Abstract
Endoplasmic reticulum (ER)-to-cytosol membrane transport is a decisive infection step for the murine polyomavirus (Py). We previously determined that ERp29, a protein disulfide isomerase (PDI) member, extrudes the Py VP1 C-terminal arm to initiate ER membrane penetration. This reaction requires disruption of Py's disulfide bonds. Here, we found that the PDI family members ERp57, PDI, and ERp72 facilitate virus infection. However, while all three proteins disrupt Py's disulfide bonds in vitro, only ERp57 and PDI operate in concert with ERp29 to unfold the VP1 C-terminal arm. An alkylated Py cannot stimulate infection, implying a pivotal role of viral free cysteines during infection. Consistent with this, we found that although PDI and ERp72 reduce Py, ERp57 principally isomerizes the virus in vitro, a reaction that requires viral free cysteines. Our mutagenesis study subsequently identified VP1 C11 and C15 as important for infection, suggesting a role for these residues during isomerization. C11 and C15 also act together to stabilize interpentamer interactions for a subset of the virus pentamers, likely because some of these residues form interpentamer disulfide bonds. This study reveals how a PDI family functions coordinately and distinctly to promote Py infection and pinpoints a role of viral cysteines in this process.
Penetration of the host membrane represents a decisive step in virus infection. For enveloped viruses that are surrounded by a lipid bilayer, such as HIV and influenza virus, this process requires fusion of viral and host membranes, resulting in the de facto delivery of the viral particle across the limiting membrane (14). In contrast, the mechanism by which nonenveloped viruses penetrate biological membranes is less clear (29). Despite this uncertainty, certain common principles have emerged from studies of their penetration mechanisms.
One of these principles is that conformational changes are imparted to the nonenveloped virus at the membrane penetration site (29). This remodeling event may generate a hydrophobic viral particle that binds and disrupts the limiting membrane, leading to the subsequent transfer of a subviral particle across the membrane. Alternatively, the conformational change can release an internal virus peptide harboring intrinsic lytic activity (often called a lytic peptide) buried inside the native virus. Disruption of the limiting membrane by this peptide enables virus transport across the membrane. In both instances, the critical trigger for virus transport is the conformational change the viral particle experiences. These structural alterations occur as a result of the concerted actions of numerous cellular factors acting on the virus. How the distinct functions of each contributing cellular factor are coordinated to produce the final penetration-competent capsid conformation remains poorly understood.
Previous studies describing the intracellular trafficking and structure of nonenveloped viruses frame our understanding of how these viruses cause infection. For example, to infect cells, the nonenveloped murine polyomavirus (Py) binds to glycolipid receptors called ganglioside GD1a or GT1b (5, 21, 28) and is transported in a retrograde manner to the endoplasmic reticulum (ER), where the virus penetrates the ER membrane to access the cytosol (30). From the cytosol, Py is transferred into the nucleus, with the ensuing transcription and replication of the viral genome leading to lytic infection or cell transformation. How Py is transported across the ER membrane from the ER lumen into the cytosol is a complicated process that recent studies have begun to unravel (6, 10, 11, 17-19).
Structurally, Py is composed of 72 pentamers of the major coat protein VP1, which encloses its DNA genome (9, 23, 24). Twelve of the pentamers are surrounded by five other pentamers (i.e., five coordinated), while the remaining 60 pentamers are surrounded by six other pentamers (i.e., six coordinated) (9). Each pentamer associates with a single copy of the minor protein VP2 or VP3 (2).
Three major forces stabilize the architecture of the viral capsid. First, the C terminus of VP1 invades a neighboring VP1 pentamer that stabilizes interpentamer interactions (9, 23). Second, intrapentamer disulfide bonds between cysteine 19 of one monomer and cysteine 114 of another monomer further stabilize the VP1 capsid (24, 26). While the X-ray structure of Py indicates that C273 and C282 do not form disulfide bonds, this structure does not provide information on the nature of the remaining C11 and C15 residues located at the VP1 N terminus (24, 26). Third, calcium ions that bind to the virus provide additional structural support (25). Thus, local unfolding of the VP1 C-terminal arm, disulfide bond disruption, and removal of calcium ions are reactions that destabilize Py structure, initiating the uncoating process that prepares the virus for ER membrane penetration. The host activities responsible for these structural disruptions, however, have not been fully defined. In the case of the related simian polyomavirus simian virus 40 (SV40), ER-resident protein disulfide isomerase (PDI) family members have been shown to disrupt the viral disulfide bonds to facilitate infection (20).
We showed previously that ERp29, a PDI family member, extrudes Py VP1 in a reaction that requires disruption of the virus disulfide bonds (11). ERp29, however, does not appear to act on SV40 (20). Here, using a combination of cell infection studies and biochemistry, we identify a network of ER-resident PDI proteins called ERp57, PDI, and ERp72 that act on Py's disulfide bonds to facilitate infection and reveal an important role of the previously uncharacterized viral C11 and C15 residues in mediating this process.
MATERIALS AND METHODS
Reagents.
Crude and purified murine Py, NIH 3T3 cells, and M1 VP1 antibody were generously provided by T. Benjamin (Harvard Medical School, Boston, MA). Polyclonal I-58 VP1 antibody was provided by R. Garcea (University of Colorado, Boulder, CO), the polyclonal antibody against ERp29 was a gift from S. Mkrtchian (Karolinska Institutet, Stockholm, Sweden), the polyclonal antibody against ERp57 was a gift from S. High (University of Manchester, Manchester, England), and the polyclonal antibody against Derlin-1 was a gift from T. Rapoport (Harvard Medical School, Boston, MA). The monoclonal antibody against BiP was purchased from BD Biosciences (San Jose, CA). The polyclonal antibody against PDI was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The polyclonal antibody against ERp72 was purchased from Assay Designs (Ann Arbor, MI). Dulbecco's modified Eagle's medium (DMEM), Optimem, Lipofectamine 2000, and 0.05% trypsin-EDTA were purchased from Invitrogen (Carlsbad, CA). Fetalclone III (FC) was purchased from HyClone (Logan, UT). Complete Mini EDTA-free protease inhibitor cocktail tablets were purchased from Roche (Indianapolis, IN). The cross-linking reagent dithiobis succinimidylpropionate (DSP) was purchased from Pierce Biotechnology (Rockford, IL). Reduced and oxidized glutathione, proteinase K, trypsin, dithiothreitol (DTT), N-ethylmaleimide (NEM), and anti-FLAG M2-agarose were purchased from Sigma (St. Louis, MO). Calmodulin was purchased from Calbiochem. Micro Bio-Spin P-30 Tris chromatography columns were purchased from Bio-Rad.
siRNA knockdown.
Stealth RNA interference (RNAi) negative-control duplexes (low or medium GC% duplex) were purchased from Invitrogen. Duplex small interfering RNAs (siRNAs) corresponding to a segment of mouse PDI (siRNA 1, 5′-GCA ACA ACU UUG AGG GUG AUU-3′ and 5′-UCA CCC UCA AAG UUG UUG CUU-3′; siRNA 2, 5′-GCA ACA ACU UUG AGG GUG AUU-3′ and 5′-UCA CCC UCA AAG UUG UUG CUU-3′), mouse ERp57 (siRNA 1, 5′-CCA GCA ACU UGA GAG AUA ATT-3′ and 5′-UUA UCU CUC AAG UUG CUG GCT-3′; siRNA 2, 5′-GCC AGC AAC UUG AGA GAU AUU-3′ and 5′-UAU CUC UCA AGU UGC UGG CUU-3′), and mouse ERp72 (siRNA 1, 5′-GCA GUU UGC UCC AGA AUA UTT-3′ and 5′-AUA UUC UGG AGC AAA CUG CTT-3′; siRNA 2, 5′-UGA CAA AGA UAC AGU GCU AUU-3′ and 5′-UAG CAC UGU AUC UUU GUC AUU-3′) were synthesized by Invitrogen. One hundred nanomolar (ERp57 siRNA 2 and ERp72 siRNA 2) or 200 nM (ERp57 siRNA 1, ERp72 siRNA 1, and PDI siRNA 1 and 2) duplexed siRNAs were transfected into 15 to 30% confluent NIH 3T3 cells using Lipofectamine 2000 according to the manufacturer's protocol. Negative-control duplexes were transfected similarly at 100 nM or 200 nM concentrations. Protein expression was assessed by SDS-PAGE and immunoblot analysis at 72 h posttransfection. Infection assays were initiated 72 h posttransfection.
XBP1 splicing assay.
Detection of XBP1 splicing was performed as described previously (31). The forward and reverse primers used were 5′ GAA CCA GGA GTT AAG AAC ACG 3′ and 5′ AGG CAA CAG TGT CAG AGT CC 3′, respectively.
Infection assay.
Cells treated with the indicated siRNA were plated on glass coverslips 48 h posttransfection at a density of 3 × 104 cells/well in a 6-well plate. At 72 h posttransfection, crude Py (100 PFU/cell) was added to the cells in fresh DMEM plus 10% FC. The infected cells were incubated at 37°C for 24 h, washed with phosphate-buffered saline (PBS), provided with fresh DMEM plus 10% FC, and allowed to incubate for an additional 24 h. The cells were then fixed and stained with a rat monoclonal antibody against murine Py large T antigen and a rhodamine-conjugated donkey anti-rat IgG (Jackson ImmunoResearch, West Grove, PA). Each condition depicted in Fig. 1C and F (also see Fig. 3B and 5A) represents the average value of at least three independent experiments. In each experiment, at least 500 cells per condition were scored for the absence or presence of nuclear large T antigen expression using standard immunofluorescence microscopy as described previously (19). A Nikon epifluoresence microscope (model Eclipse TS100; Nikon, Melville, NY) equipped with a Texas Red emission filter and a 40× objective was used.
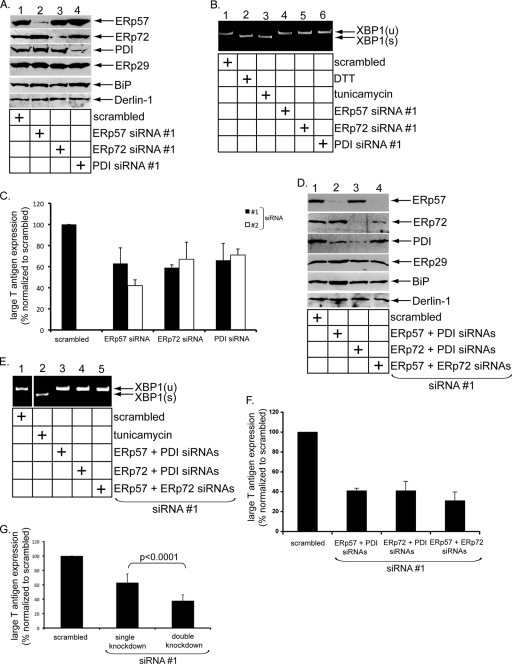
FIG. 1.
ERp57, ERp72, and PDI facilitate Py infection. (A) Knockdown of PDI family members. NIH 3T3 cells were transfected with the indicated siRNA, and lysates derived from the cells were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. (B) Induction of XBP1 splicing. Shown is RT-PCR analysis of the unspliced (u) and spliced (s) forms of the XBP1 mRNA from cells treated with DTT or tunicamycin or transfected with the indicated siRNA. (C) Py infection in knockdown cells. Cells transfected with scrambled siRNA or one of two independent siRNAs against ERp57, ERp72, and PDI (siRNAs 1 and 2) were challenged with Py (100 PFU/cell), and the large T antigen expression was analyzed by standard immunofluorescence microscopy. The values were normalized to scrambled siRNA. The data represent the means and standard deviations (SD) of at least three independent experiments. (D) As for panel A, except the indicated siRNAs were used. (E) As for panel B, except the indicated siRNAs were used. (F) As for panel C, except the indicated siRNAs were used. (G) Average infection of single- and double-knockdown cells. A two-tailed t test was used.
Production of recombinant PDI, ERp57, and ERp72.
PDI proteins were expressed and purified as described previously (4). Full-length mouse PDI, human ERp57, and mouse ERp72 containing N-terminal His6 tags were expressed from pQE30 (Qiagen) constructs in Escherichia coli strain BL21-pro (Clontech) for 2 to 4 h at 37°C upon induction with isopropyl thio-β-galactoside (1 mM; Invitrogen). Cells were lysed by incubation in buffer containing 1% Triton X-100, 300 mM potassium acetate (KOAc), 250 mM sucrose, 2 mM magnesium acetate [Mg(OAc)2], 50 mM HEPES (pH 7.5), and protease inhibitors, followed by sonication. The lysates were centrifuged, and the resulting supernatant fractions were applied to a nickel nitrilotriacetic acid-agarose column (Qiagen) in the presence of imidazole (20 mM, Sigma). The His-tagged proteins were eluted from the column with imidazole (100, 300, or 500 mM). Eluates containing purified proteins were dialyzed extensively overnight in PBS, frozen in liquid nitrogen, and stored at −80°C.
Py reduction and isomerization assay.
Reaction mixtures containing the indicated components were incubated for 1 h at 37°C. Each reaction mixture was subjected to nonreducing SDS-PAGE, followed by immunoblotting with an antibody against VP1. As shown in Fig. 2B to D, untreated purifed Py (100 ng) was incubated in the presence or absence of ERp57 (8 μM), ERp72 (6 μM), or PDI (5 μM). ERp57 (heat treated) was heated for 1 h at 95°C prior to incubation with Py. NEM-treated ERp57, ERp72, and PDI were reacted with NEM (10 mM) for 2 h at 37°C, followed by overnight dialysis against PBS to remove excess NEM. For the reactions shown in Fig. 3C to D, the reaction mixtures were treated as for Fig. 2B to D, except that the virus used was either mock treated or NEM treated as indicated. Where indicated, DTT (5 mM) was added to the reaction mixtures.
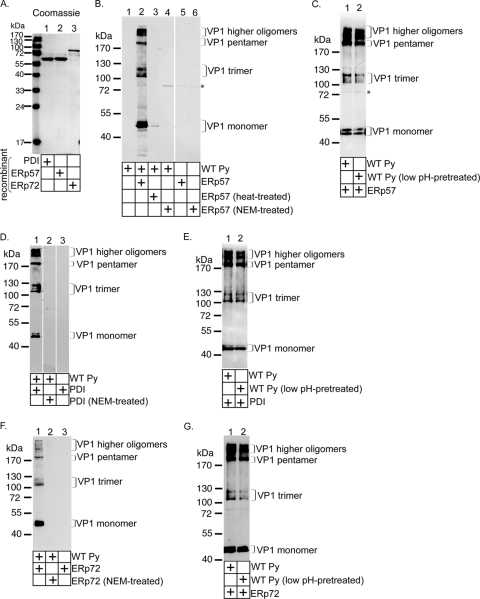
FIG. 2.
ERp57, PDI, and ERp72 disrupt Py's disulfide bonds in vitro. (A) Expression and purification of ERp57, PDI, and ERp72. N-terminally His-tagged ERp57, PDI, and ERp72 constructs were expressed in bacteria, purified to homogeneity, and subjected to Coomassie staining. (B) ERp57 acts on Py directly. Where indicated, purified Py was incubated with ERp57, heat-treated ERp57, or NEM-treated ERp57. The samples were subjected to nonreducing SDS-PAGE and immunoblotted with an I-58 VP1 antibody. The asterisk indicates a nonspecific band recognized by the VP1 antibody. (C) As for panel B, except Py was pretreated at pH 5. (D) PDI acts on Py directly. PDI or NEM-treated PDI was incubated with or without purified Py and analyzed as for panel B. (E) As for panel D, except Py was pretreated at pH 5. (F) ERp72 acts on Py directly. ERp72 or NEM-treated ERp72 was incubated with or without purified Py and analyzed as for panel B. (G) As for panel F, except Py was pretreated at pH 5.
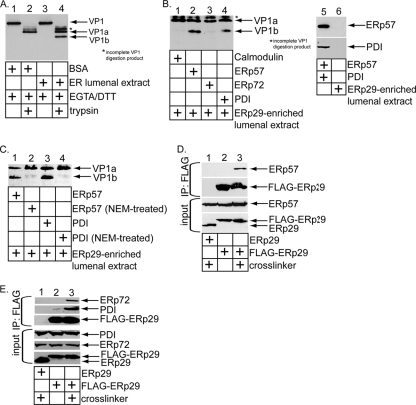
FIG. 3.
ERp57 and PDI function coordinately with ERp29 to unfold Py in vitro. (A) VP1 digestion pattern. Crude Py was incubated with DTT, EGTA, and either BSA or an ER lumenal extract, followed by trypsin addition where indicated. The samples were subjected to SDS-PAGE, followed by immunoblotting with a VP1 antibody. (B) Purified Py was pretreated with either calmodulin, ERp57, ERp72, or PDI in the presence of EGTA. The samples were then incubated with an ERp29-enriched ER lumenal extract, supplemented with trypsin, subjected to SDS-PAGE, and immunoblotted with a VP1 antibody. A 10% input for the amounts of ERp57, PDI, and the ERp29-enriched ER lumenal extract used is shown. (C) As for panel B, except NEM-treated ERp57 and PDI were used where indicated. (D) NIH 3T3 cells transfected with either a rat ERp29 or an N-terminally FLAG-tagged rat ERp29 construct were treated with the DSP cross-linker or left untreated. The resulting cell lysates were subjected to immunoprecipitation using an antibody directed against the FLAG epitope conjugated to agarose (IP:FLAG). The precipitates, as well as the cell lysates (input), were subjected to SDS-PAGE and immunoblotted with antibodies against ERp57 and ERp29. (E) As for panel D, except antibodies against ERp72 and PDI were used instead of antibodies against ERp57.
Acid pretreatement of Py.
Wild-type (WT) Py was pretreated under acidic conditions (pH 5) or neutral conditions (pH 7) as described previously (16).
Alkylation of Py.
Crude or purified Py was alkylated with NEM (10 mM) for 2 h at 37°C or mock treated. Excess NEM was removed efficiently with subsequent buffer exchange using a spin column (Bio-Rad) according to the manufacturer's protocol.
Trypsin digestion assay.
An ER lumenal extract was produced from dog pancreatic microsomes by three freeze-thaw cycles, followed by centrifugation of the microsomes at 50,000 × g for 30 min to remove the membrane material. The contents of the supernatant represented soluble proteins in the ER lumen, referred to as an ER lumenal extract. Trypsin digestion assays were performed similarly to those described previously (11). For experiments using crude virus, virus was pretreated with DTT (3 mM) and EGTA (10 mM) for 20 min at 37°C, followed by the addition of the ER lumenal extract or bovine serum albumin (BSA) (1 mg/ml) and continued incubation for 1 h at 37°C. The reaction mixtures were then treated with trypsin (0.25 mg/ml) for 30 min at 4°C or left untreated. The reaction was stopped by the addition of TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) (1 mM) for 10 min at 4°C. Samples were analyzed by reducing SDS-PAGE, followed by immunoblotting with a VP1 antibody.
For experiments using purified Py, an ER lumenal extract enriched for ERp29 was used as described previously (11). This extract was pretreated with DTT (1 mM) for 1 h at 37°C, followed by 3 to 6 h of dialysis against PBS to remove excess DTT. Calmodulin was also dialyzed against PBS to remove trace amounts of protease inhibitors. The purified Py (50 ng) was first incubated with the indicated protein (10 μM) and EGTA (10 mM) for 1 h at 37°C. An ER lumenal extract (1 mg/ml) was then added to the reaction mixtures and incubated for 30 min at 37°C. Trypsin (0.25 mg/ml) was added next, and the incubation proceeded for 30 min at 4°C. The reaction was stopped by the addition of TLCK (1 mM) for 10 min at 4°C. Samples were analyzed by reducing SDS-PAGE, followed by immunoblotting with a VP1 antibody.
Construction of N-terminally FLAG-tagged rat ERp29.
The cDNA of ERp29 was amplified by PCR using a 5′ primer containing the FLAG tag sequence with pcDNA3.1(+)-rat ERp29 as a template (18). The fragment was inserted into modified pcDNA3.1 (Invitrogen) that contained the ERp29 signal sequence.
Chemical cross-linking and coimmunoprecipitation analyses.
NIH 3T3 cells were grown to 80 to 90% confluence on a 10-cm dish before being transfected with constructs expressing rat ERp29 or N-terminally FLAG-tagged rat ERp29 using Lipofectamine 2000 according to the manufacturer's protocol. At 48 h posttransfection, the cells were harvested, pelleted, and washed twice with PBS. Where indicated, the cells were treated with the DSP cross-linker or mock treated. DSP dissolved in dimethyl sulfoxide (DMSO) (25 mM) was diluted to a concentration of 1 mM in 1.5 ml of PBS, which was used to resuspend the cells, followed by incubation for 1 h at 4°C. The cells were pelleted, and the DSP was removed. After being washed with PBS, the cells were lysed for 30 min at 4°C in a buffer containing 1% Triton X-100 for DSP-treated cells or 1% deoxyBigChap (Calbiochem) for mock-treated cells. The cells were centrifuged at 16,000 × g for 15 min, and 10% of the supernatant was taken as input. A 30-μl slurry of an anti-FLAG M2 agarose was equilibrated, added to the remaining supernatant, and incubated overnight at 4°C. The agarose was pelleted, and the supernatant was removed before extensive washing. Samples were subjected to SDS-PAGE, followed by immunoblotting with the appropriate antibody.
Mutagenesis of Py and analyses of WT and mutant viruses.
The WT Py genome (RA strain) cloned into a PBS vector was generously provided by T. Benjamin (Harvard Medical School, Boston, MA) and was used as a template for PCR-based site-directed mutagenesis with a QuikChange II site-directed mutagenesis kit from Stratagene (La Jolla, CA). The desired mutations were confirmed by sequencing. The viral genomes were removed from the PBS construct by restriction digestion and religated in a dilute reaction. Purified WT or mutant genomes were transfected into 80 to 90% confluent NIH 3T3 cells using Lipofectamine 2000 according to the manufacturer's protocol. After 24 h, the cells were washed and provided with fresh medium containing penicillin-streptomycin (Invitrogen). Medium containing viral particles was collected 5 to 7 days posttransfection and used for subsequent infection and in vitro experiments. The medium containing viral particles was subjected to reducing or nonreducing SDS-PAGE and immunoblotting with an antibody against VP1. Infection assays were performed essentially as described above, and cells were treated with equal amounts of crude WT or mutant virus as determined by VP1 signal in immunoblots.
Proteolytic analyses of WT, alkylated, and mutant Py.
Crude WT, mutant, alkylated, or mock-treated Py was incubated for 30 min at 4°C with various concentrations of proteinase K as indicated. Samples were subjected to reducing SDS-PAGE, followed by immunoblotting with an antibody against VP1.
Native agarose electrophoresis of WT and mutant Py.
Crude WT or mutant virus was mixed with sample-loading buffer without reducing agent or SDS and loaded onto a 0.4% agarose gel. Electrophoresis in 50 mM Tris-acetate (pH 8.1) was carried out at 4°C for at least 3.5 h, with the running buffer replaced frequently.
RESULTS
ERp57, ERp72, and PDI facilitate Py infection.
Using an in vitro system, we showed previously that the PDI-like protein ERp29 extrudes the C-terminal arm of VP1 in a remodeling reaction that depends on the reductant DTT (11). This finding suggests that disulfide bond reduction or isomerization of Py is required for extrusion of the VP1 C-terminal arm and implicates ER-resident reductases and/or isomerases in this reaction. While stable knockdown of PDI in the heterologous human HeLa cell line blocks Py infection (6), it is not known whether PDI or other PDI family members reduce and/or isomerize Py directly.
Three PDI family members that are highly expressed in the ER are ERp57, PDI, and ERp72. To test whether any of these factors plays a role in Py infection, we downregulated each protein in the murine fibroblast NIH 3T3 cells using two different siRNA oligonucleotides (i.e., siRNA oligonucleotides 1 and 2) against ERp57, PDI, and ERp72. Cell lysates derived from cells transfected with scrambled or ERp57-, ERp72-, and PDI-specific siRNAs were subjected to SDS-PAGE and immunoblotting. In cells incubated with the ERp57-specific siRNA 1, the ERp57 level decreased without significantly affecting the levels of ERp72, PDI, and ERp29 (Fig. 1A, top four rows, compare lane 2 to lane 1). Importantly, downregulation of ERp57 did not markedly cause the upregulation of the known unfolded protein response (UPR) markers BiP and Derlin-1 (13) (Fig. 1A, 5th and 6th rows from top, compare lane 2 to lane 1), indicating that the absence of ERp57 did not cause profound ER stress. ERp72 (Fig. 1A, second row from top, lane 3) and PDI (Fig. 1A, third row from top, lane 4) were also downregulated efficiently and specifically when cells were treated with the corresponding siRNA 1. Again, in these cases, BiP and Derlin-1 expression was also not affected (Fig. 1A, 5th and 6th rows from top, compare lanes 3 and 4 to lane 1).
To further verify that knockdown of the PDI proteins (using siRNA 1) did not induce ER stress leading to UPR stimulation, we asked whether splicing of the XBP1 transcription factor mRNA was activated and found that it was not (Fig. 1B, compare lanes 4 to 6 to lane 1), in contrast to incubating cells with the known ER stress inducers DTT and tunicamycin (Fig. 1B, compare lanes 2 and 3 to lane 1). These findings demonstrate that the three PDI family proteins can be effectively and specifically downregulated without triggering significant detectable ER stress.
Py infection, detected by expression of the virus-encoded large T antigen, was decreased by approximately 40% when the PDI family proteins were knocked down using siRNA 1 (Fig. 1C). When two PDI family members were knocked down simultaneously in all combinations using siRNA 1 (Fig. 1D, top three rows), ER stress was not triggered in any of the combinations (Fig. 1D, 4th, 5th, and 6th rows from top, and E). Under these double-knockdown conditions, infection was reduced by at least 60% in all combinations (Fig. 1F). The average infection decrease in the single- and double-knockdown experiments is shown in Fig. 1G. Cells were unhealthy when simultaneous downregulation of all three PDI family proteins was attempted.
The PDI family proteins were downregulated efficiently when another siRNA (i.e., siRNA 2) was transfected in NIH 3T3 cells (data not shown). Under these conditions, Py infection also decreased, similar to the results using siRNA 1 (Fig. 1C). We conclude that ERp57, PDI, and ERp72 play important roles in mediating Py infection.
ERp57, PDI, and ERp72 disrupt Py's disulfide bonds in vitro.
We next asked whether the three PDI family members disrupt Py's disulfide bonds in vitro. Recombinant N-terminally His-tagged mammalian PDI, ERp57, and ERp72 were expressed in bacteria and purified to homogeneity (Fig. 2A, lanes 1 to 3). We first examined the ERp57 activity. Py was incubated with or without ERp57, and the samples were subjected to nonreducing SDS-PAGE, followed by immunoblotting with an antibody against VP1. In the absence of ERp57, immunoblotting for VP1 did not detect any bands (Fig. 2B, lane 1), indicating that the native virus was stable and unable to migrate into the gel. In contrast, band patterns approximately corresponding to the sizes of the VP1 monomer (42 kDa), trimer (126 kDa), pentamer (210 kDa), and higher oligomers were observed when Py was incubated with ERp57 (Fig. 2B, compare lane 2 to lane 1). No signals were detected when ERp57 was incubated in the absence of virus (Fig. 2B, lane 5), demonstrating that the bands observed in the presence of virus were virus-derived products.
When ERp57 was heat treated prior to incubation with the virus, the various Py-derived species were not seen (Fig. 2B, lane 3). Moreover, when Py was incubated with ERp57 pretreated with the alkylating reagent NEM to block its free cysteines, no virus-derived products were generated (Fig. 2B, lane 4). We conclude that ERp57 uses its catalytic cysteines to disrupt Py's disulfide bonds directly to generate the virus-derived species. As Py is transported to the low-pH endolysosome system before reaching the ER (16), we asked whether pretreatment of Py at low pH (i.e., pH 5) affects ERp57's ability to generate the VP1-derived products and found that it did not (Fig. 2C, compare lane 2 to lane 1).
The VP1-derived products were also generated when PDI, but not NEM-treated PDI (Fig. 2D, compare lane 1 to lane 2), was incubated with (but not without) Py. PDI also acted on low-pH-treated virus with efficiency similar to that of the control virus (Fig. 2E, compare lane 1 to lane 2). Finally, the VP1-derived products were produced when ERp72, but not NEM-treated ERp72 (Fig. 2F, compare lane 1 to lane 2), was incubated with (but not without) Py. Again, pretreating Py at low pH did not significantly prevent ERp72 from acting on the virus (Fig. 2G, compare lane 1 to lane 2). Thus, similar to ERp57, PDI and ERp72 also use their cysteines to produce the virus-derived products. When Py and the PDI family proteins were incubated in a 1:1 molar concentration of oxidized glutathione (GSSG) and reduced glutathione (GSH) that mimicked the ER redox condition (20), the VP1-derived products were generated (data not shown). We conclude that the PDI family proteins can disrupt Py's disulfide bonds even under the relatively oxidizing ER conditions.
ERp57 and PDI function coordinately with ERp29 to unfold Py in vitro.
Our previous in vitro system demonstrated that ERp29 extrudes the VP1 C-terminal arm in a reaction that requires disruption of Py's disulfide bonds (11). The in vitro assay assumes that an ER activity imparts a conformational change to Py to expose a hidden proteolytic cleavage site buried in the native virus (11). Incubation of Py with an extract containing ER lumenal proteins (referred to as an ER lumenal extract) enables a general protease (such as trypsin) to digest the viral capsid proteins, which can be detected when the sample is subjected to SDS-PAGE, followed by immunoblotting.
We found that when Py was incubated with a control protein (BSA) in the presence of DTT and EGTA (to remove the virus-bound calcium), followed by trypsin addition, a VP1-derived fragment we previously termed VP1a was generated (Fig. 3A, compare lane 2 to lane 1), similar to our previous observation (11). However, when Py was incubated with an ER lumenal extract (instead of BSA), DTT, and EGTA, followed by trypsin addition, two VP1-derived tryptic digestion products, VP1a and VP1b, appeared (Fig. 3A, compare lane 4 to lane 3). Formation of VP1b (but not VP1a) requires the presence of an ER lumenal extract, EGTA, and DTT (11, 17, 18). Using mass spectrometry analysis, we previously identified VP1b as a fragment that lacks the C terminus of VP1, indicating that ER factors unfold VP1 to expose its C terminus (11). Again, using mass spectrometry, the ER factor required for VP1b generation was identified as ERp29 (11).
Can ERp57, PDI, or ERp72 functionally replace DTT? Py was pretreated with either the cysteine-less calmodulin, ERp57, ERp72, or PDI in the presence of EGTA. The samples were then incubated with an ER lumenal extract enriched for ERp29 (18), followed by trypsin addition. Our results show that VP1b was generated only when Py was pretreated with either ERp57 or PDI, but not with calmodulin or ERp72 (Fig. 3B, compare lanes 2 and 4 to lanes 1 and 3). The ERp29-enriched lumenal extract lacks ERp57 and PDI (Fig. 3B, compare lane 6 to lane 5), thus explaining the requirement for these proteins in the reaction. NEM-treated ERp57 and PDI cannot assist the lumenal extract in generating VP1b (Fig. 3C, compare lane 2 to lane 1 and lane 4 to lane 3). Thus, the catalytic activities of ERp57 and PDI replace DTT to produce VP1b. We conclude that ERp57 and PDI act coordinately with ERp29 to extrude the VP1 C-terminal arm. We note that the lumenal extract used in this reaction was pretreated with DTT, and the excess DTT in the extract was removed thoroughly by dialysis.
In cells transfected with N-terminally FLAG-tagged ERp29, immunoprecipitation of FLAG-ERp29 coprecipitated endogenous ERp57 following treatment of cells with the cross-linker DSP (Fig. 3D, top row, compare lane 3 to lane 2). Similar results were found when the FLAG-ERp29 precipitate was blotted for ERp72 (Fig. 3E, top, compare lane 3 to lane 2) or PDI (Fig. 3E, second row from top, compare lane 3 to lane 2). Thus, overexpressed ERp29 binds to endogenous ERp57, ERp72, and PDI, suggesting that they may function as a network.
ERp57 principally isomerizes Py, while PDI and ERp72 reduce the virus in vitro.
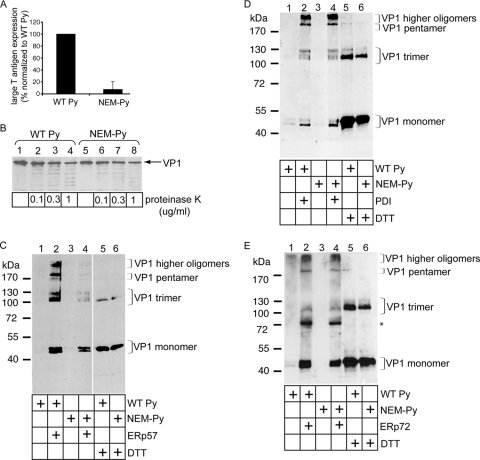
We next asked whether free cysteines in Py are necessary for infection. To this end, Py was incubated initially with NEM to alkylate these residues, and then the sample was subjected to a spin column to remove the excess NEM. We found that the infection efficiency of the NEM-treated Py was severely attenuated compared to WT Py (Fig. 4A). Limited proteolysis experiments showed that the NEM-treated Py exhibited sensitivity to proteinase K similar to that of nonalkylated WT Py (Fig. 4B, compare lanes 5 to 8 to lanes 1 to 4), suggesting that NEM did not grossly disrupt the overall structural integrity of the virus. We conclude that free cysteines in Py are critical for infection.
FIG. 4.
ERp57 principally isomerizes Py, while PDI and ERp72 reduce the virus in vitro. (A) Free cysteines in Py are required for infection. NIH 3T3 cells were incubated with either WT or NEM-treated Py (100 PFU/cell), and the infection efficiency was analyzed as for Fig. 1C. (B) NEM-treated virus is not grossly misfolded. WT and NEM-treated Py were incubated with the indicated concentrations of proteinase K, and the samples were subjected to SDS-PAGE and immunoblotted with a VP1 antibody. (C) ERp57 largely isomerizes Py. WT or NEM-treated Py was incubated with ERp57 or DTT (where indicated), and the samples were analyzed by nonreducing SDS-PAGE, followed by immunoblotting with a VP1 antibody. (D) PDI reduces Py directly. As for panel C, except PDI was used instead of ERp57. (E) ERp72 reduces Py directly. As for panel C, except ERp72 was used instead of ERp57. The asterisk indicates a nonspecific band recognized by the M1 VP1 antibody.
In disulfide bond isomerization, a free cysteine in the substrate is required to resolve the transient disulfide bond formed between the enzyme and substrate that generates a new disulfide bond in the substrate. In disulfide bond reduction, a free cysteine in the enzyme is used to resolve this transient disulfide bond. The finding that free cysteines of Py are pivotal for infection suggests that they may be involved in isomerization reactions.
To test whether free cysteines in Py are required for ERp57 to generate the VP1 species from intact virus, WT and NEM-treated viruses were incubated with ERp57. We found that generation of the VP1 higher oligomer, pentamer, trimer, and, to a lesser extent, monomer was diminished markedly when ERp57 was incubated with the NEM-treated Py compared to WT Py (Fig. 4C, compare lane 4 to lane 2). DTT treatment of WT and NEM-treated viruses produced similar VP1 monomer levels (Fig. 4C, compare lane 5 to lane 6), demonstrating that the same amount of WT and NEM-treated virus was used and that both virus types display the same sensitivity to this general reductant. We conclude that free cysteines in Py are necessary for ERp57 to generate the smaller virus-derived products. The results shown in Fig. 4 suggest that ERp57 acts principally as an isomerase in producing the virus-derived VP1 species.
In contrast to ERp57, PDI (Fig. 4D, compare lane 4 to 2) and ERp72 (Fig. 4E, compare lane 4 to lane 2) generated the Py-derived species potently regardless of whether they were incubated with WT or NEM-treated virus. The prominent 72-kDa band that appeared in Fig. 4E (lanes 2 and 4) was also present in a sample without Py (data not shown), indicating that the VP1 antibody used in this immunoblot (i.e., M1) cross-reacts with ERp72. As free cysteines in the Py were not necessary for the PDI- or ERp72-dependent reactions, these data demonstrate that PDI and ERp72 function here as reductases. We note that ERp57 also functions as an isomerase on SV40 (20).
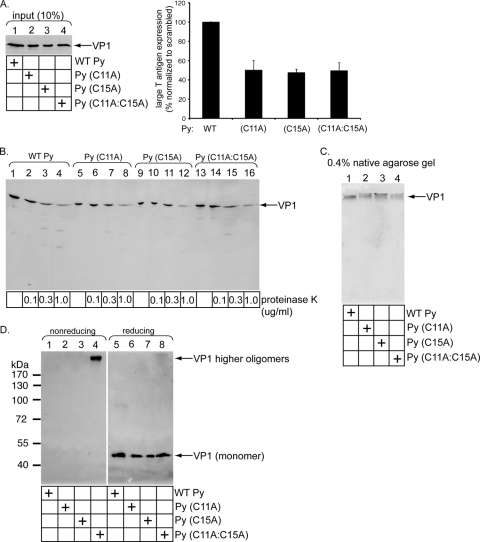
Characterization of the C11A, C15A, and C11A-C15A Py mutants.
The finding that Py's free cysteines and ERp57 may engage in an isomerization reaction to disrupt the virus disulfide bonds prompted us to examine the nature of the previously uncharacterized C11 and C15 residues. Two single mutants were generated, C11 mutated to A [Py (C11A)] and C15 mutated to A [Py (C15A)], and one double mutant was generated, C11 and C15 mutated to A [Py (C11A-C15A)]. We found that when similar levels of WT and mutant viruses were used (Fig. 5A, lanes 1 to 4), infection induced by all three mutants was decreased by approximately 50% compared to that induced by WT Py (Fig. 5A, right). Limited proteolysis demonstrated that all three mutant viruses displayed protease sensitivity patterns similar to that of WT Py (Fig. 5B, compare lanes 5 to 8, 9 to 12, and 13 to 16 to lanes 1 to 4). Moreover, these mutant viruses migrated similarly to WT Py, as analyzed by a native agarose gel system (Fig. 5C, compare lanes 2 to 4 to lane 1). These findings demonstrate that the decrease in infection caused by the mutant viruses is not due to global disruption of their structural integrity. Instead, the results indicate a specific role of C11 and C15 in infection, consistent with a requirement for viral free cysteines during ERp57-mediated isomerization.
FIG. 5.
Characterization of the C11A, C15A, and C11A-C15A Py mutants. (A) VP1 C11 and C15 are important for Py infection. NIH 3T3 cells were incubated with similar levels of crude WT Py, Py (C11A), Py (C15A), and Py (C11A-C15A), and the infection efficiency was analyzed as for Fig. 1C. (B) Mutant viruses are not misfolded globally. The viruses in panel A were subjected to limited proteolysis as for Fig. 4B. (C) Native agarose gel analyses of mutant Py. The viruses in panel A were subjected to 0.4% native agarose gel electrophoresis, transferred to a nitrocellulose membrane, and immunoblotted with an antibody against VP1. (D) VP1 C11 and C15 mediate interpentamer interaction for a subset of the pentamers. The viruses in panel A were subjected to nonreducing and reducing SDS-PAGE, followed by immunoblotting with an antibody against VP1.
Interestingly, when the WT and mutant Pys were subjected to nonreducing SDS-PAGE, a species corresponding to a VP1 higher oligomer was detected only in the Py (C11A-C15A) double mutant (Fig. 5D, compare lane 4 to lanes 1 to 3). A similar VP1 level appeared when the viruses were subjected to reducing SDS-PAGE (Fig. 5D, lanes 5 to 8). These findings indicate that C11 and C15 stabilize interpentamer interactions for at least a subset of the pentamers. This observation raises the possibility that while some C11 and C15 residues within a Py particle are in the free reduced state, other C11 and C15 residues within the same particle likely form interpentamer disulfide bonds between some of the pentamers.
DISCUSSION
Nonenveloped virus penetration of the limiting membrane represents a decisive step in virus infection. Host factors facilitate this process by imparting conformational changes to the virus to initiate membrane penetration. Although multiple host factors likely work together to mediate this process, until now, only individual cellular components responsible for these remodeling events have been identified; more complex mechanisms remain undefined. In the present study, we pinpoint a network of ER-resident proteins that act on Py distinctly and coordinately to facilitate infection. They do so by modifying the viral disulfide bonds to promote its ER membrane penetration. We also identify specific viral cysteines that play pivotal roles in this process.
Using the siRNA knockdown strategy, we demonstrate that individually downregulating ERp57, ERp72, or PDI decreased infection, while simultaneously downregulating any two of these proteins attenuated infection more severely. The fact that infection was not blocked completely is likely due to incomplete downregulation of the PDI family proteins. Additionally, compensation by other PDI family members may contribute to this effect. Under none of the knockdown conditions did we observe induction of significant ER stress, suggesting that the decrease in virus infection is not due to nonspecific effects. Instead, these results point to the possibility that the PDI family members engage Py directly. Although knockdown of PDI in a human cell line was shown previously to block Py infection (6), whether PDI interacts with Py directly is not known.
For SV40, downregulation of ERp57 and PDI attenuated infection (20), consistent with our observation for Py. However, different results using two independent siRNAs to downregulate ERp72 were observed: one enhanced infection, while the other did not have an effect (20). For Py, downregulating ERp72 using two different siRNAs decreased infection. The reason for this discrepancy is not clear, but it may be due to the different disulfide bond arrangements in the two viruses. In addition, the PDI-like protein ERp29 does not appear to act on SV40 (20), in contrast to its established role in Py infection (11). As ERp29 extrudes Py's VP1 C-terminal arm to initiate viral disassembly (11), the precise disassembly mechanisms in Py and SV40 may be different.
To test the hypothesis that ERp57, PDI, and ERp72 affect Py directly, we expressed and purified the three proteins to homogeneity. When incubated with Py, these proteins individually induced the formation of virus-derived products consisting of VP1 monomer, trimer, pentamer, and higher oligomers. VP1 monomer is likely generated when the five disulfide bonds within a pentamer (formed between C19 of one monomer and C114 of another monomer) are disrupted. Interestingly, a double band pattern corresponding to the size of the monomer was observed. This pattern could be explained by the formation of a new intramonomer disulfide bond within a subset of the monomers, causing a different mobility pattern. This doublet pattern is more obvious for the ERp57- and ERp72-mediated reactions than the PDI-triggered reaction. Precisely how this new intramonomer disulfide bond is formed is unknown. One possibility is that a free cysteine in VP1 of the native Py is used to form a new disulfide bond. Alternatively, a new free cysteine generated in VP1 after reduction may possibly catalyze intramonomer disulfide bond formation. Why PDI-mediated reduction of Py did not generate the doublet pattern as efficiently as the ERp72-dependent reaction is not clear.
How is a trimer, but not a dimer or a tetramer, generated? In principle, the trimers could be formed from three monomers within a pentamer that are disulfide bonded to each other. Alternatively, given our findings that suggest the presence of interpentamer disulfide bonds, the trimers could be formed from two monomers from a pentamer (linked to each other by a disulfide bond), with one of the monomers forming an interpentamer disulfide bond with a monomer from an adjacent pentamer.
During disassembly by the PDI family proteins, transient viral intermediates may form that expose certain disulfide bonds while rendering others less exposed. This creates a situation in which certain disulfide bonds are preferentially disrupted over others. Indeed, the intrinsic asymmetric property of Py—with 12 pentamers surrounded by 5 pentamers and 60 pentamers surrounded by 6 pentamers—favors the presence of nonequivalent disulfide bonds during viral disassembly. Although it is not entirely clear why trimers, but not tetramers or dimers, were generated in our experiments, this finding suggests that disulfide bonds within a trimer are likely less exposed than a tetramer or dimer during the course of disassembly, thereby rendering the disulfide bonds linking the trimers less easy to disrupt.
How are VP1 pentamers and higher oligomers liberated from the intact virus? One possibility is that, as the C19-C114 disulfide bonds within a pentamer clamp the invading VP1 C-terminal arm in place (24), disruption of this bond loosens the C-terminal arm, thereby generating VP1 pentamers and higher oligomers. Alternatively, interpentamer disulfide bonds may be disrupted to produce these species. The X-ray structure of Py, which did not indicate the presence of interpentamer disulfide bonds (24), did not include information on the VP1 N-terminal C11 and C15 residues. Our present analyses suggest that these residues stabilize interpentamer interactions for a subset of the pentamers, possibly by forming interpentamer disulfide bonds (see below).
Despite the finding that ERp57, PDI, and ERp72 can individually act on Py to form the virus-derived species, only ERp57 and PDI cooperate with ERp29 to extrude the VP1 C-terminal arm. We showed previously that this reaction generates a hydrophobic viral particle that binds and disrupts ER membrane integrity (11, 17), events that initiate virus penetration across the ER membrane. Although PDI can use its chaperone activity to unfold cholera toxin (CT) in the ER to initiate the toxin's translocation into the cytosol (3, 4, 27), the catalytic, but not chaperone, activity of PDI (and ERp57) is required to assist ERp29 in unfolding the VP1 C-terminal arm. As there are two pentamer types in Py (12 five coordinated and 60 six coordinated) (9), it is possible that the ERp57-PDI-ERp29 network acts on one of these two types, while ERp72 engages the other.
Our next key finding indicated that free viral cysteines play an important role in Py infection, as an alkylated virus cannot promote infection. Consistent with this observation, we found that while PDI and ERp72 function as reductases in engaging Py in vitro, ERp57 acts as an isomerase, a reaction that requires free viral cysteines. We have yet to pinpoint the precise mechanism by which ERp57, PDI, and ERp72 act on Py in cells, because less than 5% of the total internalized Py reaches the ER from the cell surface (16). However, the biochemical analyses, coupled with the infection result of the alkylated virus, point to a role of isomerization facilitated by viral cysteines as a potential critical reaction during infection. This idea is supported by our identification of a role for VP1's C11 and C15 residues in infection, as mutating C11 to A, C15 to A, or both residues to A decreased infection by 50%. These mutations did not significantly affect the global viral conformations, suggesting a specific role of these cysteines in infection. One interpretation of these data is that some of the C11 and C15 residues exist in the free and reduced state, enabling them to participate in isomerization.
Strikingly, using a nonreducing SDS-PAGE system, VP1 higher oligomers were generated when both C11 and C15 were mutated to A, but not when only a single cysteine was altered. This finding demonstrates that C11 and C15 stabilize interpentamer interactions for a subset of the pentamers, possibly by forming interpentamer disulfide bonds between some of the pentamers. If these residues stabilized all interpentameric interactions, a VP1 pentamer, but not a higher oligomer, would appear. Moreover, because mutating a single cysteine did not generate the higher oligomers, the potential interpentamer disulfide bonds likely exist between two C11 residues and two C15 residues. Py's C11 is the homolog of SV40 C9, which is implicated in C9-C9 interpentamer disulfide bonding (20). Because Py contains two types of pentamers, certain pentamers will be situated in a different local environment than others. This structural asymmetry likely allows some C11 and C15 residues to be disulfide bonded while others are in the reduced state. This situation is similar to that of C104 in SV40's VP1, where it is thought to exist in both the reduced and disulfide-bonded states (20).
While we cannot formally rule out the possibility that C11 and C15 are used to stabilize interpentamer contacts without forming interpentamer disulfide bonds, we consider this possibility unlikely based on our data provided using NEM-Py. Specifically, alkylation of these cysteine residues with NEM would presumably disrupt their ability to make non-disulfide-bond-mediated stabilizing contacts, thereby generating higher-order VP1 species. However, on nonreducing SDS-PAGE, we did not observe higher-order VP1 species containing only NEM-Py.
The double Py mutant (C11A-C15A) did not display a more pronounced decrease in infection than the single cysteine mutants. This could be because C11 and C15 are normally used to isomerize Py to generate the VP1 higher-oligomer intermediate that is critical for successful infection. Preformation of this intermediate in the double mutant thus renders these cysteine residues dispensable. However, the fact that the double mutant was nonetheless defective in infection indicates that this mutant harbors subtle structural alterations that do affect infection. The precise nature of this structural defect is not known, but it may be due to premature disassembly in the endolysosomes prior to arrival at the ER.
Although we have identified three additional PDI family members in the ER that engage and facilitate Py infection, whether additional ER components act on Py is not known. In this context, it is interesting that in the in vitro trypsin digestion assay, ERp57 or PDI added in combination with a reduced ERp29-enriched ER lumenal extract (which lacks ERp57 and PDI) generated VP1b. The fact that reducing the ERp29-enriched lumenal extract was required in this reaction suggests that ERp29 in the extract must be in the reduced form to unfold Py (ERp29 contains one cysteine). Indeed, there are precedents for the redox state of a PDI protein controlling its chaperone-unfolding activity (27). Alternatively, additional reductases/isomerases in the extract may be involved in triggering a penetration-competent viral particle. Further experiments are required to distinguish these possibilities.
In conclusion, our findings unveil a complex interplay between viral cysteine residues and host reductases, isomerases, and chaperones of the PDI family that act coordinately and distinctly on Py to facilitate infection. These reactions promote transport of the virus from the ER into the cytosol, a pivotal infection step. PDI family members not only engage endogenous cellular substrates (7, 15, 32), they are often coopted by toxins and viruses during infection (3, 8, 11, 12, 20, 22, 27). The vast number of members within this family (1), coupled with their functional versatility, thus renders them attractive targets for pathogens during entry.
Acknowledgments
We thank Emily Rainey-Barger for critical review of the manuscript.
B.T. is a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease and is supported by the NIH (RO1-AI064296). C.P.W. is funded partially by the Cellular and Molecular Biology program at the University of Michigan.
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1.Appenzeller-Herzog, C., and L. Ellgaard. 2008. The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta 1783:535-548. [DOI] [PubMed] [Google Scholar]
- 2.Chen, X. S., T. Stehle, and S. C. Harrison. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 17:3233-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forster, M. L., et al. 2006. Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. J. Cell Biol. 173:853-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forster, M. L., J. J. Mahn, and B. Tsai. 2009. Generating an unfoldase from thioredoxin-like domains. J. Biol. Chem. 284:13045-13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert, J., and T. Benjamin. 2004. Uptake pathway of polyomavirus via ganglioside GD1a. J. Virol. 78:12259-12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert, J. M., W. Ou, J. Silver, and T. Benjamin. 2006. Downregulation of protein disulfide isomerase inhibits infection by the mouse polyomavirus. J. Virol. 80:10868-10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang, K., B. Park, C. Oh, K. Cho, and K. Ahn. 2009. A role for protein disulfide isomerase in the early folding and assembly of MHC class I molecules. Antioxid. Redox Signal. 11:2553-2561. [DOI] [PubMed] [Google Scholar]
- 8.Lee, S. O., et al. 2010. Protein disulphide isomerase is required for signal peptide peptidase-mediated protein degradation. EMBO J. 29:363-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liddington, R. C., et al. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278-284. [DOI] [PubMed] [Google Scholar]
- 10.Lilley, B. N., J. M. Gilbert, H. L. Ploegh, and T. L. Benjamin. 2006. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J. Virol. 80:8739-8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnuson, B., et al. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell 28:289-300. [DOI] [PubMed] [Google Scholar]
- 12.McKee, M. L., and D. J. FitzGerald. 1999. Reduction of furin-nicked Pseudomonas exotoxin A: an unfolding story. Biochemistry 38:16507-16513. [DOI] [PubMed] [Google Scholar]
- 13.Oda, Y., et al. 2006. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 172:383-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poranen, M. M., R. Daugelavicius, and D. H. Bamford. 2002. Common principles in viral entry. Annu. Rev. Microbiol. 56:521-538. [DOI] [PubMed] [Google Scholar]
- 15.Puig, A., and H. F. Gilbert. 1994. Protein disulfide isomerase exhibits chaperone and anti-chaperone activity in the oxidative refolding of lysozyme. J. Biol. Chem. 269:7764-7771. [PubMed] [Google Scholar]
- 16.Qian, M., D. Cai, K. J. Verhey, and B. Tsai. 2009. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PloS Pathog. 5:e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rainey-Barger, E. K., B. Magnuson, and B. Tsai. 2007. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J. Virol. 81:12996-13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rainey-Barger, E. K., S. Mkrtchian, and B. Tsai. 2007. Dimerization of ERp29, a PDI-like protein, is essential for its diverse functions. Mol. Biol. Cell 18:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainey-Barger, E. K., S. Mkrtchian, and B. Tsai. 2009. The C-terminal domain of ERp29 mediates polyomavirus binding, unfolding, and infection. J. Virol. 83:1483-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schelhaas, M., et al. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516-529. [DOI] [PubMed] [Google Scholar]
- 21.Smith, A. E., H. Lilie, and A. Helenius. 2003. Ganglioside-dependent cell attachment and endocytosis of murine polyomavirus-like particles. FEBS Lett. 555:199-203. [DOI] [PubMed] [Google Scholar]
- 22.Spooner, R. A., et al. 2004. Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem. J. 383:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stehle, T., Y. Yan, T. L. Benjamin, and S. C. Harrison. 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369:160-163. [DOI] [PubMed] [Google Scholar]
- 24.Stehle, T., and S. C. Harrison. 1996. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure 4:183-194. [DOI] [PubMed] [Google Scholar]
- 25.Stehle, T., S. J. Gamblin, Y. Yan, and S. C. Harrison. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4:165-182. [DOI] [PubMed] [Google Scholar]
- 26.Stehle, T., and S. C. Harrison. 1997. High-resolution structure of a polyomavirus VP1-oligosacccharide complex: implications for assembly and receptor binding. EMBO J. 16:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai, B., C. Rodighiero, W. I. Lencer, and T. A. Rapoport. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104:937-948. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, B., et al. 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 22:4346-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai, B. 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23:23-43. [DOI] [PubMed] [Google Scholar]
- 30.Tsai, B., and M. Qian. 2010. Cellular entry of polyoma viruses. Curr. Top. Microbiol. Immunol. 343:177-194. [DOI] [PubMed] [Google Scholar]
- 31.Uemura, A., M. Oku, K. Mori, and H. Yoshida. 2009. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell Sci. 122:2877-2886. [DOI] [PubMed] [Google Scholar]
- 32.Wang, C. C., and C. L. Tsou. 1993. Protein disulfide isomerase is both an enzyme and a chaperone. FASEB J. 7:1515-1517. [DOI] [PubMed] [Google Scholar]