Abstract
Virus-specific CD8 T cells are activated when their T-cell receptors (TCRs) recognize the specific viral peptide/major histocompatibility complex (MHC) class I (pMHC) complexes present on the surface of infected cells. Antibodies able to recognize the specific pMHC can mimic TCR specificity and both represent a valuable biological tool to visualize pMHC complexes on infected cells and serve as a delivery system for highly targeted therapies. To evaluate these possibilities, we created a monoclonal antibody able to specifically recognize a hepatitis B virus (HBV) envelope epitope (Env at positions 183 to 91 [Env183-91]) presented by the HLA-A201 molecule, and we tested its ability to recognize HBV-infected hepatocytes and to deliver a cargo to a specific target. We demonstrate that this antibody detects and visualizes the processed product of HBV proteins produced in naturally HBV-infected cells, is not inhibited by soluble HBV proteins present in patient sera, and mediates the intracellular delivery of a fluorescent molecule to target cells. Additionally, compared to CD8 T cells specific for the same HBV epitope, the TCR-like antibody has both a superior sensitivity and a specificity focused on distinct amino acids within the epitope. These data demonstrate that a T-cell receptor-like antibody can be used to determine the quantitative relationship between HBV replication and specific antigen presentation to CD8 T cells and serves as a novel therapeutic delivery platform for personalized health care for HBV-infected patients.
CD8 T lymphocytes recognize neither intact viruses nor viral proteins. Rather, they are activated by the specific interaction of their T-cell receptors (TCRs) with the viral peptide/major histocompatibility complex (MHC) class I (pMHC) complex presented on the surface of infected cells. The pMHC complex is the processed product of viral proteins synthesized within the infected cells of the host. Its quantity, density, and surface localization combined with the expression of other costimulatory or inhibitory molecules shape the immunological response of CD8 T cells to their targets in a structure known as the immunological synapse (18).
Reagents with the capacity for the study of the quantity and location of the pMHC complex on infected cells are very limited; thus, the quantitative and qualitative features of the pMHC complex on infected cells are often overlooked in the study of antiviral immunity (6). Although physiologically, the ability to recognize distinct viral peptides bound to MHC class I molecules is characteristic of the alpha and beta TCRs (18), soluble TCRs have low binding affinities for their ligands (11) and thus have not been used to quantify peptide/human leukocyte antigen (HLA) complexes on the surface of infected cells. In contrast, antibodies with the ability to recognize murine peptide/MHC class I complexes were successfully produced in mice to study antigen presentation and the localization of antigen-presenting cells (4, 14). Large human antibody (Ab) phage libraries were used to select antibodies specific for the human pMHC complex, but to date, the majority of such antibodies have been used to target tumor-associated epitopes (2, 6). To our knowledge, only a single monoclonal antibody (MAb) specific for a human T-cell leukemia virus type 1 (HTLV-1) viral peptide-MHC class I complex with the ability to detect TCR ligands on virally infected cells of humans has been described (3).
Here, we describe a novel MAb specific for the Env183-91/HLA-A201 (Env183/A2) complex, and we evaluated whether this TCR-like antibody can both detect naturally HBV-infected cells and serve as a delivery system for targeted therapy. We selected the Env183/A2 complex as the target for the production of a TCR-like MAb since Env183-91-specific CD8 T cells represent dominant CD8 T-cell responses in HLA-A201-positive (HLA-A201+) HBV-infected patients (12, 17), and the envelope protein (also called hepatitis B surface antigen [HBsAg]) is produced in large quantities in HBV-infected cells (7).
MATERIALS AND METHODS
Production of Env183/A2 complexes.
Peptide/HLA-A2 complexes were produced by using a protocol similar to that described previously (1). In brief, the extracellular domain of the HLA-A201 heavy chain and β2-microglobulin were expressed as inclusion bodies in Escherichia coli cells and refolded in vitro in the presence of a 5- to 10-fold excess of the Env at positions 183 to 91 (Env183-91) peptide. After refolding, the peptide/HLA-A2 mixture was concentrated, and folded complexes were isolated from contaminants by using ion-exchange and size exclusion column chromatography methods. This complex was designated the pMHC monomer.
Generation of TCR-like antibodies.
BALB/c mice were immunized at 2-week intervals a total of four times by the intraperitoneal injection of a solution containing 25 μg of purified pMHC monomer and Freund's complete adjuvant (primary dose) or Freund's incomplete adjuvant (3 booster doses). A final booster dose in saline without adjuvant was performed by tail injection 3 days before fusion. One day before fusion, mouse peritoneal macrophages were collected from the peritoneal cavity of BALB/c mice and seeded into 96-well plates at 2 × 104 cells/well. Splenocytes from immunized mice were fused by using PEG1500 with NS1 myeloma cells at a ratio of 1:1 as previously described (10). The resulting hybridoma cell mixture was suspended in hypoxanthine-aminopterin-thymidine (HAT) selection medium and seeded into 96-well plates containing the peritoneal macrophages for 2 weeks. Wells containing hybridoma clones were scored macroscopically, and supernatants were collected for testing the appropriate MAb specificity by flow cytometry. T2 cells pulsed with 1 μM HBV Env183-91 peptide or influenza A virus matrix peptide (M1 peptide) were incubated with 50 μl of hybridoma supernatants for 30 min at 4°C, washed, and further incubated with anti-mouse IgG Alexa Fluor 488 secondary antibodies (Invitrogen) for 30 min at room temperature (RT), washed, and analyzed with a BD FACScan flow cytometer using cell quest software. Positive cultures were subcloned twice by limiting dilution and further expanded. An immunochromatography-based kit (IsoStrip; Roche) was used to determine the immunoglobulin subtype and subclass specificity of the MAb. The antibody was found to be the IgG1 subtype with the kappa light chain.
Cell lines.
T2 cells (ATCC CRL 1992) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 20 mM HEPES, 0.5 mM sodium pyruvate, minimal essential medium (MEM), nonessential amino acids, Glutamax, 5 μg/ml Plasmocin (InvivoGen), 100 U/ml penicillin, and 100 μg/ml streptomycin. HBV-expressing HepG2 (HepG2-117) cells and a vector control parent cell line (HepG2 TA2-7) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% tetracycline (Tet)-free FBS (BD Biosciences, San Diego, CA), 20 mM HEPES, 0.5 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, nonessential amino acids, 200 μg/ml G418 sulfate, and 80 μg/ml hygromycin B. Doxycycline (Dox) was added to the medium at 1 μg/ml to suppress HBV expression. HEK293 cells and NS1 myeloma cells were grown in DMEM supplemented with 10% FBS.
Liver biopsy specimens.
Liver biopsy specimens from patients with chronic hepatitis B (CHB) were collected with informed consent. Part of the biopsy specimens not used for diagnostic purposes (usually about one-third of the specimen) was treated with collagenase as previously described (8) or snap-frozen. The processed or frozen biopsy specimens were used for the detection of pMHC complexes with flow cytometry or immunohistochemical staining methods, respectively. HLA-A2 typing of the patient was performed by staining the patient's peripheral blood mononuclear cells (PBMCs) with a fluorescein isothiocyanate (FITC)-conjugated HLA-A2 antibody.
Detection of pMHC complexes on target cells with flow cytometry.
The T2 cell line is a mutant cell line that lacks the transporter associated with antigen processing 1 (TAP1) and TAP2 genes, which allows the efficient loading of peptides onto nascent HLA chains. The HepG2 cell line is an HLA-A2+ hepatoma cell line (ATCC HB-8065). Peptide-pulsed T2 cells or peptide-pulsed or HBV-transfected HepG2 cells were incubated with 0.5 μg of Env183/A2 MAb in 50 μl fluorescence-activated cell sorter (FACS) staining buffer (1% bovine serum albumin [BSA] in phosphate-buffered saline [PBS] with 0.01% sodium azide) per tube for 1 h at 4°C. The cells were washed three times and incubated with 1 μg of fluorescence-conjugated anti-mouse IgG secondary antibody in 50 μl staining buffer. After a further washing step, cells were analyzed with a BD FACSCanto flow cytometer. Where appropriate, the histograms were overlaid by using Flowjo software. Cells purified from liver biopsy specimens after collagenase treatment were stained with a modified protocol. Liver-derived cells were first blocked in 2% goat serum for 60 min at 4°C. After washing with PBS plus 1% BSA, liver cells were incubated with Env183/A2 MAb or mouse isotype control Ab for 45 min at 4°C. After washing, anti-mouse IgG-allophycocyanin (APC) secondary antibody was added at 4°C for 20 min. The cells were then fixed and permeabilized by incubation in fixation and permeabilization buffer (eBioscience) for 30 min at 4°C and then stained with rabbit anti-human albumin antibody (Sigma) or rabbit isotype (1 μg/ml) for 45 min, washed with permeabilization buffer (eBioscience), and incubated with anti-rabbit IgG-phycoerythrin (PE) secondary antibodies (20 min at 4°C). After a further washing step, cells were analyzed with a BD FACSCanto flow cytometer.
Degranulation assays of HBV-specific CD8 T cells.
CD107 PE antibody (BD Pharmingen, San Diego, CA) was added to the wells at the beginning of the 5-h incubation of CD8 T cells with target cells pulsed with different peptides or with unpulsed controls. Following incubation, the target cells were washed, stained with Cy-chrome-conjugated anti-CD8 (BD Pharmingen, San Diego, CA), washed again, and then analyzed by flow cytometry.
Lentiviral transduction of HepG2 cells.
A total of 4 × 106 HEK293 cells were plated in 10-cm tissue culture plates and cotransfected using CaCl2 with pLVX-HBsAg or pLVX and packaging vectors provided in the Lenti HT packaging mix (BD Clontech). At 24 to 48 h after transfection, the vector supernatants were collected and clarified. HepG2 cells were transduced with vector supernatants in the presence of 7 μg/ml polybrene for 24 h. Cells were selected for 2 days in 7 μg/ml puromycin and then analyzed by flow cytometry following staining with Env183/A2 MAb.
Visualization of pMHC complexes.
HepG2 and HepG2-117 cells were cultured on coverslips in DMEM with 1% dimethyl sulfoxide (DMSO). Doxycycline was removed from the medium to allow the production of HBV (16). Cells were treated with 1,000 U/ml gamma interferon (IFN-γ) for 2 days before staining with antibodies. Coverslips were washed 3 times with PBS and fixed in freshly prepared 1% paraformaldehyde (PFA) for 15 min at RT. After washing three times with PBS, the coverslips were blocked with 5% goat serum for 1 h, followed by incubation with 1 μg of Env183/A2 MAb overnight at 4°C. Coverslips were washed in PBS plus 0.05% Tween 20 (PBST) 3 times. An Alexa Fluor 647-tyramide signal amplification kit (Invitrogen) was used to reveal primary antibody binding, with the exception that an anti-mouse IgG-horseradish peroxidase (HRP) polymer (Dako) was used instead of the anti-mouse IgG-HRP provided in the kit. The rest of the protocol was performed as recommended by the manufacturer. Coverslips were mounted in Antifade gold reagent containing DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen), and images were obtained with a Carl Zeiss LSM 510 Meta upright confocal microscope.
Immunohistochemical staining of liver biopsy specimens.
Six-micrometer cryostat sections of snap-frozen liver biopsy specimens from CHB patients were stored at −80°C until use. The sections were briefly air dried, fixed in 1% paraformaldehyde for 15 min at RT, washed with PBST 3 times, and blocked with 3% BSA plus 5% normal goat serum (blocking buffer) for 1 h at RT. Following washing 3 times, sections were incubated with 1 μg of Env183/A2 MAb in a solution containing 25 μl of 0.05 M Tris buffer in 1% BSA overnight at 4°C. Another section was incubated with a control mouse EBNA/A2 MAb (specific for an Epstein-Barr virus [EBV]-derived peptide) that acts as a negative control, and sections were washed 3 times with PBST. An EnVision+ mouse-HRP kit (catalog number K4006; Dako) was used to reveal primary antibody staining using the protocol described in the instruction manual. Sections were counterstained for nuclei with hematoxylin and mounted in Clarion mounting medium (Sigma), and images were obtained.
Preparation of antibody-fluorochrome conjugate and its targeted delivery.
The Env183/A2 MAb was linked with the mouse Fab-Alexa 488 fluorochrome by Zenon labeling as described by the manufacturer (Invitrogen). The HepG2 cells either were left unpulsed or were pulsed with 1 μM Env183-91 peptide for 1 h and washed with PBS. The cells were incubated with 0.5 μg of the “MAb-fluorochrome conjugate” at 4°C for 1 h and washed twice in sterile PBS. Cells were placed onto coverslips, followed by incubation in culture medium at 37°C. At fixed time points varying from 0 h up to 16 h, coverslips were removed from the culture, washed, incubated with 5 μM DiI red for 10 min at 37°C to label plasma membrane or 50 nM LysoTracker red for 30 min at 37°C to label lysosomes, and then rinsed in PBS. Cells were fixed in 1% paraformaldehyde for 15 min, washed, placed onto Antifade gold reagent with DAPI, and mounted for confocal imaging. Images were obtained with a confocal microscope.
RESULTS AND DISCUSSION
Production and selection of a TCR-like MAb.
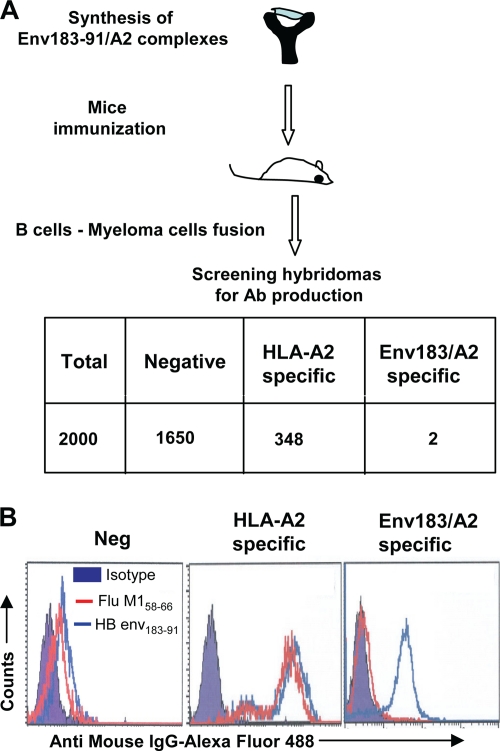
Antibodies able to recognize HBV peptide/HLA-A201 complexes were generated by the immunization of BALB/c mice with a purified complex formed by the HBV Env183-191 peptide associated with HLA-A0201 and β2-microglobulin. The correct folding of the pMHC complex and its stability were monitored by subjecting the monomer to nonreducing native PAGE and Western transfer. The blot was developed with the conformation-specific HLA-A2 MAb w6/32, which recognizes the HLA-A2 complex only when folded correctly. A single band was visible on the Western blot with w6/32 antibodies, confirming the formation of a properly folded monomer (see Fig. S1 in the supplemental material). Splenocytes from immunized animals were fused with NS1 myeloma cells at a 1:1 ratio, and supernatants of the resulting hybridomas (∼2,000 hybridomas) were screened by using HLA-A201+ T2 cells pulsed with the HBV Env183-91 peptide (FLLTRILTI) or an irrelevant HLA-A201-binding M1 peptide (influenza A virus matrix peptide at positions 58 to 66 [GILGFVFTL]). A schematic representation of the selection process is shown in Fig. 1 A. The binding of the antibody present in the supernatant to the peptide-pulsed cells was detected by using a secondary anti-mouse IgG-Alexa Fluor 488 antibody. Two hybridomas derived from mice immunized with the Env183/A2 complex produced antibodies (defined as Env183/A2 MAb) that recognized only HBV Env183-91 peptide-pulsed T2 cells (Fig. 1B). These B-cell hybridoma clones were stably expanded, the culture supernatant from individual clones was collected in bulk, and the antibodies were purified with a protein G-agarose column (data not shown).
FIG. 1.
Schematic representation of the production of Env183/A2 MAb. (A) Synthesis of HLA-A2 complexes, mouse immunization, B-cell selections, and characterization of specific antibody production (detailed in Materials and Methods). The table shows the total number of screened hybridomas and their specificities. (B) Histograms representing the different staining profiles of B-cell hybridoma supernatants. T2 cells were pulsed with 1 μM (each) Env183-91 peptide or influenza A virus matrix peptide at positions 58 to 66 (Flu M158-66) and then incubated with 50 μl of hybridoma supernatant followed by washing and incubation with anti-mouse IgG-Alexa Fluor 488 for 30 min. Cells were washed and analyzed with flow cytometry. Three different profiles (negative, specific for HLA-A2 molecules, and specific for the Env183/A2 complex) are represented. Data show representative FACS profiles.
Characterization of TCR-like MAb specificity.
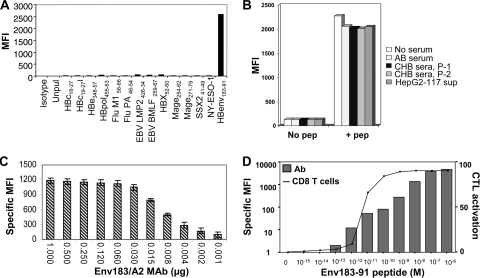
While initial hybridoma screening demonstrated the required specificity of the Env183/A2 MAb for its cognate ligand, a series of assays was performed to exclude cross-reactivity with other HLA-A201/peptide complexes. T2 cells were incubated with a variety of HLA-A201-binding peptides derived from HBV polymerase, X, envelope, and core proteins as well as peptides derived from EBV, influenza A virus, human cytomegalovirus (HCMV), and a wide variety of tumor-associated antigen epitopes, as detailed in Materials and Methods (see Table S1 in the supplemental material). We demonstrated that the Env183/A2 MAb reacted specifically only with target cells pulsed with its cognate Env183-91 peptide (Fig. 2 A).
FIG. 2.
Characterization of Env183/A2 MAb. (A) Cross-reactivity was evaluated by incubating T2 cells with 1 μM (each) the listed peptides for 1 h, and cells were stained with 0.5 μg of Env183/A2 MAb and analyzed as described above. Specific binding was observed only with the Env183-91 peptide. (B) Env183/A2 MAb is not inhibited by circulating HBV antigens. The Env183/A2 MAb was preincubated with 100 μl of sera from 2 CHB patients, the serum of healthy (AB) subjects, or culture supernatants of HepG2-117 cells. This preincubated MAb-serum mixture was used to stain Env183-91 peptide-loaded (1 μM) T2 cells. (C) Binding characterization of the TCR-like MAb. Titrated concentrations of the Env183/A2 MAb were tested with peptide-pulsed T2 cells. Bars represent the MFI values obtained at the indicated concentrations. Mean values of triplicate measurements are shown. (D) Sensitivity of Env183/A2-specific MAb and CTLs. T2 cells were incubated with the indicated concentrations of the Env183-91 peptide for 1 h and used for the binding assay of antibodies (bars) or CD8 T-cell activation (line). The binding of antibody was detected by flow cytometric analysis as described above. CD8 T-cell activation was calculated as the percentage of CD107-expressing CD8 T cells. Data represent data from one of at least two independent experiments.
Furthermore, since patients with chronic hepatitis B maintain high levels of circulating soluble envelope protein (HBsAg) (9), we also investigated whether circulating HBsAg interferes with the recognition of pMHC complexes. We preincubated Env183/A2 MAb with HBsAg-positive sera from chronically HBV-infected patients (patient 1, HBV DNA level of 108 IU/ml and HBsAg level of 67,389 IU/ml; patient 2, HBV DNA level of 108 IU/ml and HBsAg level of 45,300 IU/ml) or culture supernatants from HBV-producing HepG2-117 cells for 1 h. The resulting mixture was used to stain Env183-91 peptide-pulsed T2 cells. The binding capacity of the Env183/A2 MAb was not altered by serum HBsAg or other serum components (Fig. 2B). Similarly, we preincubated the Env183/A2 MAb with an excess of its cognate peptide and stained peptide-pulsed cells using this mixture. This did not result in an observable decrease in the staining of the specific target (data not shown), indicating that the recognition capacity of the Env183/A2 MAb is not altered by the free peptide.
To define the concentrations required for optimal staining, Env183/A2 MAb was titrated from 1-μg to 0.001-μg/ml concentrations and then used to stain T2 cells pulsed with the Env183-91 peptide (1 μM) (Fig. 2C). The specific mean fluorescence intensity (MFI) values were obtained by subtracting the MFI values of unpulsed T2 cells (control) from the MFI values obtained from peptide-pulsed T2 cells. The Env183/A2 MAb showed the ability to distinguish peptide-pulsed from unpulsed cells at working concentrations of 0.004 to 0002 μg/ml, and saturating staining was obtained at 0.06 to 0.5 μg/ml. The latter concentration (0.5 μg/ml) was used in the subsequent experiments presented unless indicated otherwise.
We next analyzed whether the Env183/A2 MAb recognized the pMHC complex with a sensitivity similar to that of CD8+ T cells specific for the same pMHC complex. T2 cells were pulsed with a synthetic peptide, washed, and then stained with Env183/A2 MAb. The Env183/A2 MAb staining intensity was proportional to the peptide concentrations used for the pulsing of T2 cells (Fig. 2D, bars), and 10−13 M (100 fM) Env183-91 peptide was sufficient to detect specific binding. We then compared the amount of peptide required to detect TCR-like MAb binding to that necessary to activate CD8+ T cells of an identical specificity. Env183-91 CD8+ T cells were cultured with peptide-pulsed T2 cells, and the activation of CD8 T cells was quantified by measuring the expression of CD107 (as a marker of antigen-specific degranulation). Interestingly, Env183-91-specific CD8+ T-cell activation required target cells pulsed with peptide concentrations higher than those necessary to detect Env183/A2 MAb-mediated T2 cell staining (10 pM versus 100 fM). These results suggest that the Env183/A2 MAb has a better sensitivity than CD8+ T cells specific for the same complex.
Ability of Env183/A2 MAb to recognize HBV-infected cells.
Any potential in vivo applications of the TCR-like antibody require that such an antibody be able to directly recognize HBV-infected hepatocytes. Hepatocytes constitutively express low levels of HLA class I molecules (8), and their antigen-processing machinery is inefficient for the generation of peptide/HLA class I complexes (15); thus, the quantity of the TCR-specific ligands expressed by HBV-infected hepatocytes may be much lower than the level in HBV peptide-pulsed T2 target cells.
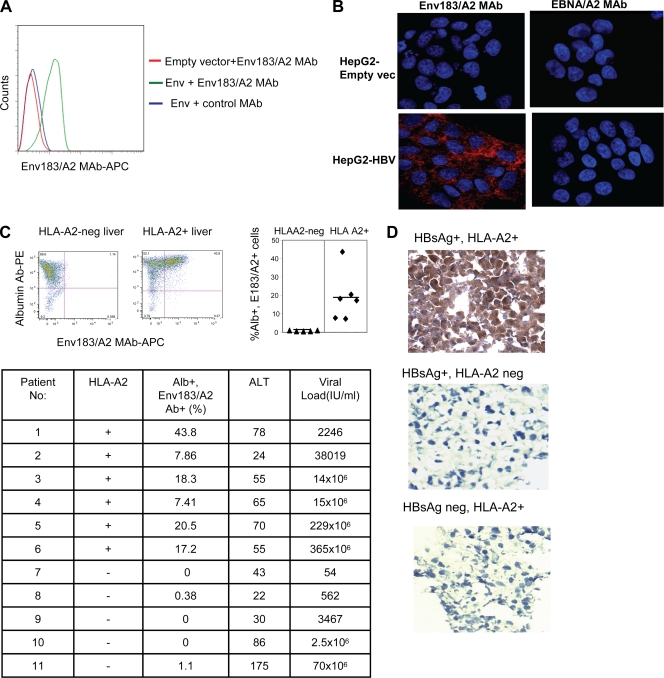
The capacity of the Env183/A2 MAb to recognize pMHC complexes produced by the physiologically active, intracellularly processed peptide in the context of HLA-A2 was first evaluated by flow cytometric analyses. For this purpose, we performed a lentivirus-mediated transduction of HepG2 cells (which are HLA-A201+) with pLVX-HBsAg plasmids expressing full-length HBsAg proteins (HBV genotype D). Transduced cells showed clear staining with Env183/A2 MAb compared to empty vector-transduced cells (Fig. 3 A), demonstrating that the Env183/A2 MAb recognizes pMHC complexes produced by the endogenous processing of the HBV envelope protein.
FIG. 3.
Recognition of pMHC complexes on HBV-infected cells. (A) HepG2 cells were transduced with empty vector or HBsAg. Forty-eight hours after transduction, cells were stained with Env183/A2 MAb or a control EBNA/A2 MAb as described above. Data show results of a typical experiment out of three experiments. (B) Visualization of the pMHC complex. HBV-producing HepG2-117 cells or control HepG2 cells were fixed with 1% PFA and stained with 1 μg of Env183/A2 MAb or a negative control EBNA/A2 MAb overnight at 4°C. An Alexa Fluor 647-tyramide signal amplification kit was used to visualize antibody binding (red). Nuclei were stained with DAPI (blue). (C) Recognition of the pMHC complex on cells obtained from liver biopsy specimens from CHB patients. Dot plots represent liver-derived cells obtained from biopsy specimens of two representative CHB patients (one HLA-A2+ and one HLA-A2 negative) stained with albumin (Alb) Ab and Env183/A2 MAb. Percentages displayed in the upper right quadrant indicate the number of cells positive for both Abs (left). The quadrant gates were set for each patient's cells stained with isotype control antibodies. The scattered diagram shows the frequency of Env183/A2 MAb and albumin Ab double-stained cells obtained from various patient biopsy specimens (right, P < 0.01). The table indicates the total number of patients tested, Env183/A2 MAb and HLA-A2 positivity, and clinical/virological features of the 11 distinct CHB patients. ALT, alanine aminotransferase. (D) Cryostat sections of liver biopsy specimens (HBV status and HLA typing are indicated) were stained with antibodies as described above (B). An EnVision Plus mouse HRP polymer kit was used to reveal primary antibody staining (brown). Nuclei were counterstained with hematoxylin (blue).
The ability of Env183/A2 MAb to recognize the corresponding pMHC on HBV-producing hepatocyte-like cells was also demonstrated by using immunohistological methods on hepatoma cell lines (HepG2) stably transfected with a tetracycline (Tet)-responsive promoter-controlled HBV genome. This Tet-controllable HBV-transfected HepG2 line (HepG2-117) produces a different quantity of HBV virions and HBcAg antigens after the removal of doxycycline (Dox), while HBsAg production is regulated by internal promoters, and thus, its expression is largely unaffected by Dox (16). HepG2-117 cells were fixed in 1% paraformaldehyde for 15 min at room temperature, washed, and then incubated with Env183/A2 MAb. A tyramide signal amplification kit was used for the visualization of antibody staining. As shown in Fig. 3B, the recognition of the pMHC complex could be clearly seen with the Env183/A2 MAb, and the staining was negative on control vector-transduced HepG2 cells and also with a control antibody (EBNA/A2 MAb), showing that the Env183/A2 MAb can directly recognize pMHC complexes present on HBV-producing hepatocyte-like cells.
To test directly whether Env183/A2 MAb could target naturally infected hepatocytes, biopsy specimens from HLA-A201-positive or -negative chronic hepatitis B (CHB) patients were mechanically treated to obtain individual-cell suspensions and then stained with anti-albumin-specific antibodies and Env183/A2 MAb (see Materials and Methods). Variable frequencies (from 44% to 7%) of albumin-positive cells derived from biopsy specimens of HLA-A2-positive patients stained with Env183/A2 MAb, while cells purified from HLA-A2-negative CHB patients were constantly negative (Fig. 3C). Note that the albumin-positive cells present a morphology compatible with that of hepatocytes (cells with elevated levels of complexity and size on forward- and side-scatter plots), and the staining of liver biopsy specimens from two additional HLA-A2+ CHB patients with a control EBNA/A2 MAb did not reveal any specific staining (not shown). Interestingly, the quantity of hepatocytes with detectable Env183/A2 complexes does not seem to correlate with the values for HBV DNA detectable in patient sera (Fig. 3C). A large study comparing HBV DNA and HBV antigen values in serum and liver with the quantity of hepatocytes expressing HBV peptide/HLA complexes will be necessary to understand the relationship between HBV viral replication and the expression of HBV peptide/MHC complexes on infected hepatocytes. It is likely that other factors, like the intrahepatic cytokine environment, which is known to modulate the hepatocyte antigen presentation capacity (15), are likely to influence such relationships.
The ability of the Env183/A2 MAb to specifically target naturally HBV-infected HLA-A2+ hepatocytes was also confirmed by using immunohistochemical methods on frozen liver biopsy specimens obtained from CHB patients. Figure 3D shows the clear positive staining of hepatocyte-like cells present in HLA-A2+ chronically HBV-infected patients (HBsAg+ anti-HBe+; HBV genotype D), while control liver samples from HLA-A2+ HBsAg-negative patients (hepatocellular carcinoma [HCC] in hepatitis C virus [HCV]-positive patients) and from HLA-A2-negative CHB patients (HBsAg+ anti-HBeAg+; HBV genotype D) were completely negative, demonstrating the ability of the Env183/A2 MAb to detect TCR-ligand-expressing cells within undisturbed tissues. Taken together, these data indicate that the Env183/A2 MAb is able to recognize pMHC complexes on naturally HBV-infected hepatocytes.
Degeneracy of TCR-like antibody recognition.
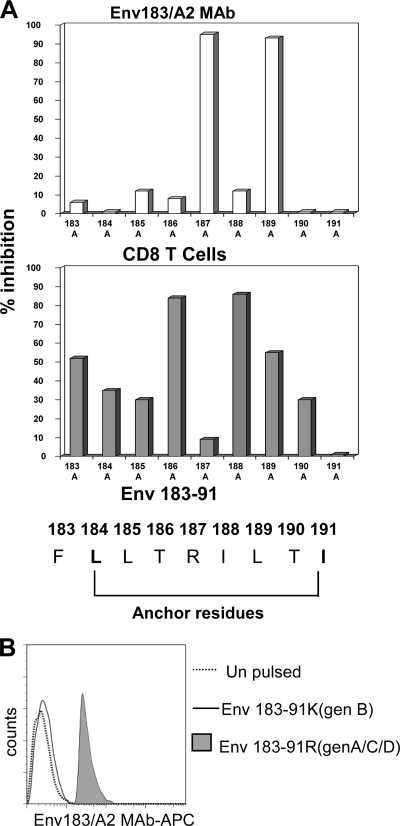
The broad applicability of the Env183/A2 MAb for HLA-A201+ patients infected with different HBV genotypes was investigated. First, we compared the recognition pattern of the Env183/A2 MAb to that of Env183/A2-specific CD8 T cells against HLA-A2+ target cells pulsed with Env183-91 peptides with single-amino-acid substitutions. Figure 4 A shows that the Env183/A2 MAb did not tolerate amino acid substitutions at positions 187 and 189, whereas CD8+ T-cell recognition was affected by mutations at positions 186 and 188. These results indicate differences in the mode of recognition utilized by the Env183/A2 MAb versus compared to that of the TCR of CD8+ T cells specific for the same complex and suggest that HBV mutations selected by envelope-specific cytotoxic T lymphocytes (CTLs) could still be recognizable by the Env183/A2 MAb. However, the inability of the Env183/A2 MAb to tolerate amino acid substitutions at position 187 suggests its exclusive specificity for the Env183-91 epitope sequence of HBV genotypes A, C, and D and not HBV genotypes B, E, and F, since the Arg-187 classically found in HBV genotypes A, C, and D is replaced with Lys-187 in genotypes B, E, and F (13). The data in Fig. 4B confirm this suggestion. The Env183/A2 MAb was not able to recognize Env183-191 (Lys-187) in the context of HLA-A201. These features might limit the applicability of this antibody to HBV genotype A-, C-, and D-infected patients, which might also be further reduced by the potential effect of HLA-A2 subtypes on TCR-like recognition. We have previously shown that A2 subtypes particularly present in Oriental populations (A203, A206, and A207) have an impact on the CD8 T-cell induction and recognition of HLA-A2-restricted HBV epitopes (17). Work is in progress to test the ability of the Env183/A2 MAb to recognize the Env183-91 epitopes presented by different HLA-A2 alleles.
FIG. 4.
Fine-mapping of Env183/A2 MAb epitope recognition. (A) Influence of amino acid mutations within the Env183-91 epitope on Env183/A2 MAb and CD8 T-cell recognition. T2 cells were pulsed with 1 μM Env183-91 peptide with or without (wild type [wt]) alanine substitutions at the indicated positions. The inhibition of TCR-like MAb binding (top) and CD8 T-cell activation (bottom) elicited by alanine substitutions on the wild-type Env183-91 peptide are indicated by bars. Percent inhibition is calculated with the following formula: MFI (or CD107 expression) values obtained with alanine-substituted peptides divided by values obtained with wild-type peptides × 100. (B) Env183/A2 MAb is exclusively specific for the Env183-91 peptide of HBV genotypes (gen) A, C, and D. T2 cells were pulsed with 1 μM Env183-91 peptides with the indicated substitutions at position 187. 187R is characteristic of HBV genotype A, C, and D isolates, and K187 is characteristic of HBV genotype B, E, and F isolates. These cells were stained with Env183/A2 MAb for 1 h and analyzed by flow cytometry. Histograms were overlaid. Results are representative of three independent experiments.
Targeted delivery of TCR-like MAb.
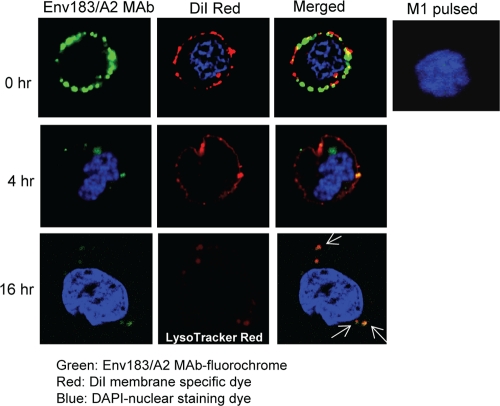
The ability of the Env183/A2 MAb to deliver a linked molecular cargo to cells expressing the correct pMHC complex was then explored. We conjugated a fluorochrome-Fab to the Env183/A2 MAb and tested if the MAb-fluorochrome conjugate is specifically delivered to target cells. Flow cytometry analysis revealed that the MAb-fluorochrome conjugate retained its parental antibody specificity, as specific binding was observed with Env183-91 peptide-loaded cells but not with cells loaded with an irrelevant peptide (data not shown). Following the incubation of Env183-91 peptide-loaded HepG2 cells with the Env183/A2 MAb-fluorochrome conjugate, the cells were incubated at 37°C. At fixed time points aliquots of cells were collected and incubated with DiI red to label the plasma membrane to visualize the internalization of the conjugate. Upon confocal imaging, cells at 0 h showed a ring-like distribution of cell surface fluorescence along with the plasma membrane-specific dye DiI red (Fig. 5, top row). At 4 h of incubation, the majority of surface fluorescence both was delivered to cell and appeared within the cell as small speckles. Cells pulsed with an irrelevant M1 peptide showed no binding of MAb-fluorochrome (indicated by the absence of surface fluorescence).
FIG. 5.
Targeted delivery of immunofluorochrome. HepG2 cells were pulsed with the Env183-91 peptide, incubated with the Env183/A2 MAb-fluorochrome conjugate (green) for 1 h, and then washed, placed onto coverslips, and incubated at 37°C. At different time points after incubation, coverslips were collected and incubated with DiI red to label the plasma membrane (red) or LysoTracker red dye to trace lysosomes in the cells (red, bottom row). Cell nuclei were stained with DAPI (blue). Cells on coverslips were fixed with 1% PFA and mounted for confocal imaging. Cells pulsed with an irrelevant M1 peptide were used as negative controls. Data represent those typical of two independent experiments.
As a vehicle of drug delivery, the nature of cellular localization defines the nature of therapeutic utility. An antibody which binds to a target with slow internalization kinetics provides different setup options than does an antibody targeted to a rapidly recycled membrane component (5). For example, an antibody designed for delivery to endosomal Toll-like receptors (TLRs) must follow a route that results in the delivery of the antibody-ligated molecule to an endosome. It was previously shown that many MAbs are internalized through receptor-mediated endocytosis, which carries the MAb into lysosomes. To address whether the TCR-like antibody follows a similar route, MAb-fluorochrome-stained cells incubated for 16 h were labeled with LysoTracker red to determine whether the internalized antibody costained with lysosomes. Analyses of the results confirmed the colocalization of MAb-fluorochrome and lysosomes (16 h) (Fig. 5). Similarly, the Env183/A2 MAb is able to internalize into HBV-producing HepG2 cells (see Fig. S2 in the supplemental material).
In conclusion, this is the first report to describe the priming, expansion, and selection of B cells producing antibodies that specifically recognize an HBV peptide/MHC complex not only on peptide-pulsed cells but also on hepatocytes naturally infected with HBV. The sensitivity of the Env183/A2 TCR-like antibody is high: not only can it detect the specific ligand on T2 cells pulsed with pM concentrations of the specific peptide, compared to CD8 T cells specific for the same HBV epitope, the TCR-like antibody also has a superior sensitivity. These characteristics, added to the fact that the HBV envelope protein is produced in high quantities on HBV-infected cells, allow this TCR-like antibody to recognize pMHC complexes on the surface of naturally infected hepatocytes despite their very low levels of HLA class I molecules expression (8) and poor antigen-processing activity (15).
Thus, as a diagnostic reagent the Env183/A2-specific TCR-like antibody might allow us to define the relationship between HBV replication and the quantity of antigen presented to specific CD8 T cells. Preliminary experiments performed on HepG2-117 cells show that cells containing approximately 130 HBV DNA copies/cell express ∼1,000 Env183/A2 complexes on their surface (data not shown). The performance of such a quantification with biopsy specimens of CHB patients over the natural course of HBV infection might allow us to correlate the quantity of HBV viremia with the quantity of complexes recognized naturally in vivo by HBV-specific CD8 T cells. This information, in parallel with the functional and quantitative study of the corresponding CD8 T cells, might help us to better understand the mechanisms responsible for the tolerance, deletion, and altered function of HBV-specific CD8 T cells in chronic HBV infection.
Additionally, the combination of the specificity of the TCR-like antibody for cells expressing the correct pMHC complex, the inability of circulating HBV antigens to inhibit this specific binding, and the ability of the antibodies to deliver its cargo into and onto cells expressing the correct pMHC complex encourage the use of humanized versions of the TCR-like antibodies as therapies as well as diagnostics. In the upcoming era of increasingly personalized medicine, TCR-like antibodies may be utilized with HLA class I-matched patients to both visualize the quantity of the infected targets and deliver antiviral drugs or cytokines directly to virus-infected cells.
Supplementary Material
Acknowledgments
We thank Michael Nassal at University Hospital Freiburg for providing the HBV-expressing HepG2-117 cells and Francis Chisari at the Scripps Research Institute for providing the HBsAg plasmid.
This work was supported by the Agency of Science Technology and Research (A*STAR), Singapore.
Footnotes
Published ahead of print on 15 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bjorkman, P. J., et al. 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329:506-512. [DOI] [PubMed] [Google Scholar]
- 2.Cohen, C. J., et al. 2002. Direct detection and quantitation of a distinct T-cell epitope derived from tumor-specific epithelial cell-associated mucin using human recombinant antibodies endowed with the antigen-specific, major histocompatibility complex-restricted specificity of T cells. Cancer Res. 62:5835-5844. [PubMed] [Google Scholar]
- 3.Cohen, C. J., et al. 2003. Direct phenotypic analysis of human MHC class I antigen presentation: visualization, quantitation, and in situ detection of human viral epitopes using peptide-specific, MHC-restricted human recombinant antibodies. J. Immunol. 170:4349-4361. [DOI] [PubMed] [Google Scholar]
- 4.Dadaglio, G., C. A. Nelson, M. B. Deck, S. J. Petzold, and E. R. Unanue. 1997. Characterization and quantitation of peptide-MHC complexes produced from hen egg lysozyme using a monoclonal antibody. Immunity 6:727-738. [DOI] [PubMed] [Google Scholar]
- 5.Dubowchik, G., and M. Walker. 1999. Receptor-mediated and enzyme-dependent targeting of cytotoxic anticancer drugs. Pharmacol. Ther. 83:67-123. [DOI] [PubMed] [Google Scholar]
- 6.Engberg, J., M. Krogsgaard, and L. Fugger. 1999. Recombinant antibodies with the antigen-specific, MHC restricted specificity of T cells: novel reagents for basic and clinical investigations and immunotherapy. Immunotechnology 4:273-278. [DOI] [PubMed] [Google Scholar]
- 7.Ganem, D., and H. Varmus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 8.Gehring, A. J., et al. 2007. The level of viral antigen presented by hepatocytes influences CD8 T-cell function. J. Virol. 81:2940-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaroszewicz, J., et al. 2010. Hepatitis B surface antigen (HBsAg) levels in the natural history of hepatitis B virus (HBV)-infection: a European perspective. J. Hepatol. 52:514-522. [DOI] [PubMed] [Google Scholar]
- 10.Kohler, G., and C. Milstein. 1975. Continuous culture of fused cells secreting antibody of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 11.Matsui, K., et al. 1991. Low affinity interaction of peptide-MHC complexes with T cell receptors. Science 254:1788-1791. [DOI] [PubMed] [Google Scholar]
- 12.Nayersina, R., et al. 1993. HLA-A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J. Immunol. 150:4659-4671. [PubMed] [Google Scholar]
- 13.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 14.Porgador, A., J. W. Yewdell, Y. Deng, J. W. Bennink, and R. N. Germain. 1997. Localization, quantitation, and in situ detection of specific peptide-MHC class 1 complexes using a monoclonal antibody. Immunity 6:715-726. [DOI] [PubMed] [Google Scholar]
- 15.Shin, E. C., et al. 2006. Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. J. Clin. Invest. 116:3006-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, D., and M. Nassal. 2006. Stable HepG2- and Huh7-based human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus. J. Hepatol. 45:636-645. [DOI] [PubMed] [Google Scholar]
- 17.Tan, A. T., et al. 2008. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J. Virol. 82:10986-10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yewdell, J. W. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533-543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.