional initiation site on the ORF50 promoter is occupied by a nucleosome to repress ORF50 expression and prevent lytic replication (55). In B cells, Zp can be repressed by MEF2D through the recruitment of class II histone deacetylases (23). Moreover, our study provides additional evidence that the expression of the EBV immediate-early gene, Zta, seems to be inhibited by histone deacetylation and nucleosome occlusion in the promoter region.
To date, only limited information suggests that PKCδ plays an important role during virus infections. In Kaposi's sarcoma herpesvirus infection, PKCδ is required for TPA-triggered lytic cycle activation (16). However, the overexpression of PKCδ alone is not sufficient to induce lytic reactivation of KSHV because it can activate c-Fos only partially. There is a requirement for another PKC isoform that induces the phosphorylation of c-Jun. c-Fos and c-Jun can then form an active AP-1 complex and induce KSHV reactivation (13, 16). Notably, overexpression of PKCδ alone can induce Zp activity and EBV reactivation in our situation. Our previous study demonstrated that, among the PKC isoforms, PKCδ is essential for TSA-induced EBV reactivation. The signs of PKCδ activation, such as phosphorylation at Thr 505 in the activation loop, nuclear translocation, and protein cleavage, can all be observed after TSA treatment and correlated with EBV reactivation (31). It is of interest that Zp activity (Fig. 1B) and Sp1 phosphorylation (Fig. 5B and C) induced by TSA are dependent on PKCδ kinase activity. Moreover, PKCδ kinase inhibition by Rottlerin has an effect on the interaction between Sp1 and HDAC2 (Fig. 7C) but not on the association of PKCδ and Sp1 bound to Zp (Fig. 6). Therefore, the interaction of PKCδ and Sp1 is indispensable for kinase activity. These findings suggest that the disruption of Sp1 and HDAC2 interaction is dependent on Sp1 modification by PKCδ kinase activity but not via direct competition for the binding site on Sp1 between PKCδ and HDAC2. Therefore, PKCδ phosphorylation and Sp1 modification seem to be required for EBV reactivation by HDAC inhibitors, which is a common feature of cellular gene expression but, to our knowledge, is the first to be reported in viral gene regulation.
Acknowledgments
We thank Tim J. Harrison of UCL Medical School, London, United Kingdom, and Scott C. Schuyler of Department of Biomedical Science, Chang Gung University, Tao-Yuan, Taiwan, for reviewing the manuscript critically.
This study was supported by the National Science Council (grants NSC 99-3112-B-002-022 and NSC 97-2320-B-002-003-MY3 to C.-H.T. and NSC98-2320-B-182-038-MY3 to S.-J.L.) and the National Health Research Institute (grants NHRI-EX-99-9726BI and NHRI-EX100-10031BI to C.-H.T.).
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1.Al-Sarraj, A., R. M. Day, and G. Thiel. 2005. Specificity of transcriptional regulation by the zinc finger transcription factors Sp1, Sp3, and Egr-1. J. Cell Biochem. 94:153-167. [DOI] [PubMed] [Google Scholar]
- 2.Borras, A. M., J. L. Strominger, and S. H. Speck. 1996. Characterization of the ZI domains in the Epstein-Barr virus BZLF1 gene promoter: role in phorbol ester induction. J. Virol. 70:3894-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, L. K., and S. T. Liu. 2000. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 28:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S. S., et al. 2008. Critical role of p53 in histone deacetylase inhibitor-induced Epstein-Barr virus Zta expression. J. Virol. 82:7745-7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y., et al. 2006. Induction of the early growth response 1 gene by Epstein-Barr virus lytic transactivator Zta. J. Virol. 80:7748-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y., et al. 1999. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J. Virol. 73:8857-8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. C., S. Illenye, and N. H. Heintz. 2001. Cooperation of E2F-p130 and Sp1-pRb complexes in repression of the Chinese hamster dhfr gene. Mol. Cell. Biol. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien, Y. C., et al. 2001. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N. Engl. J. Med. 345:1877-1882. [DOI] [PubMed] [Google Scholar]
- 9.Chou, C. W., and C. C. Chen. 2008. HDAC inhibition upregulates the expression of angiostatic ADAMTS1. FEBS Lett. 582:4059-4065. [DOI] [PubMed] [Google Scholar]
- 10.Chu, S., and T. J. Ferro. 2005. Sp1: regulation of gene expression by phosphorylation. Gene 348:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Chua, H. H., et al. 2007. Role of the TSG101 gene in Epstein-Barr virus late gene transcription. J. Virol. 81:2459-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 72:2615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, A., C. Brodie, and R. Sarid. 2006. An essential role of ERK signaling in TPA-induced reactivation of Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 87:795-802. [DOI] [PubMed] [Google Scholar]
- 14.Daibata, M., S. H. Speck, C. Mulder, and T. Sairenji. 1994. Regulation of the BZLF1 promoter of Epstein-Barr virus by second messengers in anti-immunoglobulin-treated B cells. Virology 198:446-454. [DOI] [PubMed] [Google Scholar]
- 15.Davies, A. H., R. J. Grand, F. J. Evans, and A. B. Rickinson. 1991. Induction of Epstein-Barr virus lytic cycle by tumor-promoting and non-tumor-promoting phorbol esters requires active protein kinase C. J. Virol. 65:6838-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutsch, E., et al. 2004. Role of protein kinase C delta in reactivation of Kaposi's sarcoma-associated herpesvirus. J. Virol. 78:10187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flamand, L., and J. Menezes. 1996. Cyclic AMP-responsive element-dependent activation of Epstein-Barr virus zebra promoter by human herpesvirus 6. J. Virol. 70:1784-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemington, E., and S. H. Speck. 1990. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, J., et al. 2009. Loss of NECL1, a novel tumor suppressor, can be restored in glioma by HDAC inhibitor-trichostatin A through Sp1 binding site. Glia 57:989-999. [DOI] [PubMed] [Google Scholar]
- 21.Gao, X., K. Ikuta, M. Tajima, and T. Sairenji. 2001. 12-O-tetradecanoylphorbol-13-acetate induces Epstein-Barr virus reactivation via NF-κB and AP-1 as regulated by protein kinase C and mitogen-activated protein kinase. Virology 286:91-99. [DOI] [PubMed] [Google Scholar]
- 22.Glaser, G., M. Vogel, H. Wolf, and H. H. Niller. 1998. Regulation of the Epstein-Barr viral immediate-early BRLF1 promoter through a distal NF1 site. Arch. Virol. 143:1967-1983. [DOI] [PubMed] [Google Scholar]
- 23.Gruffat, H., E. Manet, and A. Sergeant. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate-early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 3:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, W., et al. 2005. Trichostatin A induces transforming growth factor beta type II receptor promoter activity and acetylation of Sp1 by recruitment of PCAF/p300 to a Sp1.NF-Y complex. J. Biol. Chem. 280:10047-10054. [DOI] [PubMed] [Google Scholar]
- 25.Itoh, Y., et al. 2007. 17beta-estradiol induces IL-1alpha gene expression in rheumatoid fibroblast-like synovial cells through estrogen receptor alpha (ERα) and augmentation of transcriptional activity of Sp1 by dissociating histone deacetylase 2 from ERα. J. Immunol. 178:3059-3066. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, G., A. Espeseth, D. J. Hazuda, and D. M. Margolis. 2007. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 81:10914-10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juan, L. J., et al. 2000. Histone deacetylases specifically downregulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 28.Kim, T. Y., et al. 2008. Transcriptional induction of DLC-1 gene through Sp1 sites by histone deacetylase inhibitors in gastric cancer cells. Exp. Mol. Med. 40:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, Y. H., J. H. Lim, T. J. Lee, J. W. Park, and T. K. Kwon. 2007. Expression of cyclin D3 through Sp1 sites by histone deacetylase inhibitors is mediated with protein kinase C-delta (PKC-δ) signal pathway. J. Cell Biochem. 101:987-995. [DOI] [PubMed] [Google Scholar]
- 30.Lavens, S., et al. 2004. Identification of protein tyrosine kinases required for B-cell-receptor-mediated activation of an Epstein-Barr Virus immediate-early gene promoter. J. Virol. 78:8543-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, H. H., et al. 2008. Essential role of PKCδ in histone deacetylase inhibitor-induced Epstein-Barr virus reactivation in nasopharyngeal carcinoma cells. J. Gen. Virol. 89:878-883. [DOI] [PubMed] [Google Scholar]
- 32.Lin, C. T., et al. 1993. Characterization of seven newly established nasopharyngeal carcinoma cell lines. Lab. Invest. 68:716-727. [PubMed] [Google Scholar]
- 33.Lin, J. C., et al. 2004. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N. Engl. J. Med. 350:2461-2470. [DOI] [PubMed] [Google Scholar]
- 34.Lin, Y. C., et al. 2008. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 68:2375-2383. [DOI] [PubMed] [Google Scholar]
- 35.Liu, S., A. M. Borras, P. Liu, G. Suske, and S. H. Speck. 1997. Binding of the ubiquitous cellular transcription factors Sp1 and Sp3 to the ZI domains in the Epstein-Barr virus lytic switch BZLF1 gene promoter. Virology 228:11-18. [DOI] [PubMed] [Google Scholar]
- 36.Liu, S., P. Liu, A. Borras, T. Chatila, and S. H. Speck. 1997. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J. 16:143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacCallum, P., L. Karimi, and L. J. Nicholson. 1999. Definition of the transcription factors which bind the differentiation responsive element of the Epstein-Barr virus BZLF1 Z promoter in human epithelial cells. J. Gen. Virol. 80(Pt. 6):1501-1512. [DOI] [PubMed] [Google Scholar]
- 38.Manet, E., et al. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional transactivators. EMBO J. 8:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald, C., C. Elgueta Karstegl, P. Kellam, and P. J. Farrell. 2010. Regulation of the Epstein-Barr virus Zp promoter in B lymphocytes during reactivation from latency. J. Gen. Virol. 91:622-629. [DOI] [PubMed] [Google Scholar]
- 40.Minucci, S., and P. G. Pelicci. 2006. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 6:38-51. [DOI] [PubMed] [Google Scholar]
- 41.Newton, A. C. 2001. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 101:2353-2364. [DOI] [PubMed] [Google Scholar]
- 42.Petersohn, D., and G. Thiel. 1996. Role of zinc-finger proteins Sp1 and zif268/egr-1 in transcriptional regulation of the human synaptobrevin II gene. Eur. J. Biochem. 239:827-834. [DOI] [PubMed] [Google Scholar]
- 43.Piovan, E., et al. 2003. Tumor outgrowth in peripheral blood mononuclear cell-injected SCID mice is not associated with early Epstein-Barr virus reactivation. Leukemia 17:1643-1649. [DOI] [PubMed] [Google Scholar]
- 44.Safe, S., and M. Abdelrahim. 2005. Sp transcription factor family and its role in cancer. Eur. J. Cancer 41:2438-2448. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu, N., and K. Takada. 1993. Analysis of the BZLF1 promoter of Epstein-Barr virus: identification of an anti-immunoglobulin response sequence. J. Virol. 67:3240-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 47.Tan, N. Y., and L. M. Khachigian. 2009. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell. Biol. 29:2483-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarodi, B., T. Subramanian, and G. Chinnadurai. 1994. Epstein-Barr virus BHRF1 protein protects against cell death induced by DNA-damaging agents and heterologous viral infection. Virology 201:404-407. [DOI] [PubMed] [Google Scholar]
- 49.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 50.Tsai, S. C., et al. 2009. EBV Zta protein induces the expression of interleukin-13, promoting the proliferation of EBV-infected B cells and lymphoblastoid cell lines. Blood 114:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Y. C., J. M. Huang, and E. A. Montalvo. 1997. Characterization of proteins binding to the ZII element in the Epstein-Barr virus BZLF1 promoter: transactivation by ATF1. Virology 227:323-330. [DOI] [PubMed] [Google Scholar]
- 52.Wierstra, I. 2008. Sp1: emerging roles-beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 372:1-13. [DOI] [PubMed] [Google Scholar]
- 53.Won, J., J. Yim, and T. K. Kim. 2002. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 277:38230-38238. [DOI] [PubMed] [Google Scholar]
- 54.Wu, F. Y., et al. 2004. CCAAT/enhancer binding protein alpha binds to the Epstein-Barr virus (EBV) ZTA protein through oligomeric interactions and contributes to cooperative transcriptional activation of the ZTA promoter through direct binding to the ZII and ZIIIB motifs during induction of the EBV lytic cycle. J. Virol. 78:4847-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye, J., D. Shedd, and G. Miller. 2005. An Sp1 response element in the Kaposi's sarcoma-associated herpesvirus open reading frame 50 promoter mediates lytic cycle induction by butyrate. J. Virol. 79:1397-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi, T., et al. 2007. Trichostatin A-mediated upregulation of p21(WAF1) contributes to osteoclast apoptosis. Exp. Mol. Med. 39:213-221. [DOI] [PubMed] [Google Scholar]
- 57.Yokota, T., et al. 2004. Histone deacetylase inhibitors activate INK4d gene through Sp1 site in its promoter. Oncogene 23:5340-5349. [DOI] [PubMed] [Google Scholar]
- 58.Zalani, S., A. Coppage, E. Holley-Guthrie, and S. Kenney. 1997. The cellular YY1 transcription factor binds a cis-acting, negatively regulating element in the Epstein-Barr virus BRLF1 promoter. J. Virol. 71:3268-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1995. The Zif268 cellular transcription factor activates expression of the Epstein-Barr virus immediate-early BRLF1 promoter. J. Virol. 69:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zalani, S., E. A. Holley-Guthrie, D. E. Gutsch, and S. C. Kenney. 1992. The Epstein-Barr virus immediate-early promoter BRLF1 can be activated by the cellular Sp1 transcription factor. J. Virol. 66:7282-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y., M. Liao, and M. L. Dufau. 2006. Phosphatidylinositol 3-kinase/protein kinase Cζ-induced phosphorylation of Sp1 and p107 repressor release have a critical role in histone deacetylase inhibitor-mediated derepression of transcription of the luteinizing hormone receptor gene. Mol. Cell. Biol. 26:6748-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]