Abstract
Epstein-Barr virus (EBV)-encoded EBNA3C is one of the latent proteins essential for the efficient transformation of human primary B lymphocytes into continuously proliferating lymphoblastoid cell lines (LCLs) in vitro through manipulation of a number of major cellular pathways. Although it does not have direct DNA-binding activity, EBNA3C plays a central role in the transcriptional modulation of a wide range of both viral and cellular genes during latent infection. Recently, we showed that EBNA3C can directly bind to the tumor suppressor protein p53 and repress its functions, in part by blocking its transcriptional activity as well as facilitating its degradation through stabilization of its negative regulator, Mdm2. In this study, we further showed that EBNA3C can negatively regulate p53-mediated functions by interacting with its regulatory proteins, the inhibitor of growth family proteins ING4 and ING5, shown to be frequently deregulated in different cancers. Functional mapping revealed that both ING4 and ING5 bound to N-terminal domain residues 129 to 200 of EBNA3C, which was previously demonstrated to associate with p53 and is also essential for LCL growth. In addition, we showed that a conserved domain of either ING4 or ING5 bound to both p53 and EBNA3C in a competitive manner, suggesting a potential role for EBNA3C whereby the ING4 or -5/p53 pathway is modulated in EBV-infected cells. Subsequently, we demonstrated that EBNA3C significantly suppresses both the ING4- and ING5-mediated regulation of p53 transcriptional activity in a dose-dependent manner. A colony formation assay as well as an apoptosis assay showed that EBNA3C nullified the negative regulatory effects on cell proliferation induced by coupled expression of p53 in the presence of either ING4 or ING5 in Saos-2 (p53−/−) cells. This report demonstrates a possible role for the candidate tumor suppressor ING genes in the biology of EBV-associated cancers.
Epstein-Barr virus (EBV) was discovered over 4 decades ago, and its association with malignant and nonmalignant diseases in both immunocompetent and immunocompromised individuals remains indisputable (13). EBV is the main cause of infectious mononucleosis and has been linked to a wide spectrum of human malignancies, including nasopharyngeal carcinoma and other hematologic cancers, like Hodgkin's lymphoma, Burkitt's lymphoma (BL), B-cell immunoblastic lymphoma in HIV patients, and posttransplant-associated lymphoproliferative diseases (13). EBV can transform resting B lymphocytes into indefinitely growing lymphoblastoid cell lines (LCLs) in vitro (3, 13). These latently infected LCLs express only a small subset of genes which encompasses six nuclear antigens (EBNA1, -2, -3A, -3B, -3C, and -LP), three membrane-associated proteins (LMP1, -2A, and -2B), and several noncoding and microRNAs (13, 24). Genetic studies with recombinant EBV have demonstrated that EBNA2, EBNA3A, EBNA3C, and LMP1 are essential for transforming primary B cells in vitro (13, 24).
EBNA3C was originally identified as a transcriptional regulator that can regulate the transcription of both viral and cellular genes (12, 13, 27). EBNA3C regulates Notch receptor-induced transcription through association with RBP-Jκ, which plays a central role in EBV-induced cell growth (4, 9, 26). Furthermore, EBNA3C interacts with a vast range of transcriptional modulators, including PU.1, Spi-B, HDAC1, CtBP, DP103, p300, prothymosin-α, Nm23-H1, SUMO1, and SUMO3 (15, 22, 23). EBNA3C also plays a critical role in deregulating the mammalian cell cycle by targeting several cellular oncoproteins and tumor suppressors (1, 7, 8). Recently, we have shown that EBNA3C directly binds to p53 tumor suppressors and represses their apoptotic and transcriptional activities (23). In addition, EBNA3C facilitates p53's degradation by stabilizing its negative regulator Mdm2 (15). Other pathways by which EBNA3C may regulate p53 functions are yet to be elucidated; nonetheless, the mechanisms by which EBNA3C targets critical tumor suppressors and cell cycle regulators is well documented.
The ING4 and ING5 genes are two of the five members of the inhibitor of growth (ING) family of type II tumor suppressors (14, 17). ING1, the first member of this family, was discovered by a technique designed for isolating tumor suppressor genes that is based on PCR-mediated subtractive hybridization (14, 17). Subsequent studies have shown that ING1 may be crucial in contributing to different cellular functions, such as p53-dependent and -independent apoptosis, senescence, and regulation of cell cycle and DNA damage repair machineries (14, 17). Sequence homology with ING1 identified four additional members of the ING family of proteins in humans: ING2, -3, -4, and -5 (14, 17). In contrast to ING1, other ING proteins, including ING1b (a splice variant of ING1), ING2, ING4, and ING5, can induce p53-dependent apoptosis in response to DNA damage or in multiple cancer cell lines via mechanisms that involve increasing p53 acetylation, inhibiting Mdm2-mediated degradation of p53, and enhancing the expression of p53-responsive genes at both the transcriptional and posttranslational levels (14, 17).
The precise mechanism by which p53 functions are influenced by ING family proteins remains a matter of debate. However, it is evident that both ING4 and its functional homolog ING5 act synergistically with p53 to enhance its activity, possibly through regulating its acetylation status (14, 16, 17, 20). Like other ING family proteins, both ING4 and ING5 possess a plant homeodomain (PHD) finger motif, which is the most highly conserved domain among ING proteins and is found in proteins that are associated with chromatin remodeling activities (2, 14, 17). In fact, both ING4 and ING5 have been found to be associated with both histone acetyltransferase (HAT) and histone deacetyltransferase (HDAC) (2, 14, 16). However, the precise strategy utilized by the ING proteins to recruit these modifying enzyme complexes to chromatin still remains undefined.
The nuclear localization of ING proteins was previously shown to be important in the regulation of their function (14, 25). For example, the nuclear localization signal (NLS) domain of ING4 was shown to be critical for p53 binding and p53-mediated function (25). In addition, ING4 can also regulate the functions of other transcription factors, including NF-κB and hypoxia-inducible factor (HIF) (10, 11). Overall, these observations suggest a broad picture of ING functions whereby they target modifying enzymes for histone to specific regions of the cellular genome via interaction with a range of proteins known to bind DNA (10, 11, 14).
Although the deregulation of ING family proteins has been extensively reported in many types of human cancers (14, 20), the link has yet to be successfully demonstrated in virus-mediated tumorigenesis. In this study, we wanted to determine the effects of EBV-encoded latent antigen EBNA3C on ING4- and ING5-regulated p53 tumor suppressor activities. Earlier, we showed that EBNA3C blocks p53 functions in a dose-dependent manner and facilitates the degradation of p53 via stabilization of Mdm2 (15, 23). Here, we further delineate the potential role of EBNA3C in the deregulation of p53 function. We demonstrate that EBNA3C nullifies the positive regulation of both ING4 and ING5 in the tumor suppressor activity of p53. Our results thus provide a foundation for future studies which will describe the involvement of the ING family of proteins in the development of EBV-mediated human cancers.
MATERIALS AND METHODS
Plasmids, antibodies, and cell lines.
Myc-, Flag-, glutathione S-transferase (GST)-, and green fluorescent protein (GFP)-tagged EBNA3C constructs have been previously described (15, 23). A Myc-tagged p53 construct (pA3M-p53) also has been described previously (15, 23). pFlag-CMV-2-ING4 and pFlag-CMV-2-ING5, pCDNA3.1-ING4, and pCDNA3.1-ING5 were kind gifts from Curtis Harris (Center for Cancer Research, National Cancer Institute, NIH). pCDNA3.1-ING4 and pCDNA3.1-ING5 were PCR amplified to generate vectors encoding either wild-type ING4 and ING5 or proteins with different truncations inserted into either modified pGEX-2TK or modified pA3F vectors (1) at EcoRI/NotI restriction sites.
Rabbit polyclonal antibody reactive to ING4 and goat polyclonal antibody against ING5 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and Abcam, Inc. (Cambridge, MA), respectively. Mouse monoclonal antibodies against the myc epitope (9E10), the flag epitope (M2), and EBNA3C (A10) have been described previously (15, 23).
HEK 293, HEK 293T, Saos-2 (p53−/−), MEF (p53−/− Mdm2−/−), and U2OS cells were maintained as described previously (15, 23). BJAB cells, BJAB cells that stably express EBNA3C (E3C7 and -10), and the two EBV-transformed lymphoblastoid cell lines (LCL1 and -2) have been previously described (15, 23).
IP, Western blotting, and GST pulldown assays.
Immunoprecipitation (IP), Western blotting, and GST pulldown assays were essentially performed as described previously (15, 23).
Immunofluorescence.
Immunofluorescence assays were performed as described previously (15, 23) with few modifications. Briefly, U2OS cells plated on coverslips were transfected with expression vectors, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. After 36 h of transfection, cells were fixed with an ice-cold acetone-methanol mixture (1:1) for 10 min at −20°C. LCLs were air dried and fixed similarly. Ectopically expressed ING4 and ING5 were detected using M2 antibody, and EBNA3C (residues 1 to 365) was detected by GFP fluorescence. In LCLs, endogenously expressed ING4, ING5, and EBNA3C were detected using specific antibody. The slides were examined with a FluoView FV300 confocal microscope (Olympus, Inc., Melville, NY).
Reporter assays.
All reporter assays were performed as previously described (15, 23) with few modifications. Briefly, 1 × 106 Saos-2 (p53−/−) cells were transfected by Lipofectamine with 2 μg of a wild-type p21WAF1/CIP1 promoter construct (a generous gift from Wafik S. El-Deiry, Penn State Hershey Cancer Institute, Hershey, PA) and increasing amounts of either myc-tagged p53 (0, 1, 2, 4, and 8 μg), untagged EBNA3C (0, 2, 5, and 10 μg), flag-tagged ING4 and ING5 (0, 5, 10, and 20 μg), or combinations of plasmids. After 36 h of transfection, cells were harvested and lysed, and the luciferase activity was measured. The results are shown as a representation of duplicate experiments.
Colony formation assay.
Saos-2 (p53−/−) cells (10 × 106) were typically transfected using electroporation with different combinations of expression plasmids, as shown in the text. Transfected cells were cultured in the selection medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 5 mg/ml G418). After a 2-week selection, cells were fixed on the plates with 4% formaldehyde and stained with 0.1% crystal violet. The area of the colonies (pixels) in each dish was calculated by Image J software (Adobe, San Jose, CA). The data are shown as the averages and standard deviations of results from two independent experiments.
Detection of apoptosis.
Saos-2 cells (2 × 106) were plated and cotransfected using the Ca3(PO4)2 precipitation method with 5 μg of myc-p53, 10 μg of flag-ING4, 10 μg of flag-ING5, and 10 μg of untagged EBNA3C, as described in the text. Cells were additionally transfected with 5 μg of the pEGFP-C1 vector (Clontech Laboratories, Inc., Mountain View, CA) as a marker for transfection efficiency. After 24 h, posttransfection cells were subjected to serum starvation using 0.1% serum for 12 h. Transfected cells were collected after 36 h, fixed in cold 70% ethanol for 2 h at −20°C, washed with 1× phosphate-buffered saline (PBS), and stained with propidium iodide (PI) staining buffer (10 mM Tris [pH 7.5], 0.2 mg/ml RNase A, and 50 mg/ml propidium iodide) for 2 h in the dark at room temperature. The stained cells were subsequently analyzed using FACScan (BD Biosciences, San Jose, CA) and FlowJo (Tree Star, Inc., Ashland, OR) software.
RESULTS
EBNA3C can form a p53-independent complex with both ING4 and ING5.
ING4 and ING5 are type II suppressors and were shown to form complexes with both p53 and p300 (16). Both of these tumor suppressors have been shown to be frequently deregulated in a variety of cancer types and associated with enhanced apoptosis in a p53-dependent manner (14, 16, 20). Recently, we have shown that EBV nuclear antigen EBNA3C directly binds to p53 and blocks its functional activity (23). Herein we further aim to investigate whether EBNA3C can block ING4- and ING5-regulated p53 tumor suppressor activities.
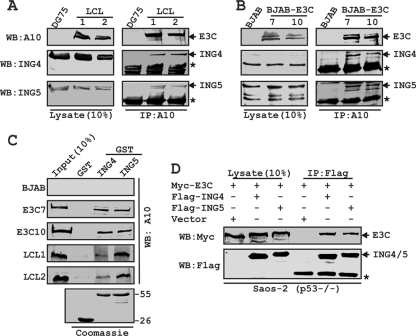
First, we determined whether EBNA3C interacts with either ING4 or ING5 in human cells. Endogenously expressed EBNA3C was immunoprecipitated from either EBV-transformed cell lines (LCL1 and LCL2) or BJAB cells stably expressing EBNA3C (E3C7 and E3C10), and coimmunoprecipitation (co-IP) of both ING4 and ING5 was monitored by immunoblotting using specific antibodies (Fig. 1 A and B). EBV-negative Burkitt's lymphoma (BL) lines DG75 and BJAB were used as controls (Fig. 1A and B). The results demonstrated that EBNA3C formed a stable complex with both ING4 and ING5 in human cells (Fig. 1A and B). To further corroborate this association, an in vivo GST pulldown assay was performed in which recombinant GST-ING4 and -ING5 were incubated separately with lysates prepared from either EBV-transformed cells (LCL1 and LCL2) or EBNA3C-positive cells (BJAB cell line clones E3C7 and E3C10), with BJAB cells as a negative control (Fig. 1C). The results showed that EBNA3C interacted strongly with both GST-tagged ING4 and ING5 but not with the GST control protein (Fig. 1C). A parallel gel with Coomassie staining showed the amount of recombinant GST proteins employed in this interaction study (Fig. 1C). However, all these B cells are p53+/+, so we could not rule out the possibility that p53 could serve as a bridge between EBNA3C and either the ING4 or ING5 interface.
FIG. 1.
EBNA3C forms a p53-independent complex with both ING4 and ING5. BJAB cells and two BJAB cell stable clones expressing EBNA3C (E3C7 and 10) (A) and DG75 cells and two LCL clones (LCL1 and 2) (B) were subjected to IP with EBNA3C specific antibody (A10). Samples were resolved by SDS-PAGE and detected by Western blotting (WB) for the indicated proteins by stripping and reprobing the same membrane. (C) Either purified GST or GST-ING4 or -ING5 beads were incubated with cell lysates, and EBNA3C was detected by WB with A10. Coomassie staining of GST-labeled proteins is shown in the bottom panel. Numbers at the right of the bottom blot are molecular masses (in kilodaltons). (D) Saos-2 (p53−/−) cells were cotransfected with myc-EBNA3C in the presence of either the vector control, flag-tagged ING4, or flag-tagged ING5, as indicated. At 36 h posttransfection, cells were harvested, lysed, and immunoprecipitated with flag antibody. Samples were subjected to WB with the indicated antibodies. The asterisks indicate the immunoglobulin bands.
In order to confirm whether the interaction between INGs and EBNA3C is dependent on p53, we performed an in vivo IP experiment using Saos-2 cells, a p53-deficient cell line. Saos-2 (p53−/−) cells were subsequently cotransfected with myc-EBNA3C and either flag-ING4 or -ING5 and an empty vector as a control. EBNA3C was strongly coimmunoprecipitated with both ING4 and ING5 pulldown using Flag antibody but not with the vector control (Fig. 1D). Analysis of the ectopic expression data strongly demonstrated that both ING4 and ING5 form a p53-independent complex with EBNA3C.
EBNA3C residues 129 to 200 interact with both ING4 and ING5.
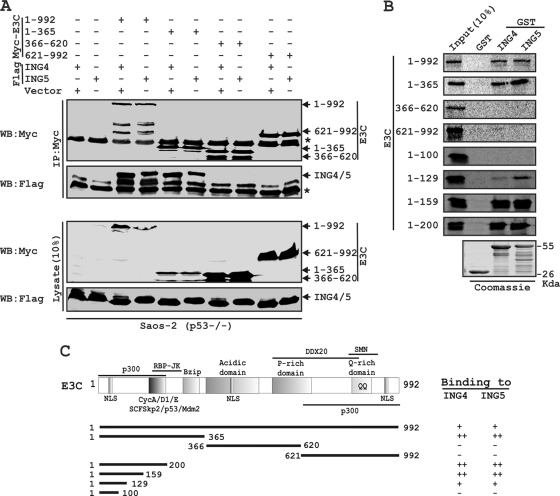
Next, we wanted to determine the functional residues of EBNA3C that specifically interact with ING proteins. Two successive binding assays were performed with different truncated polypeptides of EBNA3C. First, p53 null Saos-2 cells were transfected with either ING4 or ING5 in combination with either the control vector or different myc-EBNA3C expression constructs covering the entire length of the molecule. The results showed that both ING4 and ING5 were coimmunoprecipitated with either full-length EBNA3C (residues 1 to 992) or residues 1 to 365 of the amino-terminal domain (Fig. 2 A). However, no co-IP was noticed with either the vector control or vectors encoding EBNA3C residues 366 to 620 or EBNA3C residues 621 to 992 (Fig. 2A).
FIG. 2.
A critical N-terminal domain of EBNA3C interacts with both ING4 and ING5. (A) Saos-2 (p53−/−) cells were cotransfected with either flag-ING4 or -ING5 in the presence of different myc-EBNA3C constructs, as indicated. Cells were harvested, lysed, and immunoprecipitated with anti-myc antibody. Samples were resolved by a 7-to-15% gradient SDS-PAGE and detected by Western blotting (WB) for the indicated proteins by stripping and reprobing the same membrane. (B) The wild-type plasmid or plasmids of EBNA3C expressing different truncated mutant proteins were in vitro translated in the presence of [S35]Met. After preclearing of all S35-radiolabeled translated proteins with GST beads for 1 h at 4°C, samples were subjected to GST pulldown assay by incubating them with either a GST control, GST-ING4, or GST-ING5 as indicated. Reactions were resolved by appropriate SDS-PAGE, exposed to a phosphorimager plate, and scanned using the Storm 850 imaging system. Coomassie staining of a parallel SDS-PAGE gel resolved purified GST proteins, seen in the bottom panel. (C) The schematic illustrates different structural and interaction domains of EBNA3C and summarizes the results of studies of the binding between different domains of EBNA3C and ING4 and ING5. ++, strong binding; +, moderate binding; −, no binding. NLS, nuclear localization signal; Bzip, basic leucine zipper domain; SMN, survival of motor neurons. The asterisk indicates the immunoglobulin bands.
We further mapped the binding residues of EBNA3C within the amino-terminal region (residues 1 to 365) by GST pulldown assay. Recombinant GST-fused ING4 and ING5 were incubated with different in vitro-translated, radiolabeled fragments of EBNA3C. The result showed that EBNA3C residues 1 to 129, 1 to 159, and 1 to 200, along with full-length EBNA3C and residues 1 to 365, strongly bound with both GST-ING4 and -ING5 (Fig. 2B). However, the remaining residues of EBNA3C polypeptides starting from 1 to 100 and 200 to 992, along with a GST control, showed little or no binding, indicating the specificity of this binding experiment (Fig. 2B). A parallel Coomassie blue-stained gel showed the amounts of recombinant GST proteins employed in this interaction study (Fig. 2B). The results showed that the residues of EBNA3C, which was previously shown to interact with p53 (23), can also directly bind to both ING4 and ING5 in a p53-independent manner (Fig. 2C).
EBNA3C colocalizes with both ING4 and ING5 in EBV-transformed cells.
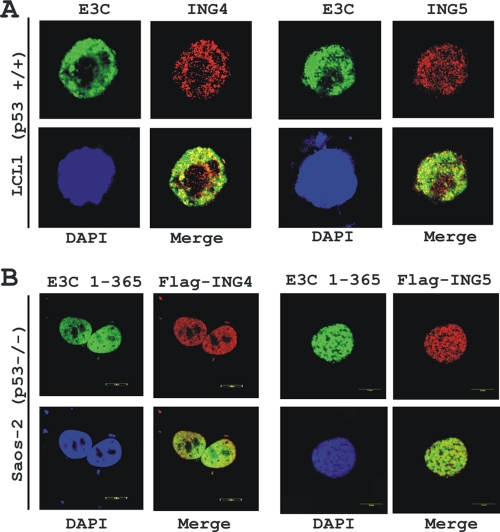
For additional support of the binding data described above and to visualize the subcellular localization pattern of both ING4 and ING5 with EBNA3C, colocalization experiments were conducted with an EBV-transformed cell line, LCL1 (Fig. 3 A). Immunofluorescence staining using antibodies specific to EBNA3C and ING proteins demonstrated that both proteins had distinctive nuclear staining with a speckled pattern (Fig. 3A). The results showed that both ING4 and ING5 clearly colocalized with EBNA3C in human cells, as visualized by yellow fluorescence (Fig. 3A). The colocalization study was further extended using ectopically expressed flag-tagged ING4 and ING5 with the GFP-tagged, amino-terminal, interacting domain of EBNA3C (residues 1 to 365) in Saos-2 cells. The results showed that both ING4 and -5 clearly colocalized with the amino-terminal domain of EBNA3C, except in the nucleoli, as indicated by yellow fluorescence signals (Fig. 3B). Further, these data suggested that the colocalization among EBNA3C and either ING4 or ING5 was exclusively p53 independent.
FIG. 3.
EBNA3C colocalizes with both ING4 and ING5. (A) EBV-transformed LCL1 cells were plated and air dried onto slides. (B) Saos-2 (p53−/−) cells plated on coverslips were transfected with GFP-tagged EBNA3C (residues 1 to 365) in the presence of either flag-ING4 or -ING5 using Lipofectamine 2000. Endogenously (A) and ectopically (B) expressed ING4 and ING5 were detected using specific antibodies and M2 antibody, respectively, followed by specific anti-Alexa Fluor 594 20 antibody (red). (A) Endogenous EBNA3C in EBV-positive LCLs was detected using an EBNA3C-reactive mouse monoclonal antibody (A10 ascites) followed by anti-mouse Alexa Fluor 488 (green). Ectopically expressed GFP-EBNA3C (residues 1 to 365) was detected by GFP fluorescence. The nuclei were subsequently stained with DAPI (4′,6-diamino-2-phenylindole), and the images were captured using an Olympus confocal microscope. All panels are representative pictures from approximately 50 cells of five different fields of three independent experiments.
EBNA3C competes with p53 for binding with both ING4 and ING5.
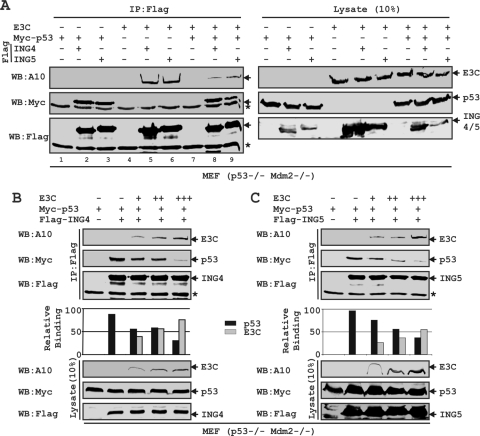
co-IP as well as GST pulldown assays demonstrated that both ING4 and ING5 bind to the same N-terminal region of EBNA3C (residues 129 to 200) which was earlier shown to interact with p53 (23). In an attempt to check whether these molecules can form a ternary complex, both p53 and Mdm2-null MEF cells (p53−/− Mdm2−/−) were cotransfected with different combinations of heterologous expression systems for EBNA3C, myc-p53, and both flag-tagged ING4 and ING5, as shown in Fig. 4 A. The reason for using Mdm2-deficient cells was to exclude the possibility of Mdm2 binding in this context, as we showed earlier that Mdm2 also interacted with the same binding region of EBNA3C (15). The results showed that both p53 and EBNA3C were strongly coimmunoprecipitated with flag IP of either ING4 or ING5 (Fig. 4A, lanes 3, 4, 5, and 6). The data also indicated that, separately, both ING4 and ING5 possibly form a transient ternary complex with p53 and EBNA3C (Fig. 4A, lanes 8 and 9). However, interestingly, the interaction of EBNA3C with either ING4 or ING5 was markedly diminished in the presence of p53 (Fig. 4A, compare lanes 5, 6, 8, and 9), which signified that p53 has a higher binding affinity than EBNA3C to either ING4 or ING5. This result also strongly suggested that these proteins share a binding region.
FIG. 4.
EBNA3C competes with p53 for binding with both ING4 and ING5. (A to C) MEF (p53−/− Mdm2−/−) cells were cotransfected with different combinations of untagged EBNA3C, myc-p53, and either flag-ING4 or -ING5 as indicated. (B and C) EBNA3C-expressing constructs were transfected in increasing amounts (0, 1, 5, and 15 μg). Transfected cells were harvested, lysed, and subjected to IP with anti-flag antibody. Samples were resolved by 7-to-15% gradient SDS-PAGE and detected by Western blotting (WB) using the indicated antibodies by stripping and reprobing the same membrane. The asterisks indicate the immunoglobulin bands.
The sharing of a common binding region among these proteins led us to investigate further whether an increasing amount of EBNA3C can block the association of p53 with either ING4 or ING5. MEF (p53−/− Mdm2−/−) cells were cotransfected with increasing amounts of an untagged-EBNA3C-expressing construct and a constant amount of myc-tagged p53, plus either flag-tagged ING4 or ING5 (Fig. 4B and C, respectively). Indeed, the results showed that escalating the amount of EBNA3C caused displacement of p53 binding from both ING4 and ING5 in a dose-dependent manner (Fig. 4B and C, respectively).
Both EBNA3C and p53 bind to the same region of either ING4 or ING5.
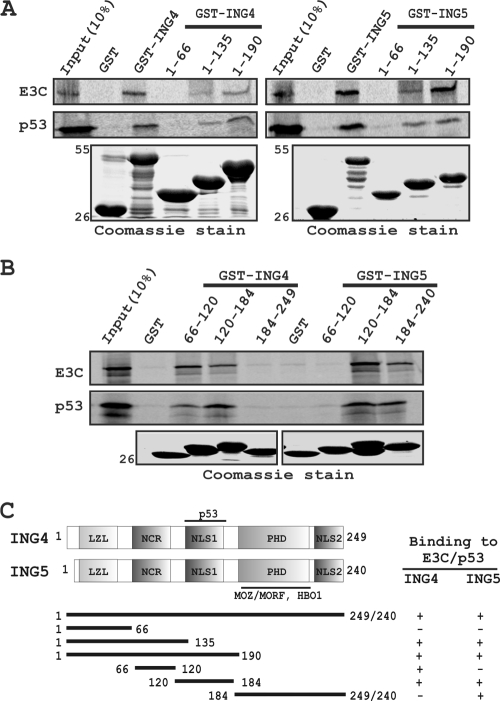
Competitive binding assays further led us to map the precise binding domain of both ING4 and ING5 responsible for their interaction with either EBNA3C or p53. A series of GST-fused mutants of both ING4 and ING5 with C-terminal deletions were generated and further tested for their binding activity with both EBNA3C and p53 by in vitro GST pulldown assays. The results showed that both the full-length proteins and the carboxy-terminal domains of GST-ING4 and -ING5 bind to both 35S-radiolabeled EBNA3C and p53 (Fig. 5 A). No binding was detected either with the leucine zipper-like (LZL) N-terminal domains of both ING4 and ING5 or with the GST control (Fig. 5A), providing a level of specificity for this experiment. A parallel gel with Coomassie staining showed the levels of the GST proteins used in the binding assay (Fig. 5A).
FIG. 5.
Both EBNA3C and p53 bind to the same region of either ING4 or -5. (A and B) Both S35-radiolabeled, in vitro-translated EBNA3C and p53 were subjected to binding reactions with either a GST control or different GST-fused, truncated proteins of both ING4 and ING5. Precipitated proteins were resolved by 10% SDS-PAGE, dried, exposed to a phosphorimager plate, and scanned. (A and B) Coomassie stains of SDS-PAGE-resolved purified GST-fused proteins are shown in the bottom panels. Numbers at the left indicate molecular masses in kilodaltons. (C) The schematic illustrates different structural and interaction domains of both ING4 and ING5 and summarizes the results of studies of the binding of ING4 and ING5 with p53 and EBNA3C. +, binding; −, no binding. LZL, leucine zipper-like motif; NCR, novel conserved region; NLS, nuclear localization signal; PHD, plant homeodomain.
To map in more detail the domains located within the C-terminal sites of both ING4 and ING5 required for interaction, additional constructs containing residues within the C-terminal domains of both ING4 and ING5 were generated. In vitro GST pulldown experiments showed that residues 66 to 184 of ING4 and residues 120 to 240 of ING5 predominantly bind to both EBNA3C and p53 (Fig. 5B and C). These results are consistent with a mechanism of competitive binding where increasing amounts of EBNA3C disrupted the interaction of p53 with both ING4 and ING5.
EBNA3C blocks both ING4- and ING5-mediated p53 transcriptional activity.
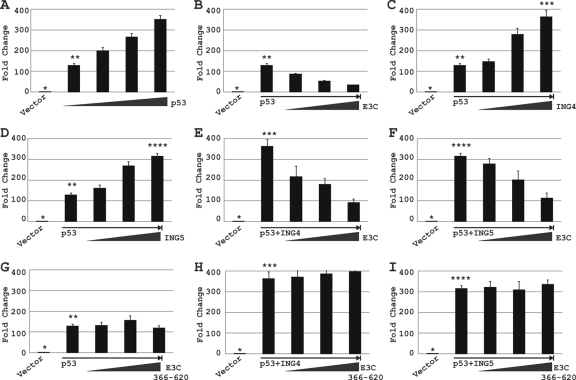
Previous studies have shown that both ING4 and ING5 increased p53-mediated p21WAF1/CIP1 promoter activity, possibly through modulation of the acetylation status of p53 (16). Moreover, recently we have shown that EBNA3C blocks p53-mediated transactivation (23). In order to determine whether EBNA3C can also block ING4- and ING5-regulated p53 transcriptional activities, a wild-type p21WAF1/CIP1 reporter construct was transiently cotransfected into Saos-2 cells with different combinations of expression constructs, as indicated in Fig. 6. As expected, p53 increases the p21WAF1/CIP1 promoter activity in a dose-dependent manner (Fig. 6A). In the presence of increasing amounts of the EBNA3C-expressing plasmid, p53-mediated transactivation was gradually reduced (Fig. 6B). Moreover, as expected, both ING4 and ING5 increased p53-mediated p21WAF1/CIP1 promoter activity in a dose-dependent manner (Fig. 6C and D). Next, an increasing amount of EBNA3C showed repression of both ING4- and ING5-mediated p53 transactivation in a dose-dependent fashion (Fig. 6E and F). However, interestingly, the nonbinding domain of EBNA3C (residues 366 to 620) had no significant effect on either p53 alone or p53 plus ING4/ING5-dependent promoter activity (Fig. 6G, H, and I). Overall, these data suggest that EBNA3C can repress ING4- and ING5-mediated p53 transcriptional activity, possibly by interfering with the interaction between p53 and either ING4 or ING5, as described in the legend of Fig. 4B and C.
FIG. 6.
EBNA3C blocks ING4- and ING5-triggered p53 transcriptional activities. (A to I) Saos-2 (p53−/−) cells were cotransfected with 2 μg of a wild-type p21WAF1/CIP1 promoter construct and different combinations of plasmids expressing untagged EBNA3C, myc-p53, and either flag-tagged ING4 or flag-tagged ING5 as described in Materials and Methods. At 36 h posttransfection, cells were harvested and lysed in reporter lysis buffer. Total amounts of proteins were normalized by the Bradford assay. The bars plot the means of the results of two independent experiments. Error bars represent standard deviations (SD). Different numbers of asterisks represent identical bars in panels A to E. A single asterisk represents the p21WAF1/CIP1 promoter plus the vector control in panels A to I; two asterisks represent the p21WAF1/CIP1 promoter plus 1 μg of a myc-tagged p53-expressing vector in panels A to D and G; three asterisks represent the p21WAF1/CIP1 promoter plus 20 μg of a flag-tagged ING4-expressing vector in panels C, E, and H; and four asterisks represent the p21WAF1/CIP1 promoter plus 20 μg of a flag-tagged ING5-expressing vector in panels D, F, and I.
EBNA3C nullifies the negative regulation of cell proliferation mediated by p53 together with either ING4 or ING5.
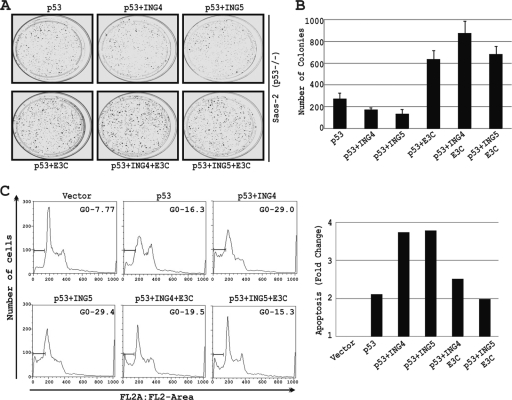
Earlier studies have demonstrated that both ING4 and ING5 negatively controlled cell growth in a p53-dependent manner (16). Similarly, we investigated the effect of EBNA3C on p53 as well as either ING4- or ING5-mediated cell growth using a colony formation assay of Saos-2 cells. The results showed that coexpression of either ING4 or ING5 with p53 significantly reduced the colony formation of Saos-2 cells compared to that produced with p53 alone (Fig. 7 A and B). In contrast, cotransfection with an EBNA3C expression vector markedly increased (approximately 4-fold) the colony formation of Saos-2 cells in the presence of either p53 alone or p53 plus ING4 or -5 (Fig. 7A and B).
FIG. 7.
EBNA3C reverses the negative regulation of cell proliferation mediated by p53 coupled with either ING4 or -5. (A to B) Saos-2 (p53−/−) cells were electroporated with expression plasmids for myc-p53 with either a vector control, flag-ING4, or flag-ING5 or in combinations with EBNA3C, as indicated. Transfected cells were cultured in the selection medium (DMEM supplemented with 1 mg/ml G418). After a 2-week selection, cells were fixed on the plates with 4% formaldehyde and stained with 0.1% crystal violet. The area of the colonies (pixels) in each dish was calculated by Image J software. (B) The bar diagram represents the averages of data from two independent experiments. (C) Saos-2 cells were transfected with different combinations of expression constructs, as indicated, using the Ca3(PO4)2 precipitation method. Cells were collected at 36 h posttransfection after a 12-h serum starvation and fixed. Levels of cells undergoing apoptosis (sub-G0 phase) in individual PI-stained samples were analyzed by flow cytometry, and the data were analyzed by FlowJo software. The bar diagram represents the fold differences in sub-G0 value from that of the vector control sample. All graphs are from at least three similar repeated experiments.
It was earlier demonstrated that ectopic expression of both ING4 and ING5 can lead to apoptosis in a p53-dependent manner (16). Moreover, we have shown previously that EBNA3C significantly blocks p53-mediated apoptosis in Saos-2 cells (23). To determine whether EBNA3C expression also affected the ability of both ING4 and ING5 to trigger p53-dependent apoptosis, the levels of cells undergoing apoptosis (sub-G0 phase) in Saos-2 cells were examined (Fig. 7C). Further, to verify the transfection efficiency as well as the viability of the cells, a GFP expression plasmid was introduced to each sample, which revealed approximately 80% transfection efficiency (data not shown). The results showed that about 16% of p53-expressing Saos-2 cells underwent apoptosis, which was approximately 2-fold higher than that of vector control cells (Fig. 7C). As expected, both ING4 and ING5 enhanced apoptosis when they were coexpressed with p53; compared to apoptosis with p53 alone, coexpression increased apoptosis about 2-fold, and compared to the level with the vector control, coexpression increased apoptosis 4-fold (Fig. 7C). However, in the presence of EBNA3C, both ING4- and ING5-induced p53-mediated apoptosis was drastically inhibited (Fig. 7C). These data clearly demonstrated that EBNA3C significantly protects cells from both ING4- and ING5-enhanced p53-dependent apoptosis (Fig. 8).
FIG. 8.
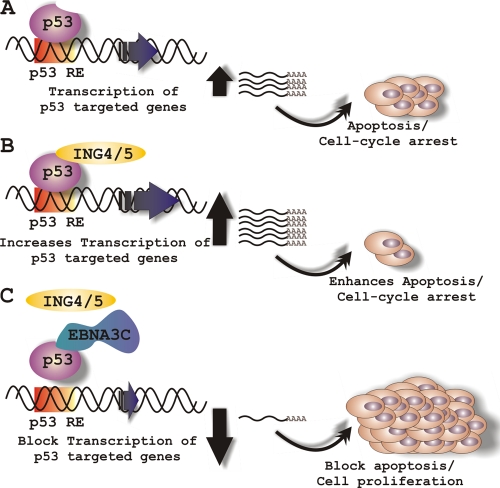
Schematic representation of EBNA3C's role in modulating both ING4- and ING5-triggered p53 activity. (A) In response to genotoxic stress, p53 activates its downstream target genes, which subsequently leads to cell cycle arrest and apoptosis. Both ING4 and ING5 trigger a p53-mediated effect on cell proliferation. However, in EBV-positive cells, EBNA3C potentially inhibits both ING4- and ING5-mediated p53 transcriptional activity via displacing the interaction among p53, ING4, and ING5.
DISCUSSION
The inhibitor of growth (ING) family of genes is rarely mutated in human cancer, although their expression is frequently downregulated in several cancer types (14, 17, 20). A number of functional analyses of the ING family of proteins provided a link to p53 signaling pathways, supporting their involvement in cellular processes such as growth arrest, senescence, apoptosis, and DNA repair (14, 17, 20). A physical association between different ING proteins and p53 has been reported in several studies (14, 17, 20). Like other members of the ING family proteins, both ING4 and ING5 were also previously shown to complex with p53 and to positively modulate its functional activity (16, 20).
A large number of earlier studies have successfully established that EBNA3C, an indispensable oncoprotein required for EBV-mediated B-cell transformation, strongly interferes with the functions of many important cellular proteins which are principally involved in cell cycle checkpoints at both the G1-to-S and G2-to-M transitions (1, 6, 7, 22). Recently, in an initial study, we clearly showed that EBNA3C directly interacts with p53 and efficiently blocks its transcriptional as well as apoptotic activities (23). Here, studies were undertaken to explore the possibility that EBNA3C can also block both ING4- and ING5-triggered p53-mediated functions.
An interaction study was first performed to demonstrate whether EBNA3C was able to form a complex with either ING4 or ING5. Indeed, binding experiments showed that EBNA3C can form a stable complex with both ING4 and ING5 in EBV-transformed LCLs. Moreover, binding experiments using p53 null cell lines clearly suggested that the interaction between EBNA3C and either ING4 or ING5 was not p53 dependent. Interestingly, functional mapping of the binding region of EBNA3C demonstrated that it associated with both ING4 and ING5 through residues 129 to 200 at the amino-terminal site, which was previously shown to interact with p53, along with numerous other critical cellular factors, such as Mdm2, RBP-Jκ, SCFSkp2, pRb, cyclin A, and c-Myc (9, 15, 23). The addition of both ING4 and ING5 to this group further accentuates the requirement of these critical residues of EBNA3C for circumventing various cell cycle checkpoints in EBV-transformed cells. Recently, a genetic study using recombinant EBV expressing conditionally active EBNA3C showed that deletion of this particular domain could not support the cell proliferation of EBV-transformed LCLs, demonstrating the importance of this domain of EBNA3C (9).
We also demonstrated a possible transient ternary-complex formation with EBNA3C, p53, and either ING4 or ING5 molecules. However, the interaction with EBNA3C was significantly less in the presence of p53 than in the presence of EBNA3C alone, also suggesting potential dimeric complexes of ING proteins with p53 or EBNA3C. Thus, either p53 blocked the interaction between EBNA3C and ING4 or ING5, or p53 and EBNA3C share a binding region on ING4 and ING5. Competitive binding assays also showed that an increasing amount of EBNA3C blocked p53 association with both ING4 and ING5 in a dose-dependent manner, further suggesting that both EBNA3C and p53 may have identical binding surfaces on either ING4 or ING5. In fact, mapping experiments clearly demonstrated that both EBNA3C and p53 bound to the same region of either ING4 or ING5. Like other ING proteins, both ING4 and ING5 contain several highly conserved domains, including a leucine zipper-like (LZL) motif, two nuclear localization signal domains (NLS1 and NLS2), and a C-terminal plant homeodomain (PHD) (Fig. 5C) (14, 17, 20). The PHD motif is a zinc finger domain that binds to histone H3 in a methylation-sensitive fashion and is a key structural component of the ING proteins (14, 17, 20). This domain is required for the function of ING proteins in chromatin remodeling through their interaction with both HAT and HDAC complexes (14, 17, 20). ING5 has also been shown to interact with different chromatin remodeling factors, such as monocytic leukemia zinc finger protein (MOZ)/MOZ-related factors (MORF) and HBO1 HAT complexes, via its PHD zinc finger domain (2). Interestingly, we now show that ING5 also interacts with both EBNA3C and p53 via the same conserved PHD domain. This not only corroborates the importance of this domain but also raises the possibility of modulating ING5 functions by EBNA3C and p53. Unlike with ING5, our data showed that ING4 bound to both EBNA3C and p53 through the bipartite NLS1 domain. A second, nonfunctional NLS2 domain is located at the C terminus (residues 184 to 249) (21). ING4 was previously shown to interact with p53 via its nuclear localization domain (NLS1) (25). Interestingly, even though ING4 and ING5 showed a high degree of similarity in sequence homology (∼70%) as well as in functional activity (16), they can be regulated via different mechanisms, as observed in our binding results. However, it can be hypothesized that, by blocking the interaction interface between p53 and either ING4 or ING5, EBNA3C may interfere with both ING4- and ING5-prompted p53-mediated deregulations. Furthermore, we demonstrated that EBNA3C efficiently rescues both ING4- and ING5-mediated activation of p53 functions, as evidenced by the reporter and apoptosis assays. Both ING4 and ING5 have been shown earlier to positively regulate p53 activity, perhaps by enhancing the acetylation of p53 via recruiting p300 HAT activity (16), although the precise mechanism involved remains elusive. Interestingly, EBNA3C was shown to interact with both p300 and HDAC1 (5, 18, 19). It is possible that the interaction of EBNA3C with p300 and HDAC1 may result in tempering the acetylation status of both the p53 and ING proteins, which eventually defines their functions in regulating cell growth.
In summary, our findings suggest that the recently discovered type II tumor suppressor proteins ING4 and ING5 are potential targets of EBV-encoded oncoprotein EBNA3C and provide the first evidence that the ING family of tumor suppressor genes could be implicated in virally induced oncogenesis. The study has provided us further insight into understanding the possible mechanisms by which EBV-mediated transformation is associated with human cancers. Understanding the role of EBNA3C in ING protein deregulation in EBV-infected cells provides clues into the mode of inactivation by ING proteins and thus the possibility that restoration of ING protein expression or functions may be used as a form of therapeutics. A large body of evidence has revealed that many tumor suppressor genes, including those for p53 and pRb, are crucial targets of a number of oncogenic human viruses. Our work has raised the question of whether ING4 and ING5 may also be possible targets for many other oncogenic viruses.
Acknowledgments
We thank Curtis Harris (Center for Cancer Research, National Cancer Institute, NIH) and Wafik S. El-Deiry (Penn State Hershey Cancer Institute, Hershey, PA) for generously providing reagents.
The project is supported by NCI grants CA137894-02 and CA138434-02 to E.S.R. E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1.Bajaj, B. G., et al. 2008. Epstein-Barr virus nuclear antigen 3C interacts with and enhances the stability of the c-Myc oncoprotein. J. Virol. 82:4082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyon, Y., et al. 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21:51-64. [DOI] [PubMed] [Google Scholar]
- 3.Halder, S., et al. 2009. Early events associated with infection of Epstein-Barr virus infection of primary B-cells. PLoS One 4:e7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johannsen, E., C. L. Miller, S. R. Grossman, and E. Kieff. 1996. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJkappa in Epstein-Barr virus-transformed B lymphocytes. J. Virol. 70:4179-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight, J. S., K. Lan, C. Subramanian, and E. S. Robertson. 2003. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J. Virol. 77:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight, J. S., and E. S. Robertson. 2004. Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J. Virol. 78:1981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 102:18562-18566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. SCFSkp2 complex targeted by Epstein-Barr virus essential nuclear antigen. Mol. Cell. Biol. 25:1749-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruo, S., et al. 2009. Epstein-Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc. Natl. Acad. Sci. U. S. A. 106:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozer, A., and R. K. Bruick. 2005. Regulation of HIF by prolyl hydroxylases: recruitment of the candidate tumor suppressor protein ING4. Cell Cycle 4:1153-1156. [DOI] [PubMed] [Google Scholar]
- 11.Ozer, A., L. C. Wu, and R. K. Bruick. 2005. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF). Proc. Natl. Acad. Sci. U. S. A. 102:7481-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radkov, S. A., et al. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe et al. (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 14.Russell, M., P. Berardi, W. Gong, and K. Riabowol. 2006. Grow-ING, age-ING and die-ING: ING proteins link cancer, senescence and apoptosis. Exp. Cell Res. 312:951-961. [DOI] [PubMed] [Google Scholar]
- 15.Saha, A., et al. 2009. Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J. Virol. 83:4652-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiseki, M., et al. 2003. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 63:2373-2378. [PubMed] [Google Scholar]
- 17.Soliman, M. A., and K. Riabowol. 2007. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem. Sci. 32:509-519. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian, C., et al. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian, C., J. S. Knight, and E. S. Robertson. 2002. The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration. Front. Biosci. 7:d704-d716. [DOI] [PubMed] [Google Scholar]
- 20.Unoki, M., K. Kumamoto, S. Takenoshita, and C. C. Harris. 2009. Reviewing the current classification of inhibitor of growth family proteins. Cancer Sci. 100:1173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unoki, M., J. C. Shen, Z. M. Zheng, and C. C. Harris. 2006. Novel splice variants of ING4 and their possible roles in the regulation of cell growth and motility. J. Biol. Chem. 281:34677-34686. [DOI] [PubMed] [Google Scholar]
- 22.West, M. J. 2006. Structure and function of the Epstein-Barr virus transcription factor, EBNA 3C. Curr. Protein Pept. Sci. 7:123-136. [DOI] [PubMed] [Google Scholar]
- 23.Yi, F., et al. 2009. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology 388:236-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, X., et al. 2005. Nuclear localization signal of ING4 plays a key role in its binding to p53. Biochem. Biophys. Res. Commun. 331:1032-1038. [DOI] [PubMed] [Google Scholar]
- 26.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jkappa. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]