Abstract
Adenovirus type 5 (Ad5) inactivates the host cell DNA damage response by facilitating the degradation of Mre11, DNA ligase IV, and p53. In the case of p53, this is achieved through polyubiquitylation by Ad5E1B55K and Ad5E4orf6, which recruit a Cul5-based E3 ubiquitin ligase. Recent evidence indicates that this paradigm does not apply to other adenovirus serotypes, since Ad12, but not Ad5, causes the degradation of TOPBP1 through the action of E4orf6 alone and a Cul2-based E3 ubiquitin ligase. We now have extended these studies to adenovirus groups A to E. While infection by Ad4, Ad5, and Ad12 (groups E, C, and A, respectively) cause the degradation of Mre11, DNA ligase IV, and p53, infection with Ad3, Ad7, Ad9, and Ad11 (groups B1, B1, D, and B2, respectively) only affects DNA ligase IV levels. Indeed, Ad3, Ad7, and Ad11 cause the marked accumulation of p53. Despite this, MDM2 levels were very low following infection with all of the viruses examined here, regardless of whether they increase p53 expression. In addition, we found that only Ad12 causes the degradation of TOPBP1, and, like Ad5, Ad4 recruits a Cul5-based E3 ubiquitin ligase to degrade p53. Surprisingly, Mre11 and DNA ligase IV degradation do not appear to be significantly affected in Ad4-, Ad5-, or Ad12-infected cells depleted of Cul2 or Cul5, indicating that E1B55K and E4orf6 recruit multiple ubiquitin ligases to target cellular proteins. Finally, although Mre11 is not degraded by Ad3, Ad7, Ad9, and Ad11, no viral DNA concatemers could be detected. We suggest that group B and D adenoviruses have evolved mechanisms based on the loss of DNA ligase IV and perhaps other unknown molecules to disable the host cell DNA damage response to promote viral replication.
Human adenoviruses (Ad) comprise a large family of more than 50 different serotypes, which have been classified into six groups (A to F; summarized in Supplemental Table 1 at http://www.cancersciences.bham.ac.uk/research/supplementarydata.shtml) (3, 42). Most of the viruses cause mild infections during early childhood, although much of the interest in the scientific community stems from the demonstration that members of group A viruses (e.g., Ad12) can cause tumors in newborn rodents (48). Despite these observations, the great majority of studies have been carried out on group C viruses, Ad2 and Ad5, with some interest being shown in Ad12 and Ad9.
TABLE 1.
Summary of protein degradation and localization following infection with Ad3, Ad4, Ad5, Ad7, Ad9, Ad11, and Ad12a
| Group | Ad serotype | Protein | Upreg | Transient upreg | Stable | Degraded | Tracks | VRCs |
|---|---|---|---|---|---|---|---|---|
| A | 12 | p53 | ✓ | ✓ | ||||
| Mre11 | ✓ | ✓ | ||||||
| TOPBP1 | ✓ | ✓ | ||||||
| DNA lig IV | ✓ | |||||||
| B1 | 3 | p53 | ✓ | ✓ | ||||
| Mre11 | ✓ | ✓ | ||||||
| TOPBP1 | ✓ | ✓ | ||||||
| DNA lig IV | ✓ | |||||||
| B1 | 7 | p53 | ✓ | ✓ | ||||
| Mre11 | ✓ | ✓ | ||||||
| TOPBP1 | ✓ | ✓ | ||||||
| DNA lig IV | ✓ | |||||||
| B2 | 11 | p53 | ✓ | ✓ | ||||
| Mre11 | ✓ | ✓ | ||||||
| TOPBP1 | ✓ | ✓ | ||||||
| DNA lig IV | ✓ | |||||||
| C | 5 | p53 | ✓ | ✓ | ||||
| Mre11 | ✓ | ✓ | ||||||
| TOPBP1 | ✓ | ✓ | ||||||
| DNA lig IV | ✓ | |||||||
| D | 9 | p53 | ✓ | ✓ | ||||
| Mre11 | ✓ | ✓ | ||||||
| TOPBP1 | ✓ | ✓ | ||||||
| DNA lig IV | ✓ | |||||||
| E | 4 | p53 | ✓ | ✓ | ||||
| Mre11 | ✓ | ✓ | ||||||
| TOPBP1 | ✓ | ✓ | ||||||
| DNA lig IV | ✓ |
Upreg, upregulated expression; VRCs, viral replication centers.
Among the properties of adenoviruses that have been investigated in detail recently is their ability to disrupt various cellular DNA damage response pathways (51). These studies derive from original investigations that showed that, following infection with a mutant adenovirus lacking the E4 region, viral genomes are joined end to end to form high-molecular-weight concatemers that cannot be packaged into virus capsids (50). This phenomenon probably can be attributed to the cell's recognition of viral DNA as cellular DNA that has undergone double-strand breaks (DSBs), although it is possible that it represents an antiviral defense mechanism. Additionally, the cell can inhibit viral replication by its DSB response (51). It is now clear that adenoviruses have evolved a series of mechanisms that can disable cellular DNA repair pathways. A major target appears to be the MRN complex (comprised of Mre11, Rad50, and NBS1) (9, 45). In uninfected cells this complex is involved in the initial recognition of DSBs and binds to ATM, where it is required for the full activation of the kinase; however, it is not essential for residual activity following DNA damage (28, 33). In addition, components of the MRN complex play a role in the cellular response to UV, which is taken as an initiator of single-strand breaks and stalled replication forks (30, 31). Thus, the MRN complex also may be involved in ATR activation and checkpoint signaling.
The MRN complex is inactivated by two mechanisms in the Ad5-infected cell. Through the action of the E1B55K/E4orf6 complex MRN components are targeted for proteasomal degradation (45). It is likely that the Mre11 subunit is the direct target for E1B55K/E4orf6-dependent degradation, because ATLD cells that express no detectable Mre11 but still have low levels of Rad50 and NBS1 show no further loss of these proteins after Ad5 infection (40). A second mechanism employed by Ad5 is the mislocalization of the MRN complex by E4orf3 to promyelocytic leukemia protein (PML)-containing “nuclear tracks” (1, 17). In the absence of Ad5E4, MRN is recruited to viral replication centers (VRCs). Mislocalization of MRN by Ad5 E4orf3 has been shown to prevent both ATR signaling and the formation of viral genome concatemers (10, 45).
Other proteins that play a significant role in the cellular DNA damage response also are degraded during Ad5 infection. Thus, it has long been established that p53 is targeted to the proteasome after infection by the actions of E1B55K and E4orf6 (35, 43). These viral proteins associate with a cullin-based RING E3 ubiquitin ligase (CRL) comprising Cul5, RBX1, and elongins B and C (21, 36). Ad5E1B55K has been proposed to act as a substrate receptor, binding p53, while E4orf6 is an adaptor molecule binding to E1B55K/p53 and the CRL (5, 7). The loss of p53 may be important for the viral life cycle by preventing the activation of the G1/S cell cycle checkpoint, and perhaps for the inhibition of its ability to induce apoptosis. The degradation of p53 and the MRN complex is accompanied, during Ad5 and Ad12 infection, by the proteolysis of DNA ligase IV (2). This is also dependent on E1B55K/E4orf6 and is responsible for the adenovirus-mediated inhibition of nonhomologous end joining (NHEJ) (2, 45).
Previous studies of DNA damage repair protein degradation have concentrated primarily on the action of Ad5 (2, 36, 45). It has been established that Ad12 and Ad4 also cause the degradation of p53 and Mre11 in a manner similar to that of Ad5, and this is dependent on the E4orf6 and E1B55K proteins (4, 46). However, it has been proposed that the activity of Ad5E4orf3 differs from that of Ad4E4orf3 and Ad12E4orf3, with the former protein having the ability to mislocalize the MRN complex, whereas the latter two proteins do not alter its localization (10). Further, we recently have shown that there are other marked differences between the impacts of Ad5 and Ad12 on the proteins regulating the cellular DNA damage response (4, 6). Ad12 causes the rapid degradation of TOPBP1 in the early stages of infection, whereas Ad5 does not. This has the effect of inhibiting the ATR activation of Chk1 during Ad12 infection; in contrast, the inhibition of ATR by Ad5 has been attributed to the relocalization of MRN (10). In addition, it has been shown that Ad12 recruits a Cul2-based CRL to degrade TOPBP1 and p53, whereas Ad5 degrades p53 by recruiting Cul5 (6).
In view of these observations, we have undertaken a widespread screen of how viruses from groups A to E interact with and affect the cellular DNA damage response pathways. We have demonstrated that the loss of MRN and p53 are not universally associated with all adenoviral infections, with Ad4, Ad5, and Ad12 causing degradation, whereas Ad3, Ad7, Ad9, and Ad11 do not. Instead, we show that Ad3, Ad7, and Ad11 induce a marked accumulation of p53, although the further analysis of the protein has suggested that it is unlikely to be transcriptionally active. Furthermore, we demonstrate that TOPBP1 is degraded only by Ad12, possibly indicating a unique cellular activity of group A oncogenic viruses, whereas DNA ligase IV probably is degraded by all viruses examined. Lastly, we show that Ad4, like Ad5, recruits a Cul5-based CRL to degrade p53. Taken together, these observations indicate that there is significant diversity in the mechanisms utilized by different adenoviral serotypes to inactivate host cell stress response pathways.
MATERIALS AND METHODS
Cell lines and viruses.
HeLa, A549, and H1299 cells were grown in Dulbecco's modified Eagle's medium supplemented with 8% fetal calf serum. Viruses used in this study are summarized in Supplemental Table 1 at http://www.cancersciences.bham.ac.uk/research/supplementarydata.shtml. Ad3, Ad4, Ad5, Ad7, Ad11, and Ad12 either were obtained from the ATCC or were a gift from Joe Mymryk. Ad9 was a gift from Ron Javier. The construction of the Ad5E4orf3/orf6-negative mutant, H5pm4155, is described in the supplementary data. Cells were infected with viruses at a multiplicity of infection (MOI) of 20, such that viral structural proteins were visible at 72 h following wild-type (WT) infection. For most time courses of infection, cells were harvested after approximately 24, 48, 72, and 96 h; however, for some assays additional time points were included.
RNA interference (RNAi).
Control short interfering RNA (siRNA; LacZ) was purchased from Ambion, and specific siRNAs targeting Cul2 and Cul5 (SMARTpool siRNA) were purchased from Dharmacon. HeLa cells (4 × 105) were plated out, left for 24 h, and transfected with siRNA using Oligofectamine transfection reagent (Invitrogen). Cells were split the following day and infected with the appropriate viruses the day after. Cells were harvested for Western blotting as indicated.
p53 reporter assays.
The effect of infection on p53 transcriptional activity was determined using a dual-luciferase reporter assay system (Promega). H1299 cells were grown in 6-cm dishes and transfected with p53 cDNA and a reporter construct containing p53 binding sites upstream of the luciferase gene, together with the Renilla luciferase gene as a transfection control. Transfection was performed using Lipofectamine LTX according to the manufacturer's instructions (Invitrogen). Cells were infected after 24 h with Ad3 and Ad7, harvested after a further 24 h, and lysed using passive lysis buffer (Promega). The luciferase activity was measured using a luminometer according to the manufacturer's instructions (Promega).
Semiquantitative reverse transcriptase PCR (RT-PCR).
Total RNA was extracted from cell lysates using the RNeasy mini kit (Qiagen). Total RNA (1 μg) was converted into single-strand cDNA using the reverse transcriptase system (Promega). Semiquantitative PCR was performed using the following primers: p21 forward, 5′-CGACTGTGATGCGCTAATGG-3′; reverse, 5′-CCGTTTTCGACCCTGAGA-3′; gapdh forward, 5′-ACCCCTTCATTGACCTCA; reverse, 5′-CAGCGCCAGTAGAGGCAG-3.
Western blotting and antibodies.
SDS-PAGE and Western blotting were carried out as described previously (4, 6). Primary antibodies were purchased from Abcam (Cul2), AbD Serotec (DNA ligase IV), Bethyl Laboratories (Cul5, KAP1, pKAP1-S824, and TOPBP1), Cell Signaling Technology (pChk1-S345), Genetex (Mre11), Merck (RPA32), Santa Cruz Biotechnology (Chk1, PML), and Sigma-Aldrich (β-actin). Rabbit polyclonal DNA ligase IV antibody was a generous gift from Stephen Jackson. Mouse monoclonal antibodies recognizing MDM2 (3G5 and 4B2) and Ad5E1B55K (2A6) were kind gifts from Arnold Levine, and p53 (DO-1) was a gift from David Lane. Ad12E1B55K was detected using mouse monoclonal antibody XPH9 (29).
PFGE.
HeLa cells were infected with viruses at an MOI of 30 and left for 48 h. Cells were washed and harvested. A single-cell suspension was set in an agarose plug that was, in turn, suspended in proteinase K (1 mg/ml) in 0.5 M EDTA, 10 mM Tris, pH 9.5, 1% N-lauroyl sarcosine for 48 h at 50°C essentially as described previously (19). Agarose plugs were subjected to pulsed-field gel electrophoresis (PFGE) for 22 h in a 1% agarose gel. DNA was visualized by ethidium bromide staining.
Immunofluorescence microscopy.
Cells were grown on 12-well multispot glass microscope slides at a density of 4 × 104 cells/well. Cells were infected with the different virus serotypes and left for the appropriate times. Slides were gently washed twice in phosphate-buffered saline (PBS), prepared by treatment in ice-cold preextraction buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 20 mM NaCl, 0.3 M sucrose, 3 mM MgCl2, 0.5% Triton X-100] for 5 min, and fixed in 4% paraformaldehyde in PBS for 10 min. Primary antibodies were diluted with 8% fetal calf serum (FCS) in PBS and incubated with the cells for 1 h at room temperature. Slides then were washed and incubated for a further hour with fluorescence Alexa Fluor secondary antibodies (Invitrogen) diluted with 8% FCS in PBS at room temperature in the dark. Finally, slides were washed and mounted in Vectashield mounting medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole (DAPI) and protected with glass coverslips. Cells were viewed and images acquired using an LSM 510 Meta confocal laser-scanning microscope (Carl Zeiss) and processed with the associated software.
RESULTS
In the past few years it has become apparent that Ad5 and Ad12 disable various aspects of the cellular DNA damage response pathways by initiating the rapid degradation of various key components, such as p53, members of the MRN complex, and DNA ligase IV (2, 36, 45). However, it has never been clear to what extent other virus serotypes adopt a similar stratagem, particularly as we have recently demonstrated that TOPBP1 is degraded during Ad12 but not Ad5 infection (6). In this study, we investigate the fate of a series of DNA damage proteins following infection with different adenovirus serotypes (see Supplemental Table 1 at http://www.cancersciences.bham.ac.uk/research/supplementarydata.shtml).
Expression of p53, Mre11, TOPBP1, and DNA ligase IV following viral infection.
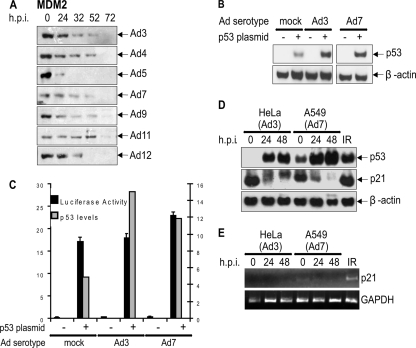
HeLa cells were infected at an MOI of 20, which was sufficient to produce viral structural proteins at 72 h (see Supplemental Fig. 1 at http://www.cancersciences.bham.ac.uk/research/supplementarydata.shtml), harvested at various times after infection, and processed for Western blotting. It can be seen from Fig. 1 that Mre11 and DNA ligase IV were degraded following infection with Ad5 (group C), as has been reported previously (2, 45). Similarly, infection with Ad12 (group A) and Ad4 (group E) also led to rapid protein degradation. However, Ad3, 7 (both group B1), Ad9 (group D), and Ad11 (group B2) had little effect on Mre11 levels, even at late times after infection (Fig. 1A). DNA ligase IV was degraded following infection with all virus serotypes (Fig. 1B). It is interesting, however, that the loss of DNA ligase IV tended to occur later and more gradually than was the case for Mre11. In view of the lack of antibodies against most of the viruses used in this study, confirmation that infection proceeded at a similar rate and to a similar extent was provided by the detection of viral structural proteins (see Supplemental Fig. 1A to G at http://www.cancersciences.bham.ac.uk/research/supplementarydata.shtml). (Viral replication centers also were observed in infected cells at comparable times [see Fig. 4 to 6 and see below]). However, it should be noted that there could have been differences in the rate of expression of the early region proteins for the different viruses, perhaps contributing, to a limited extent, to some of the differences seen in rates of protein degradation or phosphorylation (Fig. 1; also see Fig. 7 and 8).
FIG. 1.
Expression of Mre11, DNA ligase IV, p53, and TOPBP1 following viral infection. HeLa cells were infected with Ad3, Ad4, Ad5, Ad7, Ad9, Ad11, and Ad12. Cells were harvested at the times shown and subjected to Western blotting for Mre11 (A), DNA ligase IV (B), p53 (C), and TOPBP1 (D).
The initial observations that specific proteins are targeted for degradation during adenovirus infection were based on studies of p53 (36, 43). It can be seen in Fig. 1C that p53 was degraded during infection with Ad5, Ad4, and Ad12, as has been reported previously (36, 46). It also is apparent that there is an initial increase in protein expression after infection before degradation. However, following infection with Ad3, Ad7, Ad9, and Ad11, the expression of p53 increased dramatically, even at very late times, with no sign of protein degradation. Comparable results were obtained when the same viruses were used to infect A549 cells (data not shown), indicating that the effects seen are not dependent on an idiosyncrasy of the host cell.
We recently reported that a further substrate for Ad-initiated proteolysis is TOPBP1, which is degraded during Ad12 but not Ad5 infection (6). It can be seen (Fig. 1D) that, of the viruses examined, only Ad12 caused TOPBP1 degradation, indicating appreciable specificity for this approach to the inhibition of ATR activity by this group A virus. (The confirmation of the equal loading of samples in Fig. 1 is provided by β-actin blots shown in Supplemental Fig. 2 at http://www.cancersciences.bham.ac.uk/research/supplementarydata.shtml).
FIG. 2.
Effect of ablation of Cul2 and Cul5 expression on protein degradation during viral infection. HeLa cells were treated with nonsilencing siRNAs (non-sil) or siRNAs against Cul2 or Cul5. Cells then were infected with Ad4 (A), Ad5 (B), and Ad12 (C). Cells were harvested at the times shown and Western blotted with the indicated antibodies.
Requirements for CRLs in the degradation by adenoviruses.
The cellular E3 ubiquitin ligase, which degrades p53 during Ad5 infection, comprises Cul5, Rbx1, and elongins B/C in conjunction with Ad5E1B55K and Ad5E4orf6 (21, 36). More recently, we have shown that Cul2, rather than Cul5, is required for the degradation of both p53 and TOPBP1 during Ad12 infection, suggesting that different adenoviral serotypes utilize separate CRLs to facilitate the destruction of the desired target protein (6). There is some evidence implicating Cul5-based CRLs in the degradation of Mre11 following Ad5 infection (39, 52). To ascertain whether E3 ligase utilization varied with other adenoviral serotypes, we examined which CRL is required for the degradation of Mre11, p53, and DNA ligase IV following Ad5, Ad12, and Ad4 infection (Fig. 2). As previously reported (6), the loss of Cul5 stabilized p53 following Ad5 infection, whereas the siRNA-mediated depletion of Cul2 blocked the degradation of p53 after infection with Ad12 (Fig. 2B and C). Interestingly, the degradation of p53 by Ad4 was inhibited when the expression of Cul5 was reduced with siRNA, indicating that Ad4 utilizes mechanisms for protein degradation similar to those of Ad5 rather than those of Ad12 (Fig. 2A). Surprisingly, however, neither a reduction of Cul2 nor Cul5 expression had any significant effect on the degradation of Mre11 or DNA ligase IV following infection with Ad4, Ad5, or Ad12 in the experiments shown in Fig. 2A to C. These observations suggest that, despite current assumptions that Cul5 is required for the degradation of Mre11 and DNA ligase IV, none of the adenoviral serotypes tested primarily use a Cul5-dependent CRL to degrade these two repair proteins, implicating an as-yet unidentified cellular E3 ligase complex in the process.
Transcriptional activity of p53.
The rapid degradation of p53 induced during Ad4, Ad5, and Ad12 infection prevents it from activating the transcription of its target genes. However, as shown above, p53 is stabilized during infection with other serotypes. To evaluate the effect of the increased expression of p53 seen following infection with Ad3 and Ad7, three approaches were adopted. First, it can be seen that infection with all of the viruses resulted in the rapid reduction in levels of MDM2, a downstream target of p53 (Fig. 3A), suggesting that the high levels of overexpressed p53 are transcriptionally inactive. Significantly, MDM2 and DNA ligase IV were the only proteins that showed a decrease in expression following infection with all viruses used in this study. Second, the direct measurement of p53 transcriptional activity was undertaken. Thus, a luciferase reporter downstream of multiple copies of a p53 consensus binding site was transfected, together with a plasmid expressing p53, into H1299 cells. After 24 h the cells were infected with Ad3 and Ad7 (Fig. 3B and C). Following a further 24 h, luciferase activity and levels of p53 expression were determined. Although the level of p53 more than doubled following infection with Ad3 and Ad7, there was only a marginal increase in the transcription of the reporter (Fig. 3C), suggesting that the overexpressed p53 was transcriptionally inactive. In a third experiment, the transcriptional inactivation of p53 was confirmed by semiquantitative RT-PCR for p21, a further p53-regulated gene (Fig. 3D and E). Following the infection of HeLa and A549 cells with Ad3 and Ad7, there was a marked increase in the level of p53 expression but no increase in p21 protein level (Fig. 3D). RT-PCR showed that the reduction in p21 protein expression was due to reduced p21 mRNA (Fig. 3E), presumably as a result of the transcriptional inactivation of p53. The treatment of A549 cells with ionizing radiation as a positive control gave a moderate increase in p53 but an appreciably greater increase in p21 expression (Fig. 3D and E).
FIG. 3.
Transcriptional activity of p53 after viral infection. (A) HeLa cells were infected with Ad3, Ad4, Ad5, Ad7, Ad9, Ad11, and Ad12, harvested at the times shown, and Western blotted for MDM2. H1299 cells were transfected with a luciferase reporter construct (see Materials and Methods) and cDNA expressing p53. They then were infected with Ad3 or Ad7 and harvested after 24 h, and cells were Western blotted for p53 expression (B) and luciferase activity was determined (C). Relative p53 levels are shown in grey columns, and relative luciferase activity is shown in black columns (D and E). HeLa and A549 cells were infected with Ad3 and Ad7 as indicated. Samples were Western blotted for p53, p21, and β-actin (D) and subjected to RT-PCR for p21 (E). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is included as a loading control. The right lane in D and E shows A549 cells treated with ionizing radiation (3 Gy) after 2 h.
Localization of p53, Mre11, and TOPBP1 during adenovirus infection.
Following adenovirus infection, a number of centers of viral replication and transcription are established within the host cell nucleus; these can be visualized by fluorescence microscopy using an antibody directed to RPA32, which is relocalized into several large subnuclear inclusion bodies (46). This is particularly useful, since high-quality antibodies against proteins of most viral serotypes are not available. However, at least in the case of Ad5, these RPA32-positive replication centers have been verified by the presence of a number of viral proteins colocalizing to these sites; in particular, the single-stranded DNA binding protein (DBP), terminal protein (TP), and E1B55K protein can be seen at viral replication centers (32, 49). In addition to RPA32, a number of other cellular proteins known to be involved in coordinating the DNA damage response also have been reported to relocalize to viral replication centers during Ad5 WT infection; these include ATR, ATRIP, E1B-AP5, Rad9, and Rad17 (4, 9). Interestingly, it has been shown that, as well as the generation of viral replication centers, infection with adenoviruses results in the disruption of PML bodies and the relocalization of PML into nuclear track-like structures, which is dependent on the Ad5E4orf3 protein (11, 16). However, very little is known about whether the relocalization of cellular repair proteins to viral replication centers or the disruption of PML bodies has any impact on the viral life cycle or the function of these proteins.
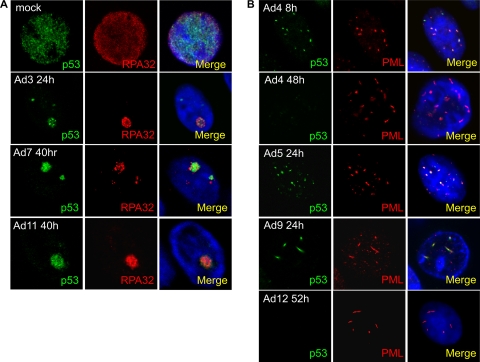
Given that we observed differences between adenoviral serotypes with regard to whether or not they specifically targeted DNA repair/checkpoint proteins for degradation, we examined the localization of Mre11, p53, and TOPBP1 during infection with the panel of adenoviruses to determine whether those serotypes that did not degrade Mre11 and p53 induced their mislocalization as a potential mechanism to inhibit their function (Fig. 4, 5 and 6; summarized in Table 1). As has been reported previously (46), we observed that Ad5 and Ad12 relocalize Mre11 to PML-containing nuclear tracks and RPA32-positive VRCs, respectively (Fig. 4A and B). However, contrary to a previous study, we found that in Ad4-infected cells Mre11 is localized to PML tracks rather than VRCs (Fig. 4C). For Ad3, Ad7, and Ad11, Mre11 localized to viral replication centers costained with RPA32 (Fig. 4B). In the case of Ad9, although Mre11 was not degraded, it still colocalized with PML in nuclear track structures (Fig. 4A). p53 colocalized with RPA32 at viral replication centers following infection with Ad3, Ad7, and Ad11 (Fig. 5A). In Ad9-infected cells, p53 colocalized with PML, although there was little or no degradation apparent by Western blotting (Fig. 5B). Similarly, p53 localized to tracks following Ad4, Ad5, and Ad12 infection prior to degradation. When infected cells were stained for TOPBP1 in the case of Ad3, Ad4, Ad5, Ad7, Ad9, and Ad11, it located to VRCs with RPA32 (Fig. 6A and B). At early times of infection by Ad12, limited TOPBP1 was present in the replication centers, but at later times no protein could be seen due to its degradation (Fig. 6B) (6). Unfortunately, it was not possible to determine the subcellular localization of DNA ligase IV due to the lack of an antibody effective in immunofluorescence microscopy.
FIG. 4.
Localization of Mre11 following adenovirus infection. HeLa cells were infected with Ad5 and Ad9 (A), Ad3, Ad7, Ad11, and Ad12 (B), and Ad4 (C) for the times shown. Cells were stained for Mre11 (green), PML (red), and RPA32 (red) as indicated and viewed by confocal microscopy as described in the text. Merged images are shown in the right column of images together with DAPI-stained DNA.
FIG. 5.
Localization of p53 following adenovirus infection. HeLa cells were infected with Ad3, Ad7, and Ad11 (A) and Ad4, Ad5, Ad9, and Ad12 (B) for the times shown. Cells were stained for p53 (green), RPA32 (red), or PML (red) and viewed by confocal microscopy as described in the text. Merged images are shown in the right column of images together with DAPI-stained DNA.
FIG. 6.
Localization of TOPBP1 following adenovirus infection. HeLa cells were infected with Ad3, Ad4, Ad5, and Ad7 (A) and Ad9, Ad11, and Ad12 (B) for the times shown. Cells were stained for RPA32 (green) and TOPBP1 (red) and viewed by confocal microscopy as described in the text. Merged images are shown in the right column together with DAPI-stained DNA.
Phosphorylation of DNA damage response proteins following viral infection.
It is now clear that adenoviruses activate various aspects of the cellular DNA damage response. Following infection with certain mutant viruses, cells respond by, for example, activating the ATM and ATR signaling pathways and producing concatemers of viral DNA (9, 45, 50). The activation of the ATM and ATR kinases is seen as the phosphorylation of appropriate downstream substrates. Proteins expressed during infection with WT Ad5 and Ad12 inhibit concatemer formation and full ATR activation. To ascertain whether all adenoviral serotypes were capable of inhibiting the cellular DNA damage response, following viral infection, phosphospecific antibodies were used to monitor ATM and ATR activation using the phosphorylation of KAP1 as a marker of ATM activity (55) and Chk1 as a marker of ATR activity (14). It can be seen in Fig. 7A and B that the phosphorylation of Chk1 is strongly inhibited during infection with certain serotypes, most obviously with Ad5, Ad9, and Ad12, such that no phosphorylation was seen up to 72 h, when viral structural proteins were being expressed (see Supplemental Fig. 1 at http://www.cancersciences.bham.ac.uk/research/supplementarydata.shtml). However, in the case of Ad3, Ad4, Ad7, and Ad11 infection, Chk1 was phosphorylated to a greater (Ad3, Ad7, and Ad11) or lesser (Ad4) extent at earlier times. The kinetics of phosphorylation varied between serotypes, with Ad11 infection causing the marked phosphorylation of Chk1 (although not as strongly as in cells treated with HU) and Ad3 causing the weak transient phosphorylation of Chk1. Similarly, the kinetics of KAP1 phosphorylation varied appreciably between the different serotypes, although all viruses examined caused some activation of ATM (Fig. 8A and B). Thus, Ad5 caused marked phosphorylation, which was visible at 8 h postinfection and was still apparent at 72 h. On the other hand, phosphorylation in the presence of Ad12 was transient, being apparent at 24 h but not at late times. However, it is clear that infection with all of the viruses used in this study resulted in the activation of the ATM signaling pathways, with no obvious correlation with Mre11 degradation.
FIG. 7.
Phosphorylation of Chk1 following adenovirus infection. HeLa cells were infected with Ad3, Ad4, Ad5, Ad7, Ad9, Ad11, and Ad12, harvested at the times shown, and Western blotted for phospho-Chk1 (Ser345) (A) and Chk1 (B). The final track in the Ad12 phospho-Chk1 blots shows HeLa cell extracts treated with hydroxyurea (HU) as a positive control.
FIG. 8.
Phosphorylation of KAP1 during adenoviral infection. HeLa cells were infected with Ad3, Ad4, Ad5, Ad7, Ad9, Ad11, and Ad12, harvested at the times shown, and Western blotted for phospho-KAP1 (Ser824) (A) and KAP1 (B).
Viral DNA concatemers.
As we could detect only limited degradation of the components of the DNA damage repair pathways during infection with Ad3, Ad7, Ad9, and Ad11, we considered the possibility that these viruses had evolved such that they might be able to tolerate the concatemerization of a proportion of their genomes and still replicate. To test this hypothesis, HeLa cells were infected for 48 h and then harvested. The composition of the viral DNA was analyzed by PFGE. Figure 9 shows the presence of viral DNA concatemers following infection with an Ad5E4orf3/orf6-deleted virus (H5pm4155) as reported previously (50). However, for Ad3, Ad7, Ad9, and Ad11, no concatemers were visible, only the monomeric viral genomes at about 35 kb. We therefore have concluded that for these viruses, the degradation of DNA ligase IV may be sufficient to incapacitate the cellular DNA damage repair pathways. It is, of course, possible that the viruses also have evolved novel mechanisms for the inhibition of the pathways to prevent the illegitimate repair of linear viral genomes during replication.
FIG. 9.
Absence of concatemer formation in viral DNA. HeLa cells were infected with Ad3, Ad7, Ad11, and Ad12 WT viruses along with the Ad5 mutants dl355 and H5pm4155. Once harvested, cells were set into agarose plugs before being subjected to prolonged digestion with proteinase K and subsequent PFGE. DNA was visualized through ethidium bromide staining. A molecular size ladder is shown in the left lane.
DISCUSSION
Until now, the examination of the relationship of adenoviruses to the cellular DNA damage repair pathways had, like the great majority of other adenovirus studies, focused on Ad5. In a limited number of cases, Ad12 and Ad4 also were investigated (4, 6, 27, 46). These three virus serotypes gave broadly similar results, causing the degradation of the DNA damage response proteins p53 and Mre11. However, even among this limited group, differences in the ways in which they deregulate the host cell DNA damage repair mechanism have been reported. Thus, only Ad12, but no other adenovirus so far examined, causes the degradation of TOPBP1 (Fig. 1D) (6). A further difference centers on the observation that Ad5E4orf3 mislocalizes the MRN complex during viral infection, whereas E4orf3 proteins from other virus serotypes, such as Ad12, do not. This is likely to contribute to the inhibition of ATR signaling by the Ad5 protein but not the Ad12 protein; differences in their properties have been linked to an amino acid sequence difference at residue 104, such that there is an isoleucine in E4orf3 of group C viruses and an arginine in group B, D, and E adenoviruses (10).
We now have extended these investigations to encompass members of adenovirus group B1 (Ad3 and Ad7), group B2 (Ad11), and group D (Ad9), as well as a more detailed examination of group A (Ad12) and group E (Ad4) viruses (summarized in Table 1). In this study, we have identified some marked differences from Ad5. Significantly, p53 is not degraded following infection with Ad3, Ad7, Ad9, and Ad11 but shows a pronounced increase in expression, presumably as a result of the action of AdE1A. This is similar to the effect seen after infection with Ad5E1B55K− and Ad12E1B55K− mutant viruses (20). Interestingly, a previous study has shown that an alanine substitution at R240 in Ad5E1B55K produces a protein that interacts poorly with p53 (41). Consequently, mutant Ad5 viruses expressing the R240A protein are unable to degrade p53. The sequence analysis of E1B55K from different serotypes reveals that this residue is conserved in Ad4, Ad5, and Ad12 proteins but not in Ad3, Ad7, Ad9, or Ad11 E1B55K (5). This correlates perfectly with our new study, in which we show that the former viruses all degrade p53 during infection, whereas the latter do not. It appears that in cells infected with Ad3, Ad7, and Ad11, p53 is sequestered to VRCs and is transcriptionally inactive (Fig. 3 and 5), probably as a result of the action of the AdE1A and/or E1B55K proteins (24, 43, 53, 54). There also is no evidence that p53 induces apoptosis in these infected cells. It has been known for some time that there is a transient increase in the level of p53 following Ad5 and Ad12 infection (Fig. 1), and it is possible that this protein has a similar, although obviously more limited, role that is seen during infection with group B and D adenoviruses. It has been suggested that p53 contributes to viral replication and increases late protein expression during Ad5 infection (38). Although the mechanism responsible is unclear, it appears that the combination of AdE1A and p53 enhances major late promoter (MLP) function. It is possible that a similar mechanism operates in the group B and D viruses, with the overexpressed p53 being required for viral replication.
Two other proteins involved in the cellular damage response, Mre11 and DNA ligase IV, were degraded in Ad4- and Ad12-infected cells, as is the case following Ad5 infection (2, 45). As has been noted previously for Ad5-infected cells, Mre11 colocalized to nuclear tracks before being degraded (10). However, at early times during Ad12 infection, Mre11 can be seen localizing with RPA32 in VRCs (Fig. 4). During Ad3, Ad7, and Ad11 infection, Mre11 also was located in VRCs (Fig. 4). Interestingly, during Ad4 infection, Mre11 can be seen colocalized with PML in nuclear tracks (Fig. 4C). It has been reported previously that NBS1 (and presumably Mre11) colocalizes to VRCs during both Ad4 and Ad12 infection (46). Differences between these observations and those shown here may reflect different multiplicities of infection and the choice of cell lines used, or perhaps they indicate a dissociation of the MRN complex from the different components localizing to alternate sites within the infected cell (17). It currently is not clear why Mre11 is recruited to VRCs or PML-containing nuclear track structures during adenoviral infection, as there does not appear to be an obvious correlation between its redistribution and whether it is targeted for degradation. Unlike p53 and Mre11, DNA ligase IV is degraded during infection with all of the viruses examined, probably preventing the illegitimate repair of viral genomes by NHEJ. Since no suitable antibody is available to detect the localization of endogenous DNA ligase IV by immunofluorescence microscopy, it is not possible to determine if it is mislocalized by these viruses to PML-containing nuclear tracks. However, previous work has shown that the presence of E1B55K and DNA ligase IV degradation are not absolutely necessary for Ad5 to inhibit NHEJ, as, in the absence of E1B55K, E4orf6 still can dissociate DNA ligase IV and XRCC4 (25) and can bind and inactivate the DNA-PK holoenzyme to prevent viral genome concatemerization (8).
DNA ligase IV was the only DNA damage response protein seen to be reduced following infection with the group B and D viruses investigated here. Since these viruses are able to promote VRC formation and produce late viral proteins, it seems likely that this is sufficient to inhibit the cellular DNA damage response unless as-yet unidentified mechanisms also are involved. It is possible that the viruses degrade or mislocalize other components of the cellular damage response pathways, as Ad12 (alone of the viruses examined here) degrades TOPBP1 (Fig. 1D) (6). To confirm that these viruses were not simply able to tolerate a degree of concatemer formation, PFGE was carried out. While multimers of viral DNA were evident following infection with the E4orf3/orf6 mutant virus H5pm4155, only the monomeric genome could be seen with Ad3, Ad7, Ad9, Ad11, and Ad12 (Fig. 9), which is consistent with these viruses being able to inactivate the host cell DNA repair machinery. The ability of the group B and D viruses to degrade other DNA damage response proteins is given indirect support by the fact that the BC boxes in E4orf6 proteins of these viruses are conserved and are similar to those in the Ad5 and Ad12 proteins. These amino acid motifs are considered to be required for the assembly of a functional E3 ligase (7, 12). It is interesting that infection with the Ad5E4orf6-negative virus (dl355) caused a limited number of concatemers (Fig. 9).
The degradation of p53 by Ad5 involves the recruitment of an E3 ubiquitin ligase complex containing Cul5, RBX1, and elongins B and C by the viral E1B55K and E4orf6 proteins acting in tandem (7, 21, 36) and subsequent proteolysis by the proteasome. As with Ad5, the ablation of Cul5 expression inhibited the ability of Ad4 to cause the degradation of p53 (Fig. 2A). With Ad12, a Cul2-based E3 ubiquitin ligase involving E1B55K and E4orf6 is necessary for p53 degradation (Fig. 2C), although this is not the case for TOPBP1 proteolysis, where Ad12E4orf6 alone is sufficient to cause protein degradation by the proteasome (6). Similar differences also have been shown very recently between Ad5 and Ad12 in relation to their cullin specificity by another group (13). It has been reported that the degradation of DNA ligase IV by Ad5 requires the activity of Cul5 (2). However, it appears from the data presented here in Fig. 2B that this may not be the case. Reasons for these differences remain unknown at present. Observations presented in Fig. 2 also indicate that the ablation of Cul5 or Cul2 expression did not affect Mre11 degradation during Ad4, Ad5, or Ad12 infection. Previous studies have suggested that Ad5 recruits a Cul5-based E3 ubiquitin ligase for the degradation of Mre11; however, it was apparent, even in those investigations, that the knockdown of Cul5 did not fully rescue Mre11 degradation (15, 39). It should, however, be borne in mind that the expression of a dominant-negative Cul5 protein has been shown to block Mre11 degradation (52). Based on the data in Fig. 2 and our previous observations, we suggest that a Cul5-based ubiquitin E3 ligase is involved in p53 degradation by Ad4 and Ad5 but is not the predominant CRL responsible for the degradation of Mre11 by any of the viruses examined here. Similarly, DNA ligase IV degradation appears not to involve cullin 2 or 5. Whether the same (as-yet unidentified) E3 ubiquitin ligase is required for Mre11 and DNA ligase IV degradation will have to await further investigation.
It is clear from our observations that the level of MDM2 is reduced following infection by all of the viruses (Fig. 3A). The regulation of MDM2 levels is complex. Its expression is dependent on transcriptionally active p53, such that the activation of p53 causes increased MDM2 expression, which, in turn, facilitates rapid p53 degradation through its activity as a RING finger E3 ligase (22, 23, 26, 34). MDM2 also is able to ubiquitylate itself, leading to its own proteolysis by the proteasome (18, 44). The ability of MDM2 to distinguish between self and target ubiquitylation appears to depend on Daxx, together with the deubiquitylating enzyme HAUSP (Usp7) (37, 47). The association of Daxx and HAUSP with MDM2 in a ternary complex ensures that autoubiquitylation by MDM2 is reduced, and its association with and degradation of p53 is enhanced (47). It has been shown that DNA damage increases MDM2 autoubiquitylation and p53 stabilization. During adenovirus infection it appears that p53 is transcriptionally inactive, such that the transcription of MDM2 is reduced (Fig. 3). Furthermore, very recent evidence indicates that Ad5 causes the degradation of Daxx (39). Therefore, it is possible that, following viral infection, Daxx is degraded, dissociating the Daxx/HAUSP/MDM2 complex and facilitating MDM2 autoubiquitylation and subsequent degradation. No changes in HAUSP levels were observed during Ad5 or Ad12 infection (data not shown). However, it should be noted that Daxx is not degraded by Ad3, Ad7, Ad9, and Ad11, although degradation occurs following Ad4 and Ad12 infection, as is the case with Ad5 (data not shown) (39). Obviously, a different series of events is likely to occur in Ad4, Ad5, and Ad12, when p53 is degraded after an initial increase in expression, from that seen in Ad3, Ad7, Ad9, and Ad11, where no p53 degradation occurs.
Although group B and D viruses (Ad3, Ad7, Ad9, and Ad11) do not degrade p53, Mre11, or TOPBP1, it is clear that they activate the cellular DNA damage response. We have demonstrated that ATM is activated following infection with Ad3, Ad4, Ad5, Ad7, Ad9, Ad11, and Ad12, although the kinetics of KAP1 phosphorylation vary somewhat between the viral serotypes. Similar activation by Ad5 and Ad12 has been reported previously (4, 9). It seems reasonable to suggest that all of the virus serotypes can cope with the activated double-strand break repair pathways, and this is not particularly deleterious to viral replication. However, it seems, at least for Ad5 and Ad12, that the partial inhibition of ATR signaling is advantageous. In the case of Ad12 this is achieved by the degradation of TOPBP1, although for the other viruses examined here TOPBP1 is recruited to VRCs, where its function remains unknown. During Ad5 infection the inhibition of ATR signaling derives from the degradation and/or mislocalization of Mre11 (9, 10). Similarly, during Ad9 infection the phosphorylation of Chk1 is inhibited, presumably because the MRN complex is localized to PML-containing nuclear tracks. During infection by Ad3, Ad4, Ad7, and Ad11, ATR appears to be activated, resulting in the phosphorylation of Chk1. Despite this, virus replication occurs (Fig. 9). In the case of Ad4, the degradation of Mre11 might be expected to inhibit ATR signaling in a manner comparable to that of Ad5, but this does not seem to be the case. Ad3, Ad7, and Ad11 promote the degradation of DNA ligase IV but also may have developed novel mechanisms for inhibiting other cellular DNA damage responses to viral DNA.
In summary, it has been shown that different adenovirus serotypes interact with the cellular DNA damage response pathways in quite distinct ways. Representatives of groups B1, B2, and D do not cause the degradation of p53, TOPBP1, or Mre11, although they, like group A, C, and E viruses, facilitate the degradation of DNA ligase IV and a rapid and complete reduction in the level of MDM2. Furthermore, marked differences are apparent in the ways in which cellular proteins are recruited to VRCs by the different virus serotypes and the extent to which ATM and ATR kinase activity is activated or inhibited. Future investigation into why the viruses inactivate the host cell stress response pathways in different ways will provide further insight into adenovirus replication and may aid our understanding of the regulation of the DNA damage response.
Acknowledgments
We are most grateful to Phil Branton and Paola Blanchette for helpful discussions and acknowledge their forthcoming information in this area of adenovirus research. We also are grateful to Kelly Townsend for invaluable advice. We thank Stephen Jackson, Arnold Levine, David Lane, Ron Javier, and Joe Mymryk for the generous gift of antibodies and viruses.
We thank the University of Birmingham College of Medicine and Dentistry and Cancer Research UK for funding.
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, T. D. Carson, V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benko, M., B. Harrach, and W. C. Russell. 2000. Adenoviridae, p. 227-235. In M. H. Van Regenmortel, C. M. Faquert, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Malinoff, M. A. Mayo, D. J. McGeooh, C. R. Pringle, and R. B. Wickner (ed.). Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, New York, NY.
- 4.Blackford, A. N., et al. 2008. A role for E1B-AP5 in ATR signalling pathways during adenovirus infection. J. Virol. 82:7640-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackford, A. N., and R. J. Grand. 2009. Adenovirus E1B-55 kilodalton protein: multiple roles in viral infection and cell transformation. J. Virol. 83:4000-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackford, A. N., et al. 2010. Adenovirus 12E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. U. S. A. 107:12251-12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette, P., et al. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34K and E411k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 9.Carson, C. T., et al. 2003. The MreII complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson, C. T., et al. 2009. Mislocalization of the MRN complex prevents ATR signalling during adenovirus infection. EMBO J. 28:652-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho, T., et al. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, C. Y., P. Blanchette, and P. E. Branton. 2007. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology 364:36-44. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, C. Y., T. Gilson, F. Dallaire, G. Ketner, P. E. Branton, and P. Blanchette. 2010. The E4orf6/E1B55K E3 ubiquitin ligase complexes of human-adenoviruses exhibit heterogeneity in composition and substrate specificity. J. Virol. doi: 10.1128/JVI.01890-10. [DOI] [PMC free article] [PubMed]
- 14.Cimprich, K. A., and D. Cortez. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Boil. 9:616-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dallaire, F., P. Blanchette, P. Groitl, T. Dobner, and P. E. Branton. 2009. Identification of integrin alpha 3 as a new substrate of the adenovirus E4orf6/E1B55 kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doucas, V., et al. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 17.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, S., J. P. Jenson, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RInG finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 19.Formstone, C. J., et al. 1993. The order and orientation of a cluster of metalloproteinase genes stromelysin 2, collagenase and stromelysin together withD115385 on chromosome 11q22-q23. Genomics 16:289-291. [DOI] [PubMed] [Google Scholar]
- 20.Grand, R. J., M. L. Grant, and P. H. Gallimore. 1994. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology 203:229-240. [DOI] [PubMed] [Google Scholar]
- 21.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteasome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 23.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 24.Hutton, F. G., A. S. Turnell, P. H. Gallimore, and R. J. Grand. 2000. Consequences of disruption of the interaction between p53 and the larger adenovirus early region 1B protein in adenovirus E1 transformed human cells. Oncogene 19:452-462. [DOI] [PubMed] [Google Scholar]
- 25.Jayaram, S., et al. 2008. E1B55K-independent dissociation of the DNA ligase IV/XRCC4 complex by E434K during adenovirus infection. Virology 382:163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 27.Lakdawala, S. S., R. A. Schwartz, K. Ferenchak, C. T. Carson, B. P. McSharry, G. W. Wilkinson, and M. D. Weitzman. 2008. Differential requirements of the C terminus of Nbsl in suppressing adenovirus DNA replication and promoting concatemer formation. J. Virol. 82:8362-8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavin, M. 2007. ATM and Mre11 complex combine to recognise and signal DNA double-strand breaks. Oncogene 26:7749-7758. [DOI] [PubMed] [Google Scholar]
- 29.Merrick, R. M., R. J. Grand, J. C. Brown, and P. H. Gallimore. 1991. The use of beta-galactoside fusion proteins encoding the early region 1 transforming proteins of adenovirus type 12 to examine the humeral response of tumour-bearing animals. J. Gen. Virol. 72:955-960. [DOI] [PubMed] [Google Scholar]
- 30.Myers, J. S., and D. Cortez. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 281:9346-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson, E., C. J. Nievera, A. Y. Lee, L. Chen, and X. Wu. 2007. The Mre11− Rad 50− Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad 3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J. Biol. Chem. 282:22939-22952. [DOI] [PubMed] [Google Scholar]
- 32.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrini, J. H., and T. H. Stracker. 2003. The cellular response to DNA double strand breaks: defining the sensors and mediators. Trends Cell Biol. 13:458-462. [DOI] [PubMed] [Google Scholar]
- 34.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 35.Querido, E., et al. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein 20 and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Querido, E., et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronai, Z. 2006. Balancing Mdm2-a Daxx-HAUSP matter. Nat. Cell Biol. 8:790-791. [DOI] [PubMed] [Google Scholar]
- 38.Royds, J. A., et al. 2006. p53 promotes adenoviral replication and increases late viral gene expression. Oncogene 25:1509-1520. [DOI] [PubMed] [Google Scholar]
- 39.Schreiner, S., et al. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz, R. A., et al. 2008. Distinct requirements of adenovirus E1b 55K protein for degradation of cellular substrates. J. Virol. 82:9043-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid-substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 75:4297-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shenk, T., and M. S. Horwitz. 2001. Adenoviridae: the viruses and their replication; adenoviruses, p. 2265-2326. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman. and S. E. Straw (ed.), Fields virology, 4th ed., Lippincott Williams and Wilkins, Philadelphia, PA.
- 43.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 44.Stommel, J. M., and G. M. Wahl. 2004. Accelerated MDM2 auto-degradation induced by DNA damage kinases is required for p53 activation. EMBO J. 23:1547-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11 Rad50 NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 46.Stracker, T. H., et al. 2005. Serotype-specific reorganisation of the Mre11 complex by adenoviral E4orf3 proteins J. Virol. 79:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, J., L. K. Qu, J. Zhang, W. Wang, J. S. Michaelson, Y. Y. Degenhardt, El-W. S. Deiry, and X. Yang. 2006. Critical role of Daxx in regulating Mdm2. Nat. Cell Biol. 8:855-862. [DOI] [PubMed] [Google Scholar]
- 48.Trentin, J. J., Y. Yabe, and G. Taylor. 1962. The quest for human cancer viruses. Science 137:835-841. [DOI] [PubMed] [Google Scholar]
- 49.Webster, A., I. R. Leith, J. Nicholson, J. Hounsell, and R. T. Hay. 1997. Role of preterminal protein processing in adenovirus replication. J. Virol. 71:6381-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. U. S. A. 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686-7696. [DOI] [PubMed] [Google Scholar]
- 52.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 54.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 55.Ziv, Y., et al. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8:870-876. [DOI] [PubMed] [Google Scholar]