Abstract
The CD4 binding site (CD4bs) on the HIV-1 envelope plays a major role in determining the capacity of R5 viruses to infect primary macrophages. Thus, envelope determinants within or proximal to the CD4bs have been shown to control the use of low CD4 levels on macrophages for infection. These residues affect the affinity for CD4 either directly or indirectly by altering the exposure of CD4 contact residues. Here, we describe a single amino acid determinant in the V1 loop that also modulates macrophage tropism. Thus, we identified an E153G substitution that conferred high levels of macrophage infectivity for several heterologous R5 envelopes, while the reciprocal G153E substitution abrogated infection. Shifts in macrophage tropism were associated with dramatic shifts in sensitivity to the V3 loop monoclonal antibody (MAb), 447-52D and soluble CD4, as well as more modest changes in sensitivity to the CD4bs MAb, b12. These observations are consistent with an altered conformation or exposure of the V3 loop that enables the envelope to use low CD4 levels for infection. The modest shifts in b12 sensitivity suggest that residue 153 impacts on the exposure of the CD4bs. However, the more intense shifts in sCD4 sensitivity suggest additional mechanisms that likely include an increased ability of the envelope to undergo conformational changes following binding to suboptimal levels of cell surface CD4. In summary, we show that a conserved determinant in the V1 loop modulates the V3 loop to prime low CD4 use and macrophage infection.
HIV-1 fusion and entry into cells is triggered by sequential interactions between the viral envelope glycoprotein with CD4 and a seven-transmembrane coreceptor. CCR5 is the main coreceptor for transmitted HIV-1 (5, 9), while CXCR4-using variants become detectable in up to ca. 50% of AIDS patients and are associated with a rapid decline in CD4+ T cells, leading to death (6). HIV-1 R5 viruses efficiently infect primary CD4+ memory T cells that express CCR5. However, it is now clear that R5 isolates and envelopes (amplified from patient tissues) confer widely divergent abilities to infect primary macrophages (16, 17, 36-38). Macrophage infectivity thus varies by at least a 1,000-fold among R5 envelopes. We and others have described envelopes derived from brain tissue that were highly macrophage-tropic (16, 17, 21, 25, 30, 36-38, 43). In contrast, many envelopes from immune tissue such as lymph nodes, infected macrophages very inefficiently (38). Macrophage infectivity correlated with the capacity to infect indicator cell lines via low levels of CD4 (17, 31, 36, 38), an observation that reflects the lower expression of CD4 on macrophages compared to T cells (2, 24, 33). Consistent with this observation, envelope determinants that control macrophage tropism have been mapped to residues within or proximal to the CD4 binding site (11, 13, 38). Thus, Dunfee et al. reported that an asparagine at residue 283 in the C2 part of the CD4bs was associated with brain envelopes and HIV-associated dementia (13). N283 also conferred increased levels of macrophage infectivity when introduced into R5 envelopes where residue 283 is usually T or I (11, 13). N283 was reported to confer an increased affinity of gp120 for CD4, probably because the asparagine may facilitate the formation of a hydrogen bond with Q40 on CD4 (13). Nevertheless, N283 only partially associates with macrophage infectivity, and we have identified further important envelope determinants that likely affect exposure of CD4 contact residues (11). Thus, determinants on the flanks of the CD4 binding loop were identified, which probably modulate exposure of the adjacent and conserved CD4 contact residues. The CD4 binding loop is likely to be the main part of the CD4bs that is exposed on the native envelope trimer and an early contact for CD4 (7). Of note, the identified CD4 binding loop flank residues also shifted sensitivity to the glycan-specific monoclonal antibody (MAb), 2G12, indicating that movement of proximal glycans could be one mechanism involved in the exposure of CD4 contact residues (12).
The CD4 binding site is also a major target for vaccine development aimed at eliciting neutralizing antibodies (32). It is therefore important to fully understand how this site is exposed or protected during viral evolution and how this affects tropism, neutralization sensitivity, and other envelope properties. Here, we describe closely related HIV-1 envelopes from a pediatric subject in late disease that differ profoundly in their capacity to infect primary macrophages. We identify a single amino acid determinant in the V1 loop that confers dramatic shifts in macrophage tropism, as well as affecting sensitivity to the V3 loop MAb, 447-52D, sCD4, and the CD4bs MAb, b12. These data are consistent with a model where the V1 determinant controls the conformation or exposure of the V3 loop, which in turn primes the envelope for low CD4 use and macrophage infection.
MATERIALS AND METHODS
Patient envelopes.
HIV-1 clade B envelopes amplified from pediatric subject P1114, as well as NA118 LN27, NA420 LN40, NA20 LN8, and JRCSF, have been described previously (21, 36, 38). The non-macrophage-tropic LN40 envelope was chimeric carrying the gp41 sequence of NA420 B33 to confer optimal infectivity (36, 38). The Indian clade C envelope, 25925-2, was obtained from the NIH AIDS Research and Reference reagent program (22). The clade D envelope, CMT19 S531, was amplified from a Cameroon semen sample, whereas the clade B envelope, FL5-2-209 was amplified from a frontal lobe autopsy sample of an AIDS patient with HIV-associated dementia that was obtained from the National NeuroAIDS Tissue Consortium. Envelopes were expressed from pSVIIIenv or from pcDNA3.1TOPO for the production of pseudovirions.
Cell cultures.
293T and HeLa TZM-BL cells were cultured as previously described (10, 11). Primary macrophages were prepared from elutriated monocytes (provided by the University of Massachusetts Medical School Center for AIDS Research) or by adherence from buffy coats as reported previously (11). Briefly, 0.5 ml of elutriated monocytes (5 × 105/ml) was plated in each well of 48-well cell culture dishes and cultured in 10% human plasma in Dulbecco modified Eagle medium (DMEM) for 5 to 7 days before infection. Alternatively, 5 × 107 PBMC from a buffy coat (Research Blood Components LLC, Brighton, MA) were plated into 14-cm bacterial culture dishes for 3 h before extensively washing away nonadherent cells, overnight culture, and repeating the washes. The adhered monocytes were then cultured for 5 to 7 days in 10% human plasma in DMEM before treatment with EDTA and transfer to 48-well tissue culture dishes the day prior to infection (42).
Preparation of Env+ pseudovirions.
Portions (1.25 μg) of pSVIIIenv or pcDNA3.1TOPO plasmids carrying complete envelope sequences were cotransfected by using calcium phosphate into 293T cells, together with 1.25 μg of pNL43 that carried a premature stop codon in the envelope (Env+) (10, 11). Pseudovirions were harvested 48 h posttransfection, clarified by low-speed centrifugation, aliquoted into 0.5-ml portions, and snap-frozen in liquid nitrogen.
Infectivity assays.
Infectivity assays were carried out as described previously (10, 11). Briefly, primary macrophages were treated with 100 μl of DEAE-dextran (10 μg/ml) prior to infection before adding an equal volume of serially diluted Env+ pseudovirions and spinoculation for 30 min in a benchtop centrifuge (34). Growth medium containing 10% human plasma was then added. After 7 days of incubation, the macrophages were fixed in cold methanol-acetone (1:1), washed, and immunostained for p24 with the MAbs 38:96K and EF7 (UK Centre for AIDS Research), followed by an anti-mouse IgG-β-galactosidase conjugate and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) substrate (0.5 mg of X-Gal/ml, 3 mM potassium ferricyanide, 3 mM potassium ferricyanide, 1 mM magnesium chloride). DEAE dextran and spinoculation enhance virus infectivity by ∼20-fold by increasing attachment (34). Infection according to this procedure does not bypass the requirement of CD4 and CCR5 for infection, which thus remains sensitive to entry inhibitors. Thus, macrophage infection conferred by envelopes described here was completely inhibited by maraviroc (data not shown).
HeLa TZM-BL cells were plated at 0.5 ml per well (5 × 105 cells/ml) in 48-well dishes the day prior to infection and infected. After 72 h, the HeLa TZM-BL cells were fixed in 0.5% glutaraldehyde in phosphate-buffered saline, and β-galactosidase-X-Gal substrate was added.
Infected macrophages and HeLa TZM-BL cells stain blue. Since Env+ pseudovirions are only capable of a single round of infection, stained infected cells are recorded as focus-forming units (FFU). The macrophage infectivity data presented were derived from FFU values averaged from two experiments using cells from different donors.
Inhibition and neutralization assays.
Inhibition and neutralization assays were carried out as described previously using HeLa TZM-BL cells as target cells (10, 11, 37). The residual infectivity was estimated by measuring the β-galactosidase activity using a luminescent readout. The β-galactosidase gene is a reporter gene under the control of an HIV long terminal repeat promoter in these cells (47).
Analysis of V1 loop phylogeny.
HIV-1 V1 loop sequences were downloaded from the HIV sequence database at Los Alamos and aligned by using CLUSTAL W2, and the frequencies of different amino acids at residue 153 were evaluated.
RESULTS
HIV-1 envelopes amplified from the plasma of a pediatric HIV-1 subject vary in their capacities to confer macrophage infection.
We previously described several HIV-1 envelopes that had been amplified by PCR from the plasma of a pediatric AIDS patient, P1114 (38). These envelopes were amplified when the patient was asymptomatic in the A2 stage of disease (in 1995 to 1996), as well as after the patient had progressed to symptomatic disease in C3 (in 1998) using the CDC categories (http://www.aids-ed.org/aidsetc?page=cm-105_disease). Single round pseudovirions carrying each envelope were titrated in infectivity assays on HeLa TZM-BL cells and monocyte-derived macrophages. All of the Envs conferred high infectivity titers on HeLa TZM-BL cells, which express high levels of CD4 and CCR5. However, only two Envs from the C3 stage of infection (C98-15 and C98-18) conferred significant infection of primary macrophages (Fig. 1).
FIG. 1.
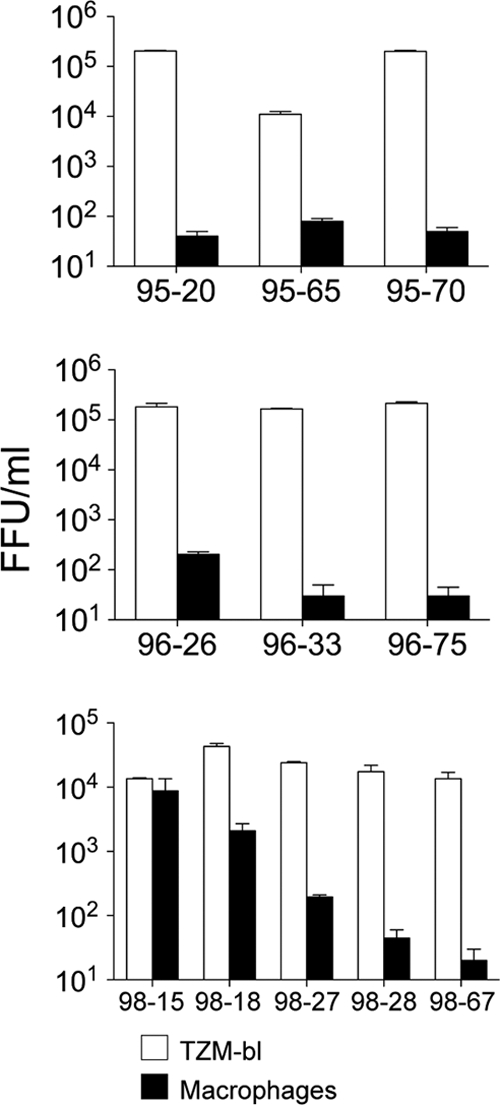
Macrophage tropic R5 viruses detected in late-stage disease of a pediatric AIDS patient. R5 envelopes were amplified from three plasma samples of a pediatric AIDS patient P1114 in 1995, 1996 (disease stage A2), and 1998 (disease stage C3). Single-round Env+ pseudovirions were prepared and used to infect HeLa TZM-BL cells and monocyte-derived macrophages. All envelopes exhibited high infectivity titers on TZM-BL cells. Two of the envelopes, 98-15 and 98-18, exhibited significant infection on macrophages.
Specific envelope residues in the V1V2 loops that segregate with macrophage infectivity.
The five envelopes from the C3 stage of infection were very closely related in sequence. Two distinct amino acid changes in the V1V2 loops were the only differences over the entire gp160 sequence that segregated with macrophage infectivity (Suppl. Fig. 1). Figure 2 shows a sequence alignment for the V1V2 loops of these envelopes. Env 98-15 is the most macrophage-tropic envelope and carries E153G and D167N substitutions in V1 and V2, respectively. Env 98-18 confers more modest levels of macrophage infectivity and possesses only the D167N change. The non-macrophage-tropic envelopes all have E153 and D167. These residues are the only candidates that could explain the differences in macrophage infectivity between these envelopes.
FIG. 2.
Specific envelope residues in the V1V2 loops segregate with macrophage infectivity. Aligned V1V2 sequences from C3 disease stage envelopes show that E153 and D167 in boxes are found in non-macrophage-tropic envelopes. 98-15 is the only envelope carrying G153 and is the most macrophage tropic. Both 98-15 and 98-18 have N167, but 98-18, which has intermediate macrophage infectivity, lacks G153.
Macrophage infectivity of envelopes mutated at residues 153 and 167.
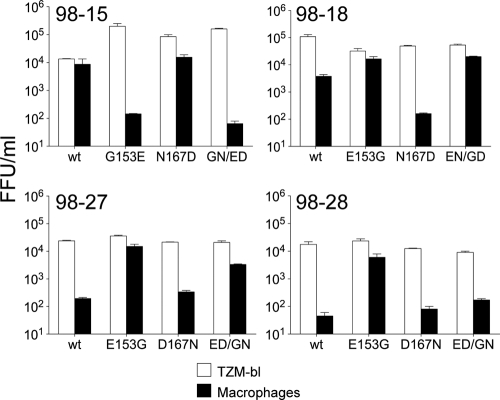
To determine whether the two polymorphisms in the V1/V2 region of these envelopes were responsible for the differences in macrophage infectivity, point mutations were made by site-directed mutagenesis and infectivity evaluated on TZM-BL cells and macrophages. A G153E substitution in 98-15 abolished macrophage tropism, which is consistent with the requirement of G153 for macrophage infectivity. In contrast, an N167D substitution in 98-15 had no effect on macrophage infectivity, while a combination of G153E and N167D substitutions conferred the same loss of macrophage infectivity as the G153E substitution alone (Fig. 3).
FIG. 3.
V1 loop residue 153 modulates macrophage tropism. A G153E substitution in macrophage-tropic R5 C98-15 reduced macrophage infectivity by at least a 100-fold, while N167D had only a modest effect (top, left panel). For 98-18 (top, right panel), E153G enhanced modest macrophage infectivity to levels similar to 98-15 wt, whereas N167D abrogated macrophage infection to levels close to background. For non-macrophage-tropic envelopes (98-27 and 98-28; bottom panels), an E153G substitution conferred >100-fold increase in macrophage infectivity. In contrast, a D167N substitution had only modest effects on these two envelopes. Double substitutions at residues 153 and 167 conferred phenotypes similar to the single change at residue 153, although this was less evident for 98-28. These data show that residue 153 alone modulates macrophage infectivity.
The reciprocal substitutions were then made in the non-macrophage-tropic envelopes, 98-27 and 98-28. For both of these envelopes, E153G caused a drastic increase in macrophage infectivity, approximately equal to the level of the 98-15 wild-type (wt) envelope. In contrast, D167N did not confer any significant increase in macrophage infectivity for 98-27 and 98-28. For 98-18, E153G enhanced the modest levels of macrophage infection conferred by this envelope to levels similar to 98-15, whereas N167D abrogated macrophage infection.
Taken together, these results indicate that G153 is the major determinant responsible for the macrophage-tropic phenotype observed for the C98-15 envelope, while N167 conferred only modest effects on macrophage infectivity for 98-18.
Residue E153 is conserved across HIV-1 clades.
We next evaluated the frequencies of E and G at position 153 for other HIV-1 clade B and non-B clades. In clades A, B, and C, residue E153 is highly prevalent, a situation similar to the non-macrophage-tropic Envs evaluated in the present study, whereas G153 is less common (Table 1). Interestingly, G153 is more prevalent in clades D and CRF02_AG, while for clade G and CRF01_AE, E153 was less prevalent but was not frequently substituted with a G. The conservation of E153 is also apparent in the recent review by Zolla-Pazner and Cardozo (51).
TABLE 1.
Conservation of V1 loop residue 153
| Clade or CRF | Prevalence (%)a at amino acid: |
Total no. of sequences | |
|---|---|---|---|
| E153 | G153 | ||
| Clades | |||
| A | 74.22 | 10.59 | 1,699 |
| B | 89.40 | 3.86 | 17,721 |
| C | 79.55 | 3.42 | 4,968 |
| D | 12.43 | 23.98 | 1,593 |
| G | 22.38 | 1.23 | 648 |
| CRF | |||
| CRF01_AE | 32.92 | 2.97 | 1,953 |
| CRF02_AG | 33.84 | 28.24 | 1,179 |
Residue E153 is conserved across clades. V1 sequences were obtained from the HIV database (http://www.hiv.lanl.gov/content/index) for clades A, B, C, D, G, AE, and 02AG. These sequences were aligned using CLUSTAL W2 and manually corrected. E153 is highly conserved across clades A, B, and C. G153 is most prevalent in clades D and 02AG.
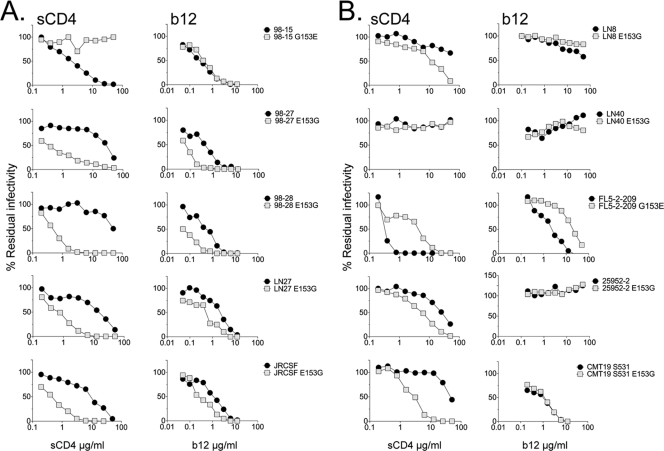
The E153G substitution confers macrophage infectivity for heterologous clade B envelopes.
Since E153 is relatively conserved, we next investigated whether the E153G substitution conferred macrophage infectivity for heterologous Envs or was limited to the pediatric envelopes described here. We introduced the E153G substitution into four heterologous clade B R5 envelopes that carry E153 in the V1 loop (NA20 LN8, NA118 LN27, NA420 LN40, and JRCSF) and that are non-macrophage-tropic. For two of the envelopes, LN27 and JRCSF (21, 36), the E153G substitution conferred substantial levels of infectivity for macrophages (Fig. 4). We next investigated the effects of the reverse G153E substitution into a highly macrophage-tropic brain-derived envelope, FL5-2-209, that naturally carried a G at residue 153. However, this change had no effect on the capacity of FL5-2-209 to infect macrophages (Fig. 4). Finally, we made E153G substitutions into clade C (25925-2) and D (CMT19 S531) envelopes. However, these envelopes remained non-macrophage-tropic. In summary, substitution of V1 loop residue E153 with G conferred macrophage tropism for two of four heterologous clade B envelopes tested.
FIG. 4.
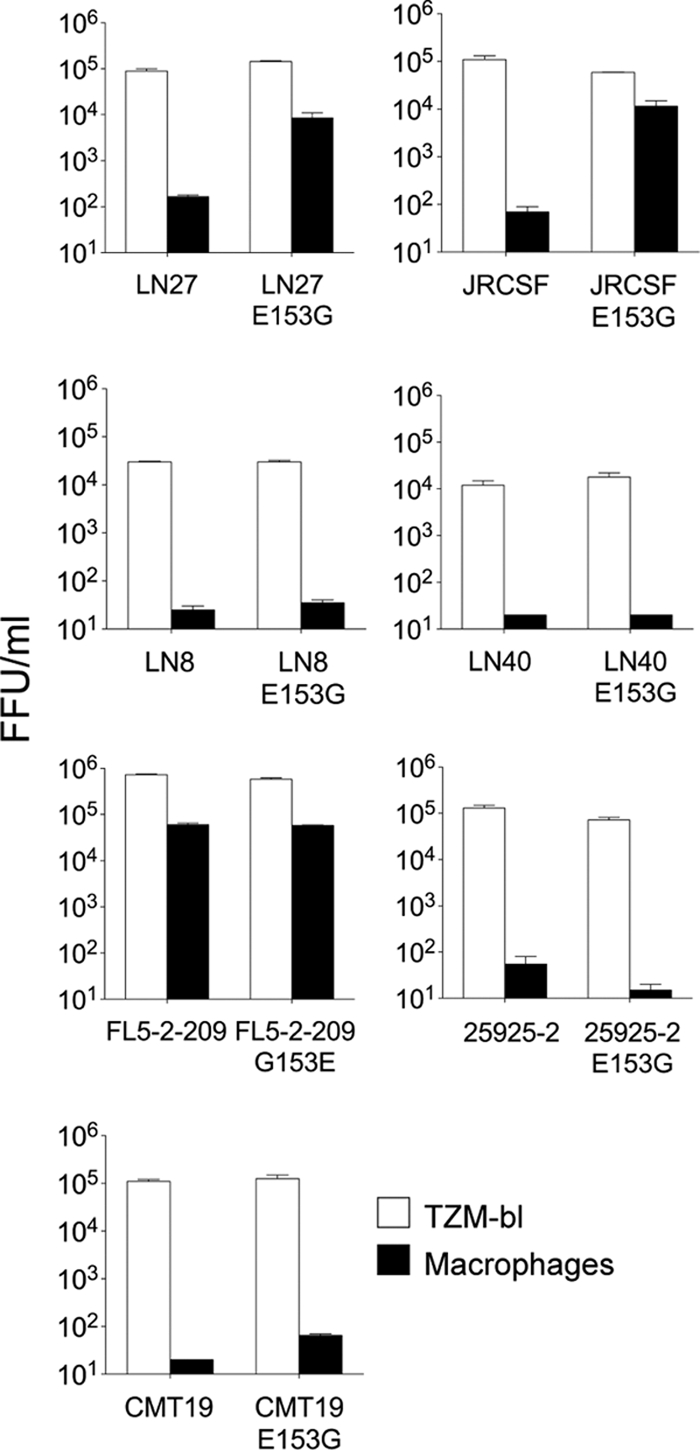
V1 loop residue 153 modulates macrophage tropism for two of seven heterologous R5 envelopes. The E153G substitution was introduced into four heterologous, non-macrophage-tropic clade B R5 envelopes. For two (LN27 and JRCSF; top panels), the substitution conferred at least a 100-fold increase in macrophage infectivity. In contrast, E153G had no effect on the lack of macrophage infection conferred by LN8 and LN40 (second panels down). The reverse substitution G153E in the highly macrophage-tropic FL5-2-209 also had no effect on the high levels of macrophage tropism conferred by this envelope (third panel down, left). Finally, E153G substitutions in clade C (25925-2) (third panel down, right) and clade D (CMT19 S531) (bottom panel) envelopes also failed to affect their lack of macrophage infection. E153G substitutions thus conferred macrophage tropism for two of seven heterologous HIV-1 R5 envelopes.
Substitutions in V1 loop residue 153 affect sensitivity to neutralizing antibodies and entry inhibitors. (i) MAb b12 and sCD4.
The capacity of HIV-1 R5 envelopes to infect macrophages correlated with infection of indicator cell lines via low levels of CD4 (17, 31, 36, 38). The ability to use low levels of CD4 for infection was shown previously to be due to changes within or proximal to the CD4bs (11, 13), which presumably confer an increased envelope avidity for cell surface CD4. Such changes in the use of CD4 are usually evident in shifts in the sensitivity of envelopes to inhibition by soluble CD4 and the CD4bs MAb, b12, with sensitivity to b12 a reasonable surrogate for the exposure of the CD4bs on many clade B envelopes. However, residue 153 is in the V1 loop, which has been proposed to sit at the apex of the envelope trimer away from the CD4bs (27). We tested whether substitutions at residue 153 affected sensitivity to sCD4 and b12. For the non-macrophage-tropic envelopes where E153G substitutions increased macrophage tropism (98-27, 98-28, LN27, and JRCSF), there were dramatic increases in sensitivity to sCD4 reflected in decreased 50% inhibitory concentrations (IC50s) of 50- to 100-fold (Fig. 5 A, left panels). We also noted more modest shifts in b12 sensitivity for these envelopes. Thus, for envelopes with E153G, IC50 shifts of ∼10-fold were observed for b12 inhibition. For the macrophage-tropic envelope 98-15, the reverse G153E substitution (which abrogated macrophage infection) conferred decreased sensitivity to sCD4 with an increased IC50 of >50-fold observed but only a minimal shift in b12 sensitivity at best. Nevertheless, these changes in sCD4 and b12 sensitivity are consistent with effects on the CD4bs mediated by residue 153 in the V1 loop.
FIG. 5.
Substitutions at residue 153 in the V1 loop modulate sensitivity to sCD4 and b12 in addition to macrophage tropism. (A) The five R5 envelopes that switched macrophage tropism following E153G (C98-27, C98-28, LN27, and JRCSF) or G153E (C98-15) substitutions showed changes in sensitivity to sCD4 and to b12. (B) The five R5 envelopes that failed to switch macrophage tropism following E153G (LN8, LN40, 25925, and CMT19 S531) or G153E (FL5-2-209) substitutions were also tested for changes in sensitivity to sCD4 and b12. Despite no change in tropism, changes in sensitivity to these reagents were detected for some but not all of the envelopes. Nonetheless, consistent changes in sCD4 and b12 sensitivity were associated with changes in macrophage tropism.
We next investigated the effects of the E153G substitution on sCD4 and b12 sensitivity for envelopes that failed to switch tropism. For LN8 and LN40, E153G had no significant effect on sCD4 or b12 sensitivity (Fig. 5B). However, for both the clade C (25925-2) and clade D (CMT19 S531) envelopes, E153G increased their sensitivity to sCD4, although there was no change for b12. The clade D envelope (CMT19 S531) stayed resistant to b12, and this may reflect the reduced cross-reactivity of b12 for this clade (3). Interestingly, the reverse G153E substitution in the highly macrophage-tropic FL5-2-209 envelope, which naturally carries G153, conferred substantial decreases in sensitivity to sCD4 and b12, even though the mutant envelope still conferred high levels of macrophage infection.
Together, these results show that the V1 loop residue at 153 frequently confers alterations in the CD4bs affecting sensitivity to sCD4 and b12. Of note, changes in sCD4 sensitivity sometimes occurred in the absence of detectable changes in macrophage tropism. Nevertheless, envelopes that did switch tropism generally showed the largest changes in sensitivity to sCD4 and b12.
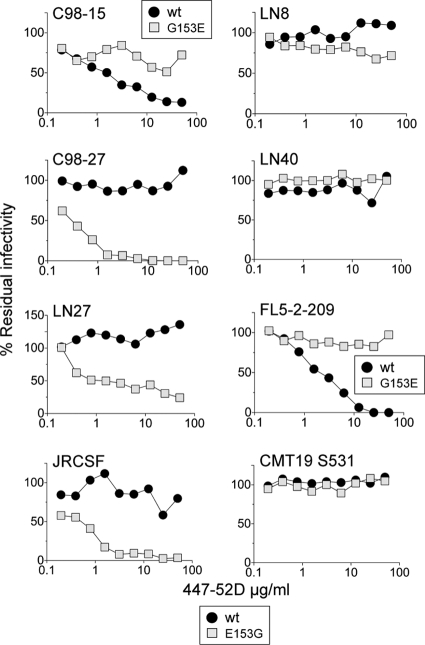
(ii) The V3 loop-specific MAb, 447-52D.
Dramatic changes in sensitivity to the V3 loop MAb, 447-52D, were also observed for envelopes that switched macrophage tropism after substitution at residue 153. For the non-macrophage-tropic envelopes where E153G conferred macrophage infection, there were large increases in sensitivity to 447-52D with at least 100-fold decreases in IC50s (Fig. 6). In contrast, for 98-15, G153 conferred resistance to 447-52D to go alongside the abrogation of macrophage infection (Fig. 6).
FIG. 6.
Sensitivity to the V3 loop-specific MAb, 447-52D, is modulated by substitutions at residue 153 and associated with switches in macrophage tropism. For R5 envelopes with altered macrophage tropism following E153G (C98-27, LN27, and JRCSF) or G153E (C98-15) substitutions, there were substantial changes in sensitivity to the V3 loop MAb, 447-52D (left panels). For three envelopes that failed to switch tropism following E153G substitution, there were no changes in 447-52D sensitivity. However, FL5-2-209 showed decreased sensitivity to 447-52D following the reverse substitution, even though this change failed to affect high levels of macrophage infectivity (right panels). In summary, substantial changes in sensitivity to the V3 loop MAb, 447-52D, were associated with changes in macrophage tropism following substitutions at V1 loop residue 153.
For LN8 and LN40, the E153G mutant envelopes remained resistant to 447-52D consistent with the lack of an effect on their non-macrophage tropism. For the macrophage-tropic brain Env FL5-2-209, G153E conferred resistance to 447-52D even though it had no effect on macrophage infectivity. Overall, these observations indicate an intriguing role for the V3 loop in R5 macrophage tropism that could be associated with effects on the CD4bs or in altered interactions with CD4 or CCR5. These possibilities are addressed below.
(iii) Other entry inhibitors and neutralizing antibodies.
The same envelopes and mutants were also tested for sensitivity to neutralizing MAbs and inhibitors that target different envelope sites or functions. Table 2 shows IC50s for envelopes tested. Overall, E153G and G153E had no substantial affect on sensitivity to the glycan-specific MAbs 2G12, 17b, maraviroc, 2F5, or T20. These observations indicate that the substitutions at residue 153 affect the CD4bs but do not confer a more global effect on other envelope functions, including interactions with CCR5 (maraviroc) or gp41 conformational changes required for fusion (2F5 and T20), nor do they result in increased exposure of the CD4i epitope (17b). Of note, the lack of change in maraviroc sensitivity suggests that alterations in the V3 loop that confer shifts on 447-52D sensitivity do not impact on the efficiency of Env-CCR5 interactions.
TABLE 2.
Sensitivity of R5 envelopes and their corresponding mutants with V1 loop residue 153 substitutions to various entry inhibitors and neutralizing antibodies
| Envelope | IC50 |
||||||
|---|---|---|---|---|---|---|---|
| 17b (μg/ml) | 2G12 (μg/ml) | Maraviroc (nM) | 2F5 (μg/ml) | T20 (nM) | PG9 (μg/ml) | PG16 (μg/ml) | |
| 98-15 | >50 | 1.05 | 5.89 | 4 | 40.59 | 0.056 | 0.0049 |
| 98-15 G153E | >50 | 1.16 | 10.2 | 3 | 65.48 | 0.02 | 0.007 |
| 98-27 | >50 | 2.55 | 12.9 | 7.41 | 58.86 | 0.05 | 0.02 |
| 98-27 E153G | >50 | 1.5 | 10.0 | 1.78 | 61.62 | 0.09 | 0.02 |
| 98-28 | >50 | 0.88 | 1.72 | 2.03 | 74.89 | <0.006 | <0.006 |
| 98-28 E153G | >50 | 1.08 | 0.92 | 3.60 | 33.43 | 0.01 | <0.006 |
| JRCSF | >50 | 1.5 | 5.12 | 7.11 | 16.62 | <0.006 | <0.006 |
| JRCSF E153G | >50 | 1.1 | 3.99 | 6.33 | 28.54 | 0.02 | <0.006 |
| LN27 | >50 | 0.59 | 3.66 | 8.2 | 32.87 | >1.56 | >1.56 |
| LN27 E153G | >50 | 0.17 | 0.72 | 5.5 | 28.41 | >1.56 | >1.56 |
| LN8 | 24 | 3.15 | 2.0 | 6.34 | 208.2 | 0.06 | 0.04 |
| LN8 E153G | 24 | 3.77 | 2.82 | 8.09 | 278.1 | >1.56 | >1.56 |
| LN40 | >50 | 1.97 | 2.01 | 2.33 | 42.37 | >1.56 | >1.56 |
| LN40 E153G | >50 | 3.53 | 2.28 | 0.84 | 97.00 | >1.56 | >1.56 |
| FL5-2-209 | >50 | >50 | 0.47 | >50 | 115.50 | >1.56 | >1.56 |
| FL5-2-209 G153E | 37.5 | >50 | 0.38 | >50 | 131.20 | >1.56 | 0.22 |
| 25925-2 cl.22 | 20 | >50 | 0.29 | >50 | 70.00 | 0.04 | 0.01 |
| 25925-2 cl.22 E153G | 23 | >50 | 0.38 | >50 | 125.40 | 0.08 | 0.008 |
| CMT 19S531 | >50 | >50 | 0.68 | 13.22 | 39.00 | 0.06 | 0.01 |
| CMT 19S531 E153G | >50 | >50 | 0.53 | 8.63 | 40.00 | 0.03 | 0.008 |
Residue 153 is proximal to conserved V1/V2 determinants that are targeted by broadly active neutralizing MAbs: PG9 and PG16.
Two human MAbs, PG9 and PG16, that were derived from a clade A-infected African donor were recently reported (46). These MAbs are able to neutralize a wide range of diverse envelopes (46). They preferentially bind to the trimeric envelope and target an epitope that involves determinants in the V2 and V3 loops (46). The proximity of the V2 determinants to V1 residue 153 in the V1 loop and the implication of the V3 loop in macrophage tropism prompted us to test whether envelopes carrying E153G or G153E substitutions differed in sensitivity to PG9 or PG16. Some shifts in sensitivity to PG9 and PG16 were noted (Table 2). However, these were mainly modest and did not segregate with macrophage infectivity.
DISCUSSION
We present data to show that a single amino acid substitution in the V1 loop modulates macrophage tropism for heterologous HIV-1 clade B R5 envelopes. The structures of the V1 and V2 loops have not yet been solved. Various studies showed that the V1 and V2 loops influenced the binding of CD4bs MAbs, suggesting that these loops may help to shield the CD4bs (28, 35, 39, 49). The identification of V1V2 stem residues as contacts for CD4 also added to the possibility that the loops may lie in the recessed space accessed by CD4 on the unliganded trimer (23, 27, 48). However, the more recent electron tomographic structure of the trimeric envelope strongly suggests that the bulk of the V1 and V2 loops sit on the apex of the trimeric envelope and thus some distance from the CD4bs (27). We therefore believe that our data are consistent with a model where the V1 loop determinant modulates the conformation or exposure of the V3 loop, which in turn affects the function of the CD4bs priming the envelope for low CD4 use and macrophage infection. Moreover, it is striking that V1 residue 153 control of the V3 loop and CD4bs was active for envelopes derived from three of six clade B-infected subjects. This mechanism is thus likely to be operative for 50% of clade B envelopes.
Previously several determinants have been reported to have major effects on macrophage infection by HIV-1 R5 envelopes. These include residue 283 in the CD4bs (11, 13, 38), V3 loop determinants and residues adjacent to the CD4 contact residues on the CD4 binding loop (11). Thus, macrophage tropism can be modulated by changes within the CD4bs, which directly impact Env-CD4 affinity, or by changes that indirectly affect the CD4bs, presumably by affecting the exposure CD4 contact residues. The identification of a distal V1 loop determinant that also confers macrophage tropism is striking and indicates that multiple envelope sites may have the potential to modulate macrophage tropism. The change from a charged glutamic acid (E) to the uncharged glycine could have profound effect on the interaction of the V1 loop with other envelope residues due to the loss of potential ionic bonds. Our initial interpretation was that the V1 determinant acted by affecting the exposure of CD4 contact residues. However, shifts in b12 sensitivity (our best indicator of CD4bs exposure) following substitutions at residue 153 ranged from extremely minor to modest at best and were substantially smaller than for sCD4 or 447-52D. It is therefore unlikely that the effects of V1 residue 153 result entirely from increased exposure of CD4bs residues. Thus, changes in the capacity of the envelope to react to suboptimal levels of cell surface CD4 (without changes in Env-CD4 affinity) and trigger conformational changes and viral entry should be considered as an alternative mechanism (see below).
For envelopes where the substitution in V1 did not affect tropism, changes in sCD4, b12, and 447-52D sensitivity were noted for some but not other envelopes. For envelopes that showed no change in sCD4, 447-52D, and b12 sensitivity, it is possible that a putative connection between the V1 and V3 loops is absent or held by alternative mechanisms, so that residue 153 substitutions have no effect on the CD4bs. Alternatively, other determinants that affect the CD4bs and macrophage tropism may simply override any effects conferred by changes in the V1 loop determinant. This latter possibility could apply to the highly macrophage-tropic brain envelope FL5-2-209. This envelope was initially highly sensitive to sCD4, 447-52D, and b12 (IC50s of 0.34, 2.0, and 2.5 μg/ml, respectively) but became substantially more resistant (IC50s of 7.5, >50, and 11.8 μg/ml, respectively) when the G153E substitution was introduced, even though no changes in macrophage infection were observed. It is thus likely that alternative envelope residues confer macrophage infectivity and simply override the effects of V1 residue 153. It is noteworthy that an E153G substitution was previously reported to be responsible for increased syncytium formation following infection of brain microglial cultures by an HIV-1 R5 variant selected in vitro (41).
The C98-18 envelope conferred modest levels of macrophage infection compared to the highly macrophage-tropic C98-15 and the non-macrophage-tropic P1114 envelopes. C98-18 carries the D167N substitution in the V2 loop but not the V1 loop E153G substitution present in the C98-15 envelope. An N167D substitution in C98-18 abrogated the modest levels of macrophage infection confirming the role of this residue (Fig. 3). In addition, N167D conferred an increase in the sensitivity of C98-18 to sCD4, reducing the IC50 from 26.62 to 8.19 μg/ml, but had no affect on the resistance to the V3 loop MAb, 447-52D (data not shown). These observations are curious since substitutions at residue 167 had no effect on the macrophage infectivity of the other P1114 envelopes tested. Presumably, there are other determinants in the P1114 envelopes that influence whether residue 167 affects macrophage infectivity. Regardless, our data indicate that V2 residue 167 is a only minor player in the macrophage tropism of the P1114 envelopes.
Other studies have also reported single or limited substitutions in the envelope that affect macrophage infectivity and/or sensitivity to neutralizing antibodies. Lynch et al. reported that the introduction of a leucine residue to replace a highly conserved isoleucine (I309) in the V3 loop of clade C envelopes affected the exposure of the CD4bs and conferred an increase in macrophage replication, although this was modest (29). Li et al. described an N197Q substitution proximal to V2, which removed a potential N-linked glycosylation site. N197Q conferred increases in sensitivity to sCD4 and b12. However, N197Q also conferred sensitivity to 17b, a CD4i MAb, indicating that more substantial structural changes had resulted that exposed sites involved in coreceptor binding (26). Similarly, Zhu et al. reported on an adjacent substitution in the β3 strand, T198P (50), while O'Rourke et al. described a D179N substitution in the LDV motif of V2 (35), both of which conferred global neutralization sensitivity and exposure of the 17b epitope. Some of these substitutions may affect the interactions of the gp120 subunits on the trimer, perhaps altering the position of the V1V2 loops, which may interact at the apex. Such changes may also result in triggering some of the conformational changes (usually associated with CD4 binding) and exposure of CD4i epitopes.
Limited changes in the extracellular segment of gp41 have also been shown to affect neutralization sensitivity. Thus, two amino acid changes in the conserved HR1 and MPER regions in gp41 together conferred increased sensitivity to sCD4, broadly neutralizing gp120 and gp41 MAbs, and to human HIV-1+ plasmas (4). In addition, Shen et al. recently reported an L669S substitution in the MPER that conferred increased sensitivity to a range of gp41- and gp120-specific neutralizing MAbs. Presumably, these substitutions also at indirectly by influencing the arrangement of gp120 subunits on the monomer and thus affecting their sensitivity to neutralizing antibodies.
In contrast, the effects of V1 residue 153 are unique in conferring large shifts in envelope phenotype that are focused specifically on the V3 loop and CD4bs without major effects on other envelope sites, as estimated by envelope sensitivity to inhibitors that targeted other stages of HIV-1 entry, including Env-CCR5 interactions and gp41 conformational changes. All envelopes also remained relatively resistant to 17b, a CD4i MAb, indicating that extensive conformational changes that are usually associated with gp120-CD4 interactions had not occurred. The changes affected by V1 residue 153 thus contrast with the substitutions described above (26).
Our data indicate that V1 residue 153 has strong affects on the V3 loop and the CD4bs, which are only partially explained by increased exposure of the CD4 contact residues. The location of the V3 loop on the unliganded envelope is not known but may sit close enough to restrict the approach of both CD4 and b12 (8, 27). It is also tempting to envisage that residue 153 modulates a gp120 conformational change to form a structure intermediate between the unliganded envelope and the CD4-bound form. Such an intermediate may be partially activated for the conformational changes required to form the coreceptor binding site and thus require less robust interactions with fewer CD4 molecules to trigger fusion and infection.
The determinant at residue 153 is proximal to sites in the V2 loop that are targeted by the recently described MAbs PG9 and PG16 (46). In fact, these MAbs target determinants in both the V2 and the V3 loops. Since the substitutions in V1 described here also had effects on the V3 loop, we hypothesized that they would impact on PG9 and PG16 sensitivity. However, only modest effects on PG9 and PG16 sensitivity were observed. This result illustrates the robustness of the PG9 and PG16 epitope and is a reassuring result for vaccine research focused on this epitope.
The conservation of glutamic acid at position 153 across several clades is intriguing and implies that this site plays an important functional role for the envelope. Our data suggest that this role may involve an interaction with the V3 loop, which in turn affects the CD4bs. The decreased frequency of E153 among clade D and CRF02_AG envelopes and increased frequency of G153 is also striking. Whether the higher prevalence of G153 associates with increased macrophage tropism for these clades is not known. An E153G substitution for the one clade D envelope that we tested (CMT19 S531) had no affect on macrophage tropism. However, a more extensive survey would be needed to establish an influence of G153 on clade D or CRF02_AG envelopes. Of note, clade D viruses have been associated with faster disease progression (1, 14, 19, 20, 45) and a higher frequency of dementia (40). In addition, CXCR4 tropism was common among CRF02_AG viruses, a sign of a mature HIV epidemic (15). It is thus possible that an increased frequency of G153 may simply reflect a high prevalence of R5 viruses with increased macrophage tropism late in disease (18, 25, 44).
In summary, we describe here a novel determinant in the V1 loop that modulates macrophage tropism conferred by HIV-1 R5 envelopes. The V1 loop determinant appears to control the exposure or conformation of the V3 loop, which in turn impacts the CD4bs to prime the envelope for low CD4 use and macrophage infection. Our data also have relevance for vaccines that aim to target the CD4bs.
Acknowledgments
Our work was supported by NIH grants R01 MH64408 and P01 AI082274. We also wish to acknowledge the University of Massachusetts Center for AIDS Research (P30-AI42845), the NIH AIDS Research and Reference Reagent Program, and the Centre for AIDS Reagents, NIBSC, United Kingdom, for services and reagents. S.Z.-P. is supported by NIH grant AI 36085.
We thank Sabine Hadulco at Roche Palo Alto for supplying T-20. We also thank the National NeuroAIDS Tissue Consortium for providing the brain tissue that gave rise to the FL5-2-209 envelope investigated here.
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1.Baeten, J. M., et al. 2007. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J. Infect. Dis. 195:1177-1180. [DOI] [PubMed] [Google Scholar]
- 2.Bannert, N., D. Schenten, S. Craig, and J. Sodroski. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophage-tropic human immunodeficiency viruses. J. Virol. 74:10984-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., et al. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blish, C. A., M. A. Nguyen, and J. Overbaugh. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington, M., M. Dean, M. P. Martin, and S. J. O'Brien. 1999. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet. 8:1939-1945. [DOI] [PubMed] [Google Scholar]
- 6.Cavarelli, M., and G. Scarlatti. 2009. Phenotype variation in human immunodeficiency virus type 1 transmission and disease progression. Dis. Markers 27:121-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B., et al. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., et al. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clapham, P. R., and A. McKnight. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809-1829. [DOI] [PubMed] [Google Scholar]
- 10.Duenas-Decamp, M. J., P. Peters, D. Burton, and P. R. Clapham. 2008. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J. Virol. 82:5807-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duenas-Decamp, M. J., P. J. Peters, D. Burton, and P. R. Clapham. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J. Virol. 83:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duenas-Decamp, M. J., et al. 2010. Variation in the biological properties of HIV-1 R5 envelopes: implications of envelope structure, transmission and pathogenesis. Future Virol. 5:417-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunfee, R. L., et al. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Easterbrook, P. J., et al. 2010. Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J. Int. AIDS Soc. 13:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esbjornsson, J., et al. 2010. Frequent CXCR4 tropism of HIV-1 subtype A and CRF02_AG during late-stage disease: indication of an evolving epidemic in West Africa. Retrovirology 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorry, P. R., et al. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorry, P. R., et al. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, L., et al. 2005. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology 337:384-398. [DOI] [PubMed] [Google Scholar]
- 19.Kaleebu, P., et al. 2002. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J. Infect. Dis. 185:1244-1250. [DOI] [PubMed] [Google Scholar]
- 20.Kiwanuka, N., et al. HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J. Acquir. Immune Defic. Syndr. 54:180-184. [DOI] [PMC free article] [PubMed]
- 21.Koyanagi, Y., et al. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 22.Kulkarni, S. S., et al. 2009. Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 385:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong, P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U. S. A. 96:5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, S., et al. 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J. Virol. 73:9741-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y., et al. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch, R. M., et al. 2010. Subtype-specific conservation of isoleucine 309 in the envelope V3 domain is linked to immune evasion in subtype C HIV-1 infection. Virology 404:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, J., C. C. LaBranche, and F. Gonzalez-Scarano. 2001. Differential CD4/CCR5 utilization, gp120 conformation, and neutralization sensitivity between envelopes from a microglia-adapted human immunodeficiency virus type 1 and its parental isolate. J. Virol. 75:3568-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Garcia, J., W. Cao, A. Varela-Rohena, M. L. Plassmeyer, and F. Gonzalez-Scarano. 2006. HIV-1 tropism for the central nervous system: brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 346:169-179. [DOI] [PubMed] [Google Scholar]
- 32.Montefiori, D. C., and J. R. Mascola. 2009. Neutralizing antibodies against HIV-1: can we elicit them with vaccines and how much do we need? Curr. Opin. HIV AIDS 4:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori, K., M. Rosenzweig, and R. C. Desrosiers. 2000. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J. Virol. 74:10852-10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Rourke, S. M., et al. 2010. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J. Virol. 84:11200-11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters, P. J., et al. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusogenicity for macrophages. J. Virol. 78:6915-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters, P. J., et al. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters, P. J., et al. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J. Virol. 80:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinter, A., et al. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sacktor, N., et al. 2009. HIV subtype D is associated with dementia, compared with subtype A, in immunosuppressed individuals at risk of cognitive impairment in Kampala, Uganda. Clin. Infect. Dis. 49:780-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shieh, J. T., J. Martin, G. Baltuch, M. H. Malim, and F. Gonzalez-Scarano. 2000. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J. Virol. 74:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmons, G., et al. 1996. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dualtropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J. Virol. 70:8355-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, E. R., et al. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuttle, D. L., et al. 2002. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res. Hum. Retrovir. 18:353-362. [DOI] [PubMed] [Google Scholar]
- 45.Vasan, A., et al. 2006. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin. Infect. Dis. 42:843-852. [DOI] [PubMed] [Google Scholar]
- 46.Walker, L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, X., et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyatt, R., et al. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 49.Ye, Y., Z. H. Si, J. P. Moore, and J. Sodroski. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J. Virol. 74:11955-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, C. B., L. Zhu, S. Holz-Smith, T. J. Matthews, and C. H. Chen. 2001. The role of the third beta strand in gp120 conformation and neutralization sensitivity of the HIV-1 primary isolate DH012. Proc. Natl. Acad. Sci. U. S. A. 98:15227-15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zolla-Pazner, S., and T. Cardozo. 2010. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 10:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]