Abstract
Syn5 is a marine cyanophage that is propagated on the marine photosynthetic cyanobacterial strain Synechococcus sp. WH8109 under laboratory conditions. Cryoelectron images of this double-stranded DNA (dsDNA) phage reveal an icosahedral capsid with short tail appendages and a single novel hornlike structure at the vertex opposite the tail. Despite the major impact of cyanophages on life in the oceans, there is limited information on cyanophage intracellular assembly processes within their photosynthetic hosts. The one-step growth curve of Syn5 demonstrated a short cycle with an eclipse period of ∼45 min, a latent phase of ∼60 min, and a burst size of 20 to 30 particles per cell at 28°C. SDS-PAGE and Western blot analysis of cell lysates at different times after infection showed the synthesis of major virion proteins and their increase as the infection progressed. The scaffolding protein of Syn5, absent from virions, was identified in the lysates and expressed from the cloned gene. It migrated anomalously on SDS-PAGE, similar to the phage T7 scaffolding protein. Particles lacking DNA but containing the coat and scaffolding proteins were purified from Syn5-infected cells using CsCl centrifugation followed by sucrose gradient centrifugation. Electron microscopic images of the purified particles showed shells lacking condensed DNA but filled with protein density, presumably scaffolding protein. These findings suggest that the cyanophages form infectious virions through the initial assembly of scaffolding-containing procapsids, similar to the assembly pathways for the enteric dsDNA bacteriophages. Since cyanobacteria predate the enteric bacteria, this procapsid-mediated assembly pathway may have originated with the cyanophages.
Cyanobacteria are ancient oxygenic photosynthetic organisms that originated about 3 billion years ago (62). Marine cyanobacteria of the genera Prochlorococcus and Synechococcus belong to the dominant picophytoplankton and, on average, contribute about half of the primary production in the oceans (22, 38). Of the two genera, Synechococcus is less abundant in oligotrophic zones but is more widely distributed in aquatic environments worldwide (53).
Viruses are the most abundant life forms on Earth, with an estimated number of 4 × 1030 in the oceans (64). Investigating their impact is an important part of understanding the biogeochemical processes in these environments (63). Bacterial viruses contribute to microbial diversity by reducing the representatives of the dominant species, as described by “killing the winner” theory (66). In the process of cell lysis, the released organic matter becomes readily available to heterotrophic organisms (44). In the marine environment, gene transfer by viruses may be a key mechanism in generating and transmitting diversity (68, 75).
The genomes of cyanophages published to date share remarkably similar features with enteric phages both in sequence homology and structural organization (12, 39-41, 45, 59, 60, 71). Cyanophages have been shown to carry and express genes specifying proteins involved in host cell photosynthesis and other aspects of host cell physiology. Thus, they appear to be inseparable players in the life cycle of these photosynthetic organisms (13, 35, 36). They also carry many genes for as-yet-unidentified functions. The known cyanophages are morphologically similar to the three groups of double-stranded DNA (dsDNA) bacteriophages isolated from common enteric bacterial species, Myoviridae (contractile long tails), Siphoviridae (noncontractile long tails), and Podoviridae (short tails) (70, 73).
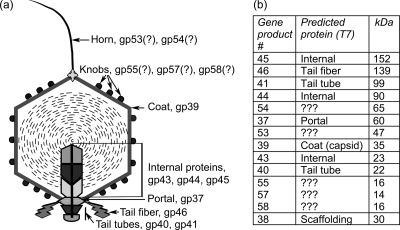
Syn5 is a dsDNA, short-tailed cyanophage isolated from the Sargasso Sea (70). Its laboratory host is the cyanobacterial strain Synechococcus sp. WH8109 (61, 70), which belongs to marine cluster A of Synechococcus, clade II, one of the most widely distributed clades in the world oceans (54, 77). Electron microscopic (EM) studies of Syn5 show an icosahedral virion with a head diameter of 60 nm and a short tail (49). Cryoelectron microscopy (cryo-EM) images reveal a mysterious long, flexible, fibrous hornlike protrusion always positioned on the capsid vertex that is opposite the tail. The horn is about 50 nm in length, 10 nm wide at its base, and 2 to 3 nm wide at its tip, and only one is observed per particle (Fig. 1a). Bacteriophages such as φX174, φ29, PRD1, and PM2 carry fibers or spike structures on their capsids, but these are usually multiple molecules attached symmetrically to a number of vertices (3, 26, 43, 65). Another cyanophage with an appendage on the vertex opposite the tail has recently been isolated (personal communication, W. Pope and R. Hendrix). Symmetrical cryo-EM reconstruction of the Syn5 capsid reveals an assembly of hexameric and pentameric units. The capsid is decorated with three knoblike proteins associated with each hexamer, as well as with distinctive curved ridges radiating from the pentamer edges. Decoration proteins are found among other bacteriophages, e.g., SPP1, λ, ɛ15, and T5, but the knob arrangement of Syn5 is unusual (5, 9, 19, 29).
FIG. 1.
Syn5 structural proteins. (a) Schematic of the Syn5 mature virion with assigned proteins based on sequence homology with phage T7. (b) Table of gene product (gp) numbers, assigned names or designations, and calculated molecular masses. In the initial report describing the protein profile of Syn5 (49), only gp58 was found in the MS analysis of the smallest protein band on SDS-PAGE. However, reanalysis revealed the presence of two additional proteins, gp55 and gp57, that were very close to gp58 in molecular mass. Scaffolding protein is included for completeness (bottom row) but is not present in the mature virion.
Syn5 has a 46,214-bp genome, similar to those of T7-like phages, with an RNA polymerase gene and short terminal repeats (49). The sequence encodes 61 predicted open reading frames (ORFs) with an organization typical of dsDNA phages, i.e., DNA replication genes clustered on the left and structural proteins on the right. The gene order shares high synteny with the order of T7 and with those of several cyanophages, especially Synechococcus bacteriophage P60 and Prochlorococcus phage P-SSP7. Photosynthetic genes are absent in Syn5, in contrast with other cyanophages (42, 55, 59, 71).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of Syn5 particles separated 11 distinct protein bands, summarized in Fig. 1b. Mass spectrometry (MS) of bands excised from the gel identified eight proteins closely resembling structural proteins in other dsDNA phages, including the coat, portal, three internal core proteins, and the tail apparatus components (49). Five structural proteins—gp53, gp54, gp55, gp57, and gp58—are novel in that their sequences do not match other proteins in the NCBI database (BLASTp E value, <0.001) (1).
Studies of cyanophages from both fresh and marine waters have focused primarily on their ecology and population genetics. Our ability to grow high-titer phage preparations of Syn5 provides the opportunity to use it as a laboratory model for studying the intracellular growth and assembly of dsDNA cyanophages.
Phage assembly has been studied in detail in many dsDNA enteric viruses, and the processes share many features. The well-characterized dsDNA phages first assemble a precursor called the procapsid or prohead, composed of a coat shell filled with scaffolding protein and a portal complex occupying a unique vertex. These capsid structures lack condensed DNA. The roles of the scaffold and portal in procapsid formation are critical, as they participate in the initiation of the process and ensure the proper organization of the coat molecules in the correct conformation (4, 20). The newly replicated DNA is then pumped into the procapsid shell as the scaffolding protein exits or is degraded. The capsid lattice expands, and the packaged genome is stabilized by the construction and attachment of the tail apparatus (30, 56).
Capsid assembly through a procapsid intermediate is not limited to bacteriophages. Eukaryotic viruses, e.g., adenoviruses (15, 18) and herpesviruses (47, 50), follow the same model. The latter have also been shown to contain a portal-occupied vertex with a role in DNA packaging (33, 46).
Examining the assembly processes of bacteriophages infecting cyanobacteria should reveal aspects of DNA packaging into the virions, which may be important for gene transfer processes in the ocean. Since cyanobacteria branch early in the phylogenetic tree, these studies will enable us to better understand the evolution of phage structure, assembly, and DNA packaging. In this work, we describe the growth cycle of Syn5 and the purification, identification, and initial characterization of the procapsid assembly intermediate.
MATERIALS AND METHODS
Host growth.
The Syn5 host used in all experiments was Synechococcus sp. strain WH8109 (kindly provided by John Waterbury), grown in artificial seawater (ASW). ASW was prepared as described by Wyman et al. (74) and modified by Lindell et al. (37), with some further adjustments. Briefly, the components, dissolved in double-distilled water (MilliQ purified), were 428 mM NaCl, 9.8 mM MgCl2·6H2O, 6.7 mM KCl, 17.8 mM NaNO3, 14.2 mM MgSO4 (anhydrous), 3.4 mM CaCl2·2H2O, 0.22 mM K2HPO4·3H2O, 5.9 mM NaHCO3, 9.1 mM Tris. The pH was adjusted to 8.0 with HCl. After autoclaving and cooling to room temperature, a trace metals mix (0.77 μM ZnSO4·7H2O, 7.0 μM MnCl2·4H2O, 0.14 μM CoCl2·6H2O, 30 μM Na2MoO4·2H2O, 30 μM citrate, 5 μM ferric citrate) was added, as well as 15 μM EDTA (disodium salt) and 100 μM Na2CO3.
The cells were grown in controlled environmental chambers (Percival Scientific) at 28°C under continuous cool white fluorescent light at an irradiance of 45 to 50 μmol m−2 s−1. The culture vessels were the same as described previously (49). Cells were counted in a Petroff-Hauser chamber with an epifluorescence microscope (Zeiss Axiostar plus) fitted with a fluorescein isothiocyanate (FITC) filter. To store the cells, an exponentially growing culture was centrifuged at 9,500 × g for 10 min and the cell pellet was resuspended and concentrated 10-fold in fresh culture medium. Dimethyl sulfoxide (7% final concentration) or glycerol (15%) was added as a cryoprotectant and equilibrated with the cells for 10 min at room temperature, and the suspensions stored at −80°C. To recover cells from frozen stocks, the suspensions were thawed on ice, transferred to ASW liquid medium, and for the first 2 days of incubation, shielded with aluminum window screens to reduce the light irradiance to about 5 to 10 μmol m−2 s−1.
Phage growth.
Syn5 was purified as described in Pope et al. (49), with some modifications. In brief, Synechococcus cells in mid-exponential phase (2 × 108 to 6 × 108 cells/ml) were infected with Syn5 with a multiplicity of infection (MOI) of 0.001. At 4 to 5 h after infection, the first signs of lysis appeared, i.e., green cell debris on the culture vessel walls and transition from the deep red color of the culture to bright pink. To obtain complete lysis, 0.1% Triton X-100 and 0.01 mg/ml of lysozyme were added and the lysate stirred at room temperature for about 1 h. The cell debris was then pelleted at 9,500 × g for 15 min at 4°C, and the supernatant stored at 4°C. To improve phage precipitation, 500 mM NaCl (final concentration, 925 mM) was dissolved in the supernatant, followed by the addition of 10% (wt/vol) polyethylene glycol (PEG) 8000 and stirring overnight to 5 days at 4°C. The phages were pelleted at 9,500 × g for 30 min at 4°C and resuspended in phage buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 100 mM MgCl2). The suspension was loaded onto a CsCl step gradient (ρ [density in g/cm3] of 1.3, 1.4, 1.5, and 1.6; each prepared in phage buffer, 50 mM Tris [pH 7.5], 100 mM NaCl, 100 mM MgCl2), followed by ultracentrifugation at 28,000 rpm for 3 h at 8°C in an SW28 rotor (Beckman). The Syn5 scattering band typically sedimented at the interface of the 1.4 and 1.5 layers. The phage band was withdrawn with a needle syringe and dialyzed stepwise against 50 mM Tris, pH 7.5, 100 mM MgCl2 buffer containing decreasing salt concentrations as follows: 2 M (1 h), 1 M (1 h), and 100 mM NaCl (2 h to overnight) at 4°C. Phage stocks were stored at 4°C.
Determination of phage titers.
Serial dilutions of phage suspensions were prepared in ASW. ASW agar (1% low-melting-point agarose; Invitrogen) was melted and held in a 32°C water bath until use. A sample of each dilution was mixed with 1 ml of exponential-phase Synechococcus cells (4 to 7 × 108 cells/ml) and, after 2 to 3 min at room temperature, 3.5 to 5 ml of ASW agar was added and the sample mixed and poured into a 50-mm petri plate. PFU were counted following overnight (16 to 20 h) incubation under the same conditions used to grow cells (see “Host growth”).
One-step growth curve.
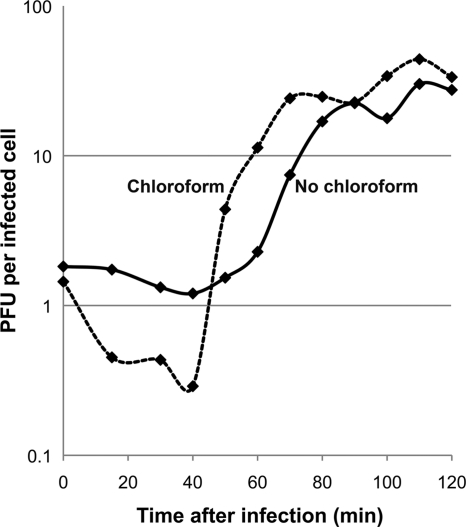
Synechococcus sp. strain WH8109 was cultured in ASW medium (as described in “Host growth”) and grown to mid-exponential phase. It was diluted 2-fold in fresh medium the night before the experiment. Cultures with cell densities of 3 × 108 to 7 × 108 cells/ml were used. Only cells with bright yellow fluorescence were counted. A 20-ml aliquot was transferred to a 125-ml Erlenmeyer flask, infected with Syn5 at an MOI of 0.01, and incubated with shaking at 400 rpm on a VWR mini shaker (orbital diameter 3 mm, catalog number 12620-938) in a Percival light chamber. Further phage attachment was interrupted at 14 min after infection by diluting the culture (2 × 104- and 2 × 105-fold) in prewarmed ASW medium in two 125-ml flasks, followed by continued incubation under the same conditions. Samples were collected from the 2 × 104-fold-diluted culture at times from 30 to 70 min and from the 2 × 105-fold-diluted culture at times from 80 to 120 min. At time 0 and after 14 min of infection, samples were collected from the original undiluted flask. Two sets of samples were collected at each time point. To determine the eclipse period, the first set of samples was treated with 1% (vol/vol) chloroform and incubated for at least 1 h before dilution and plating. To define the latent period, the second set of samples was left untreated, held on ice, and plated within a few minutes of collection. The titers of all samples were determined as described above. The experiment was repeated three times, and the mean result for each time point is plotted in Fig. 2.
FIG. 2.
One-step growth curve of Syn5. The two data sets represent samples treated with chloroform (dashed line) and samples without chloroform (solid line) after collection. The cultures (before and after infection) were incubated at 28°C under 45 to 50 μmol m−2 s−1 of light irradiation. Each curve represents average results from three experiments.
The host cells were adapted to grow under continuous light. Their growth stopped in the dark and the cultures did not recover even if illumination was resumed. However, we have yet to study the light dependence of the phage growth cycle.
Cloning, expression, and purification of Syn5 scaffolding protein (gp38).
Syn5 g38 was amplified and NdeI and BamHI restriction sites added at the 3′ and 5′ ends, respectively, by PCR. The product was inserted into cloning vector pET-15b (Novagen), which provides an N-terminal 6×His tag. Following verification of the wild-type gene sequence by full-length DNA sequencing (Harvard MGH DNA Sequencing Core Facility), the construct was transferred into Escherichia coli BL21-Gold (DE3) (Stratagene) for overexpression. The protein was expressed for 3 h at 26°C following induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). One liter of cells was pelleted, resuspended in 30 ml lysis buffer (25 mM Tris, pH 8, 300 mM NaCl, 30 mM imidazole, and 0.1% Triton X-100), and frozen at −80°C. For protein purification, thawed cells were treated with lysozyme for 20 min at room temperature (2 mg/ml) in the presence of protease inhibitors (Complete, Mini, EDTA-free, 1 tablet per 10 ml; Roche) and sonicated (5 times for 1 min each at 9 W), and the extract treated with DNase I (2 U/ml; Worthington) for 1 h at room temperature. The sample was centrifuged at 20,000 × g for 20 min, and the supernatant incubated with Ni-nitrilotriacetic acid (NTA; Qiagen) agarose for 1 h at 4°C. The scaffolding fusion protein was eluted with 25 mM Tris, pH 8, 300 mM NaCl, and 150 mM imidazole. The purity of the elution fractions was confirmed via SDS-PAGE. The protein was dialyzed against 25 mM Tris, pH 7.5, 25 mM NaCl, 2 mM EDTA and stored in the same buffer at 4°C. The protein migrated as a 40-kDa band on SDS-PAGE gel. Liquid chromatography-electrospray MS analysis was performed for molecular mass confirmation of the purified product (Proteomics Core Facility, MIT). The protein was diluted to an appropriate concentration, desalted with a ZipTipC4, and analyzed on a QSTAR Elite mass spectrometer by isocratic elution from a reversed-phase trap with acidified aqueous solvent containing a high proportion of acetonitrile. The spectra generated from the elution were averaged, and the molecular mass spectrum was generated with QSTAR BioTools software. The purified product gave a molecular mass of 32,360 Da, in agreement with the theoretical value of 32,360.8 Da for the expressed fusion product of scaffolding protein, linker peptide, and His tag sequence. This product (2 mg/ml) was used to raise polyclonal antibodies in two New Zealand White rabbits by a standard protocol (Strategic Diagnostics, Inc.).
Time course of Syn5 protein synthesis during infection.
To analyze host and phage proteins during infection, 50 ml of cells was collected at different times after infection, concentrated 100-fold by resuspension in 0.5 ml of lysis buffer (50 mM Tris, pH 8, 100 mM NaCl) at room temperature, and frozen at −20°C. The cells were thawed and treated with lysozyme (2 mg/ml) for 30 min at room temperature, sonicated 5 times for 15 s each, and treated with DNase I (1 U/ml) for 1 h at room temperature. The cell debris was pelleted at 10,000 × g for 10 min, the supernatants were decanted and retained, and the pellets were resuspended in the same volume of resuspension buffer. Pellets and supernatants were diluted 1:2, and the proteins separated by SDS-PAGE (12%) at 100 V for 2 h after boiling in reducing buffer (60 mM Tris, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, bromophenol blue for color) at 80 to 85°C for 10 min. The gels were Krypton stained (Pierce Biotechnology) and visualized on a Typhoon 9400 scanner. For Western blots, the gels were equilibrated in transfer buffer (10% ethanol, 25 mM Tris base, and 192 mM glycine) and the proteins transferred to a polyvinylidene difluoride (PVDF) membrane (0.45 μm; Millipore) overnight in the same transfer buffer at 15 V constant voltage and 4°C (Bio-Rad Criterion transfer apparatus). The membranes were washed in phosphate-buffered saline (PBS)-Tween 20 (0.1% vol/vol) and blocked with 5% Carnation nonfat dry milk in PBS (pH 7.5) overnight at 4°C. The blot-probing steps were as described in the enhanced chemifluorescence (ECF) Western blot manual (Amersham). A mixture of primary rabbit antibodies—anti-Syn5 (Covance), anti-gp53 (Strategic Diagnostics), anti-gp54 (Pacific Immunology), and anti-gp58 (Strategic Diagnostics), all diluted 1:1,000—was used for the membrane probing whose results are shown in Fig. 3b. A 1:1,666 dilution of the anti-scaffold polyclonal antibodies (Strategic Diagnostics) was used to probe the membrane in the experiments whose results are shown in Fig. 4 and 5b. The membrane shown in Fig. 5a was probed with anti-Syn5 antibodies (Covance) diluted 1:2,000. For the experiments whose results are shown in Fig. 3b and 4, the secondary antibodies were alkaline phosphatase (AP)-conjugated goat anti-rabbit antibodies (Bio-Rad) at a dilution of 1:3,000. The secondary antibodies used for the experiments whose results are shown in Fig. 5a and b were AP-conjugated goat anti-rabbit antibodies (Amersham) at a 1:10,000 dilution. The membranes were incubated for 1 h at 4°C with the primary antibodies, washed, and incubated for 45 min at room temperature with the secondary antibodies. For visualization, they were soaked for 10 min in ECF substrate (Amersham) and scanned on a Typhoon 9400 (GE Life Sciences).
FIG. 3.
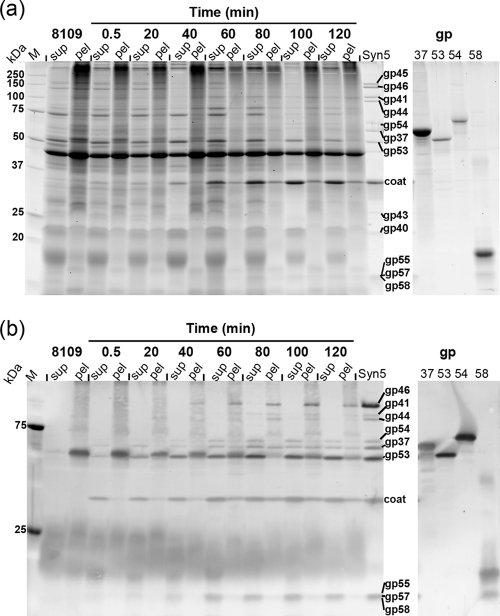
Host and phage proteins during Syn5 infection. (a) SDS-PAGE (12%, Krypton stained) of supernatants (sup) and pellets (pel) of infected cells collected at different time points from 0.5 to 120 min after infection. M, protein standard (Bio-Rad, Precision plus); 8109, uninfected Synechococcus sp. strain WH8109 host cells collected 10 to 20 min before infection; Syn5, infectious particles used in the experiment and run as control for phage structural proteins. The gp (gene product) samples (right panel) contain purified recombinant phage proteins gp37, gp53, gp54, and gp58. Protein annotation: gp45, internal virion protein; gp46, tail fiber protein; gp41, tail tube B; gp44, internal protein; gp54, novel structural protein; gp37, portal; gp53, novel structural protein; coat, capsid protein; gp43, internal virion protein; gp40, tail tube A; gp55, gp57, and gp58, novel structural proteins. (b) Western blot of a gel run in parallel to the one shown in panel a, probed with a mixture of anti-Syn5 polyclonal antibodies and rabbit serum raised against the recombinant forms of gp53, gp54, and gp58.
FIG. 4.
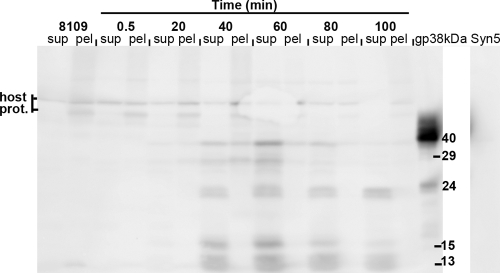
Western blot of time course of Syn5-infected host cells probed with anti-scaffold antibodies. gp38, recombinant scaffolding protein, 40-kDa and 24-kDa bands; 8109, uninfected Synechococcus cells; sup, supernatant fraction; pel, pellet fraction; host prot., host proteins recognized by the anti-scaffold antibodies.
FIG. 5.
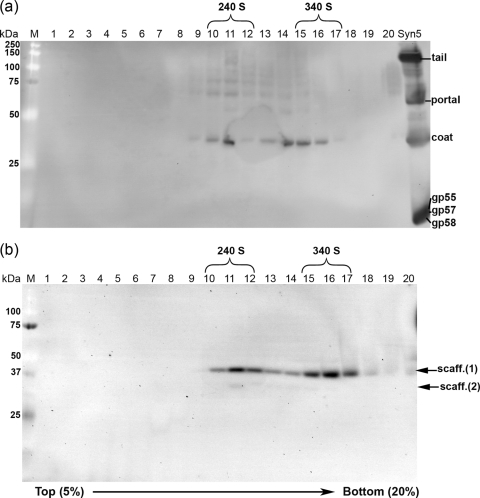
Western blot of fractionated sucrose gradients (fractions 1 to 20, 5-to-20% gradient) of the procapsid band purified in CsCl gradients. (a) Membrane probed with anti-Syn5 antibodies. Syn5, purified infectious particles (control). Fractions 10 to 12 contain the 240 S peak, and fractions 15 to 17 the 340 S peak. (b) Membrane probed with anti-scaffold serum. Scaff. (1), 40-kDa scaffolding protein band; scaff. (2), faint 29-kDa scaffolding band; M, protein standard.
Purification of procapsids.
A Synechococcus sp. strain WH8109 culture (1.4 liters) at 6 × 108 cells/ml in ASW was infected with Syn5 phages at an MOI of 2 to 3. In test experiments, samples were harvested at different times from 20 to 90 min after infection. The optimal procapsid yield was determined to be at 40 to 50 min after infection, and 50 min was used for the results described here. Samples were collected in prechilled bottles and held on ice for 10 min. The cells from 1 liter of culture were pelleted at 7,200 × g for 10 min, and the pellet was resuspended in 4 ml of 50 mM Tris, 100 mM NaCl. The lysis buffer also included protease inhibitors (Complete, Mini, EDTA-free, 1 tablet per 10 ml; Roche). Before infection, a sample of uninfected cells was collected and pelleted at 9,500 × g. All samples were frozen at −20°C.
The cell pellets were thawed and lysed for 40 min with lysozyme (2 mg/ml) at room temperature and treated with DNase I (2 U/ml; Worthington) with gentle mixing on a rocker for 1 h at room temperature. The cell debris was pelleted at 20,000 × g for 10 min at 4°C, and the supernatants were loaded onto a CsCl step gradient. To separate procapsids from mature phages, the following CsCl layers were found to be optimal for 1.5-ml samples concentrated 250-fold in 17.5-ml centrifuge tubes (Beckman): 2 ml with ρ of 1.2, 3.5 ml with ρ of 1.25, 4 ml with ρ of 1.3, 4 ml with ρ of 1.4, 1 ml with ρ of 1.5, and 1 ml with ρ of 1.6. The gradient was spun at 28,000 rpm (SW28 rotor; Beckman) for 3 h at 8°C. Procapsids migrated as an opalescent band at the interface of the layers with ρ of 1.25 and 1.3, whereas the Syn5 DNA-filled particles migrated to the interface of the layers with ρ of 1.4 and 1.5.
The initial CsCl-purified samples were contaminated with outer membrane vesicles. These contained host porins, as determined by mass spectrometry analysis. In early experiments, the vesicles did not separate well from the procapsids on sucrose gradients, and their amounts varied substantially. Mild treatments to lyse infected cells, such as using lysozyme alone and, most importantly, eliminating the sonication step, decreased the concentration of vesicles.
The procapsid band was harvested with a needle and dialyzed against phage buffer (50 mM Tris, pH 7.5, 100 mM MgCl2) with a stepwise decrease of the NaCl concentration as follows: 2 M NaCl (1 h), 1 M NaCl (1 h), and 100 mM NaCl (overnight at 4°C). The procapsids were further purified on a 5-to-20% sucrose gradient at 42,000 rpm (SW55 Ti rotor; Beckman) for 1 h at 8°C. Twenty fractions were collected using a gradient fractionator (BioComp Instruments).
Electron microscopy.
Phage and procapsid samples were blot dried on glow-discharged Formvar/carbon copper grids (Ted Pella) and negatively stained with 1% uranyl acetate. The grids were observed under a JEOL 1200 transmission electron microscope at 60 kV. Images were recorded with an Advanced Microscopy Techniques (AMT) XR41S side-mounted charge-coupled-device (CCD) camera and saved as TIFF files.
RESULTS
One-step growth.
To characterize the growth of Syn5 in Synechococcus sp. strain WH8109, a one-step growth experiment was performed with cells in mid-exponential growth phase in ASW, at 28°C and irradiated under continuous light, at an MOI of 0.01 phages per cell (Fig. 2). The presence of infectious particles was assayed by a plaque assay on WH8109 lawns incubated overnight at 28°C.
Under these experimental conditions (Fig. 2), the first infectious particles appeared about 45 min postinfection (eclipse period; chloroform-treated samples, dashed line) and cell lysis began at around 60 min (latent period; no chloroform, solid line). The phage growth cycle was considerably shorter than the doubling time of the host, which was about 15 h under the same conditions. The growth cycle of Syn5 is the most rapid among cyanophages reported to date (39, 40, 48, 52, 72). The average burst size in the triplicate data shown in Fig. 2 was about 24 infectious particles per cell, five to six times lower than that of the T7 group (69) but comparable to those of other marine cyanophages (72).
Host and phage proteins during infection.
To monitor the dynamics of the major phage structural proteins and major host proteins during infection, Synechococcus sp. strain WH8109 cells were collected by centrifugation at various times after Syn5 infection. The infected cells were lysed and fractionated into supernatant and low-speed pellet fractions, and the resulting proteins analyzed by SDS-PAGE and Western blotting (Fig. 3).
Comparison of the protein patterns from uninfected and infected cells on SDS gels revealed that at least some host proteins remained relatively constant after infection; in particular, the strong bands at 45, 53, and 70 kDa were still present 60 min after infection (Fig. 3a). In addition, the two lower-molecular-mass groups at about 18 and 22 kDa persisted through the infection. The 18-kDa protein band contained the antenna pigments of the host, identified by their color in the gel. In comparing protein levels before and after infection, it should be noted that infected cells lysed more efficiently than uninfected cells under the same conditions.
In the analysis of Syn5 proteins present in the mature virions (Fig. 3a, last lane) the coat protein and at least eight minor bands were visible. In the infected samples, the coat protein was present at 0.5 min after infection in the supernatant fraction. Although the 0.5-min sample was collected immediately following infection, some phage attachment to cells occurred during the centrifugation step. Thus, the phage proteins in the supernatants at early times (0.5 to 20 min after infection) probably reflect the presence of input phage released from cell membranes during lysis. The increase in coat protein at later times presumably represents new protein synthesized in the infected cells. The remaining viral proteins could not be unambiguously identified on the gel, partly due to interference by host proteins.
To more clearly follow the expression of the phage proteins in the infected cells, Western blots were carried out using a mixture of anti-Syn5 serum and polyclonal antibodies raised against the recombinant protein products of novel genes 53, 54, and 58 (Fig. 3b). The anti-Syn5 antibodies, raised against whole-virion particles, would detect epitopes of proteins exposed at the surface of virions. In the Syn5 sample (Fig. 3b, last lane), the coat, tail fiber, and portal were very well recognized, as were several minor proteins—the two tail tubes (gp40 and gp41), the five novel proteins gp53, gp54, gp55, gp57, and gp58, and interestingly, one of the internal proteins, gp44. It may be that gp44 lies in close proximity to the portal in the internal core and, hence, was more exposed to the immune system of the rabbit than the other two internal proteins, gp43 and gp45. All of the virion protein bands were absent from the supernatant of uninfected WH8109 cells, while the pellet contained a protein of unknown function that migrated at a position similar to that of gp53.
As in the SDS-PAGE gel, the phage coat produced the strongest band in the Western blot (Fig. 3b). It was present most abundantly in the early supernatant fraction; however, at 60 min and thereafter, it was recovered in the pellets as well. This may represent aggregated or misfolded coat protein, newly assembled phage adsorbed to cell debris, and particles in unlysed cells. In addition to the coat, most of the other proteins were resolved as of 40 min and clearly visible at 60 min. Other proteins with intensity patterns similar to that of the coat were the portal (gp37) and gp53. gp54 and the three small novel proteins, gp55, gp57, and gp58, were predominantly in the supernatant, clearly present as of 60 min, and increased over time. The tail fiber (gp46) and one of the tail tubes (gp41) were present in the pellets (40 min and later).
In summary, it is evident that as the host proteins were fading, the phage proteins were increasing in intensity.
Expression of the recombinant putative scaffolding protein.
Scaffolding proteins, while required for assembly in dsDNA enteric bacteriophages, are absent from the mature virion. Gene 38 from the Syn5 genome was tentatively designated as the scaffolding protein gene based on sequence homology with genes of T7 and other bacteriophages. If Syn5 assembly is similar to that of the majority of the commonly studied dsDNA bacteriophages, it is to be expected that the scaffolding protein would not be present in the mature virion. As seen in the Western blot (Fig. 3b), no proteins of the size expected for the scaffolding protein were detected using anti-Syn5 antibodies raised against whole virions. To examine whether gp38 is the scaffolding protein, its open reading frame was cloned and expressed and the resulting protein purified.
The product of gene 38 has a theoretical mass of 30.3 kDa and was predicted by Jpred 3 (14) to be an α-helical protein with a single β-sheet in the N-terminal region. The recombinant gp38 His-tagged fusion had a total calculated molecular mass of 32,360.8 Da (excluding the N-terminal Met), which was confirmed by liquid chromatography-electrospray MS analysis. Unexpectedly, it migrated with an apparent molecular mass of approximately 40 kDa (about 30% larger than expected) on 12% SDS-PAGE (see Fig. S1 in the supplemental material). Unboiled and boiled preparations of the protein were electrophoresed to examine whether there would be any change in behavior. However, as seen in Fig. S1 in the supplemental material, its mobility was not affected. Interestingly, this aberrant migration resembles that of the T7 scaffold (10, 58). The two proteins share about 17% amino acid identity, in the central and, especially, in the C-terminal region, when aligned using Clustal W2 (34).
Syn5 assembly involves a scaffolding protein.
To track whether the Syn5 scaffolding protein is expressed during the infection process, polyclonal antibodies were raised against the recombinant gp38 and used to probe the time course of Syn5 infection (Fig. 4). Controls for the specificity of the antibodies were uninfected WH8109 cells, the recombinant protein itself, and Syn5 infectious particles. No bands corresponding to the scaffolding protein were detected in the uninfected WH8109 supernatants, at 0.5 min after Syn5 infection (Fig. 4), or in the mature Syn5 virions (Fig. 4, right panel). The polyclonal serum recognized a few host proteins weakly, but they were predominantly in the pellet fraction and of higher molecular mass than the scaffolding protein. In the recombinant protein control lane, there were two protein products, one with a size of 40 kDa and another at about 24 kDa, possibly a degradation product due to prolonged storage.
The anti-gp38 antibodies recognized five protein bands, predominantly in the supernatant of the infected cells (Fig. 4). Their molecular masses range from 40 to 13 kDa. The 40-kDa band, together with the 29- and the 15-kDa products, were detected as early as 20 min after infection, peaking at 60 min and decreasing at later times. In addition, the other two bands, the 23 and the 13 kDa, appeared later (40 min) and persisted longer. The lower-molecular-mass bands are likely to be degradation products of the 40-kDa scaffolding protein. This may be caused by the scaffold's sensitivity to proteases resistant to inhibitors used in the experiment. Well-studied scaffolding proteins in other bacteriophages have been found to be highly protease sensitive (16).
Do Syn5 virions assemble through a procapsid intermediate?
The evidence for scaffolding protein expression during Syn5 infection implies that the virus proceeds through an assembly pathway involving procapsids empty of DNA, similar to the dsDNA enteric bacteriophages, such as P22, T7, and T4. If scaffolding-associated procapsids are precursors of Syn5 virions, they should form early in the infectious cycle.
In an effort to isolate procapsids, infected host cells were sampled at 30, 40, 50, 60, and 70 min after infection and lysed. To distinguish capsids lacking DNA from DNA-filled virions, the lysate supernatants were loaded onto a CsCl density gradient. In these gradients, a band representing DNA-containing particles was visible between the layers with ρ of 1.4 and 1.5, and a second, bluish-white scattering band was present in the layer with ρ of 1.25. The two bands were resolved clearly. The intensities of both bands increased in samples from later time points. The time chosen as optimal for procapsid purification was 50 min after infection at 28°C. While the intensity of the band in the layer with ρ of 1.25 was stronger at later times (60 to 70 min), SDS gels of these particles indicated significant contamination with ghosts (phages which have ejected or lost their DNA). Earlier times (30 to 40 min) yielded very faint bands in the layer with ρ of 1.25.
To further separate capsids from cell structures, the band in the layer with ρ of 1.25 was recovered and sedimented in a sucrose density gradient (5 to 20%). The gradient was fractionated, and the fractions were electrophoresed through SDS-PAGE and analyzed by Western blotting to assess the presence of virion proteins and the scaffolding protein.
The blot probed with anti-Syn5 antibodies (Fig. 5a) revealed two rapidly sedimenting populations of coat protein structures at 240 S (fractions 10 to 12) and 340 S (fractions 15 to 17). The results also suggest that the portal might be present in both peaks, since a few faint bands are seen closer to the expected size of the portal. However, none of them aligns perfectly with the portal, so further investigation is needed. The presence of the portal is to be expected if the Syn5 assembly pathway is similar to that of other dsDNA phages. The Western blot analysis showed that the structures in both peaks lacked the tail apparatus proteins, as expected. It was not possible to reliably identify whether the other minor internal proteins (gp45, gp44, and gp43) were present in the procapsids.
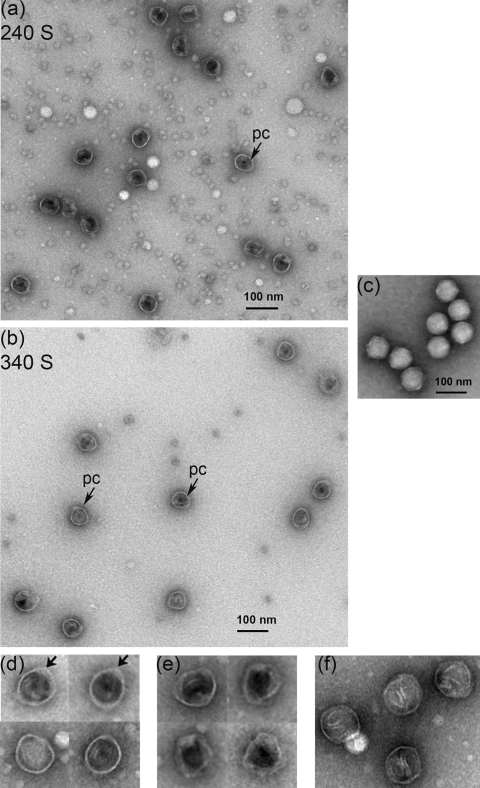
Figure 5b shows the blot probed with anti-scaffold serum. The 40-kDa scaffolding species was present in both peaks. It cosedimented with the coat protein in both peaks (240 S and 340 S). In addition, there was a very faint 29-kDa scaffolding species. The data indicated that the scaffolding protein was associated with the coat in rapidly sedimenting large complexes. The fractions containing both types of particles (240 S and 340 S) were concentrated after the sucrose was dialyzed out, negatively stained with uranyl acetate, and examined under transmission electron microscopy. In both peaks, the images revealed spherical particles about 50 nm in diameter without taillike appendages (Fig. 6a and b). Most were internally electron dense due, presumably, to the presence of the scaffolding protein and will be referred to as procapsids (Fig. 6d). In some of them, the density filled the shells completely, while in others it lined the inner capsid surface, leaving a small empty area in the middle of the shell. Empty capsids lacking internal density and appearing more collapsed, instead of spherical, were present as well (Fig. 6e).
FIG. 6.
EM images of negatively stained Syn5 procapsids. (a) Micrograph of particles in the 240 S sucrose gradient fraction showing procapsid particles (pc, arrow) filled with protein density (most likely the scaffolding protein, making them impermeable to stain) and empty, stain-permeable capsids. (b) Procapsid particles (pc, arrows) in the 340 S fraction were similar to those in the 240 S peak, with diameters of about 50 nm. Empty shells were observed more rarely. (c) Syn5 mature virions for size and shape comparison. (d) Panel of enlarged procapsidlike particles found in both peaks. The arrows indicate areas of higher density at the coat wall. (e) Enlarged empty capsids present in both fractions. (f) Icosahedral capsids found in the 340 S peak. All samples were negatively stained with 1% uranyl acetate and observed at a magnification of ×120,000.
The particles in both peaks were very similar in appearance. The 240 S peak (Fig. 6a) contained about 70% filled procapsids with a regular spherical shape containing internal protein densities, presumably representing the scaffolding protein. About 30% were stain-permeable empty capsids (Table 1). These might be procapsids, which have lost the scaffold within the cells or during purification (31). The 240 S peak also contained a background of stain-excluding 5- to 10-nm structures. We have not yet been able to identify the composition of this material. It may be derived from the extensive membrane system of the cyanobacterial host.
TABLE 1.
Table summarizing the different types of particles in the procapsid peaks (240 S and 340 S)a
| Fraction | Particle species | No. counted | % of total |
|---|---|---|---|
| 240 S | Procapsids | 104 | 70 |
| Empty capsids | 46 | 30 | |
| Icosahedral capsids | 0 | 0 | |
| 340 S | Procapsids | 127 | 85 |
| Empty capsids | 15 | 10 | |
| Icosahedral capsids | 8 | 5 | |
| Total counted | 150 |
Particles were counted from the electron micrographs of each peak and grouped based on their appearance. Figure 6 (d to f) contains enlarged images of each species.
About 85% of the capsids in peak 340 S (Fig. 6b) resembled the filled procapsids in the 240 S peak (Table 1). About 10% resembled empty capsids (Fig. 6e). Particles with a more angular shape (less than 5%) were present (Fig. 6f). Although somewhat icosahedral, they lacked the tightly packed structure of the mature virion and did not exhibit tails.
In some of the procapsid particles, a localized feature of stain-excluding density in the coat shell was visible (Fig. 6d, arrows). This might be the core, composed of the portal and, possibly, the internal virion proteins. Since Syn5 has a horn structure, the density could also be the base of the horn. It could also be the scaffolding leaving the procapsids.
Critical factors for the success of the procapsid purification protocol were the elimination of Triton X-100 in the lysis buffer and lowered NaCl concentration. In the presence of detergent, empty shells of the coat protein were abundant as seen by EM, and no scaffolding protein was present in sucrose gradient fractions with these particles. Thus, in the presence of 0.1% Triton X-100 with 300 mM NaCl, the scaffold was apparently released from the procapsids. When Triton X-100 was eliminated from the lysis buffer and the NaCl concentration was lowered to 100 mM, the particles described above were obtained.
As noted above, the procapsid shells isolated were smaller in size than the 60-nm infectious mature Syn5 virions (Fig. 6c). This suggests that the Syn5 virions may expand after DNA packaging, as in T7, P22, and λ (17).
DISCUSSION
Synechococcus sp. strain WH8109 is a member of marine cluster A, clade II (51), of the Synechococcus photosynthetic family, which is widely distributed in the world's oceans (77). The ancestors of these cyanobacteria are likely to be among the earliest photosynthetic prokaryotes to have evolved in the marine environment. Enteric bacteria, populating the metazoan gut, are believed to represent a later branch of the prokaryotic tree. It is therefore likely that phages of cyanobacteria are ancestral to phages of enteric bacteria.
Syn5 had a life cycle that was remarkably short, with a latent period of 60 min, in comparison with the life cycles of other cyanophages studied under relatively similar experimental conditions (continuous light irradiation in the range of 15 to 50 μmol m−2 s−1 and a temperature of 25 to 30°C). The latent periods of other cyanophages published to date are 9 h for S-PM2 infecting Synechococcus sp. strain WH7803 (72), 7 h for the Nostoc virus N-1 (48), 8.5 h for the AS-1 virus of Synechococcus elongates (52), and 5 and 4 h for the two freshwater cyanophages Pf-WMP3 and Pf-WMP4, respectively (39, 40). The burst size of Syn5 (20 to 30 PFU/cell) was comparable to those reported for marine cyanophage S-PM2. Another property of our system was the extensive lysis of the host cells, accompanied by obvious changes of pigmentation. These features, together with the plaque assay, make Syn5 a very promising model system to study in depth the physiology and molecular biology of the infectious process in cyanophages.
The anomalous electrophoretic migration of the scaffolding protein is not unusual. Such behavior has also been described for the scaffolds of bacteriophages λ (76), the external scaffold for P4 (16), and as mentioned earlier, the scaffolding protein of T7. These three proteins also possess abnormal gel filtration patterns, eluting at higher than expected apparent molecular weights, which may be due to elongated shape. In the case of the Syn5 and T7 scaffolding proteins, both have highly negatively charged sequences with theoretical pI values of 4.16 and 4.31, respectively. Excessive charge has been shown to alter migration (25), and replacing acidic amino acids with basic residues in highly negatively charged proteins can result in the predicted SDS-PAGE migration behavior (2). Another explanation for anomalous migration is the presence of intrinsically disordered regions (27), and a few such sequences were predicted to be present in both proteins by PrDOS (28).
The multiple protein bands detected with anti-scaffold antibodies raise the question of the fate of the scaffolding protein after building the procapsids. It is possible that the Syn5 scaffold is cleaved after phage assembly, as in T4 (6) and λ (24). On the other hand, these degradation products may reflect the unstable nature of scaffolding proteins in general (16) or autoproteolytic activity possessed by gp38. The subunits may be recycled, as is the case for the scaffolding protein of P22 (31).
A distinctive feature of the intracellular assembly of dsDNA phages and viruses is the formation of a procapsid shell, with the assistance of a scaffolding protein and containing a portal vertex, for DNA packaging and ejection (11, 21). Upon DNA packaging into the procapsids, scaffold exits from the procapsids or is proteolysed and is replaced by tightly packed genomic DNA in an expanded mature capsid (8, 67).
For dsDNA phages and viruses, the exit of the scaffolding subunits and the entry of the DNA is a complex process. This process is generally disrupted by cell lysis, resulting in particles which have lost the scaffolding protein but have not yet been stably filled with DNA. These species often have different sedimentation coefficients from the precursor procapsids. This was shown in the analysis of P22, where 240 S precursor procapsids are resolved from 170 S empty capsids, which are derived from intermediates in DNA packaging (32). We presume that the two peaks of Syn5 particles (240 S and 340 S) observed here might be the precursor and a stage further along within the intracellular DNA packaging process. By analogy with P22, it seems that the 240 S peak is the breakdown product and the 340 S fraction is the precursor.
The classes of particles shown in Fig. 6e and f, lacking scaffolding and tightly packed DNA, resemble particles observed in lysates of P22-infected cells. Using pulse-chase experiments, together with mutants blocking late steps in DNA packaging and particle maturation, the P22 particles were shown to be derived from procapsids which had initiated DNA packaging but had not completed it at the time of lysis (32, 57). The Syn5 icosahedral particles (Fig. 6f) may have been derived from capsids that had packaged DNA in vivo, with the assistance of the Syn5 terminase (gp60), and gone through the procapsid-to-capsid lattice transition but had not been stabilized by the addition of the tail proteins that close the portal.
These experiments did not resolve the question of whether the horn of Syn5 is assembled on the procapsid or on the mature virion. These and other details of the assembly pathway are under continuing investigation.
The early appearance of scaffolding-containing procapsids in the infected cells and the absence of the scaffolding protein in mature virions indicate that these procapsids are precursors to the mature virion. The evidence suggests that dsDNA cyanophage Syn5 shares a procapsid assembly pathway similar to the pathway described for the dsDNA bacteriophages (most of them enteric) (8). Given the high frequency of lateral gene transfer among dsDNA phages (7, 23), it is possible that although the evolution of the host cyanobacterium precedes the emergence of enteric bacteria, the phages may have evolved in enterics and then spread in more recent times to cyanobacteria. However, since cyanobacteria precede enteric bacteria in evolution, it is reasonable to propose that the enteric bacteriophages inherited and preserved the assembly pathway originally evolved in the cyanophages.
Supplementary Material
Acknowledgments
We thank John Waterbury for the kind donation of Syn5 and its host strain Synechococcus sp. strain WH8109 and for ongoing advice on laboratory growth conditions. We are grateful to Jeannie Chew and Althea Hill for help with phage production and culture vessel cleaning and maintenance. Many special thanks to Peter Weigele for helpful and inspirational discussions, as well as for notes on the manuscript, to Matt Sullivan for providing advice on cyanophage plaque assays, and to Welkin Pope for the artificial seawater medium recipe. Ioannis Papayannopoulos and Rick Schiavoni at the MIT Biopolymers Core Facility performed the MS analysis.
This work was partially supported by NIH grants GM17980 and AI075208 awarded to J.A.K. and by the Department of Biology, Northeastern University.
Footnotes
Published ahead of print on 22 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, D. J., and A. Roman. 1993. The anomalous electrophoretic behavior of the human papillomavirus type 16 E7 protein is due to the high content of acidic amino acid residues. Biochem. Biophys. Res. Commun. 192:1380-1387. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, J. K., and D. H. Bamford. 1990. Capsomer proteins of bacteriophage PRD1, a bacterial virus with a membrane. Virology 177:445-451. [DOI] [PubMed] [Google Scholar]
- 4.Bazinet, C., and J. King. 1985. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 39:109-129. [DOI] [PubMed] [Google Scholar]
- 5.Becker, B., et al. 1997. Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 268:822-839. [DOI] [PubMed] [Google Scholar]
- 6.Black, L. W., and M. K. Showe. 1983. Morphogenesis of the T4 head, p. 219-245. In C. K. Matthews, E. M. Kutter, G. Mosig, and P. B. Berget (ed.), Bacteriophage T4. American Society for Microbiology, Washington, DC.
- 7.Canchaya, C., G. Fournous, S. Chibani-Chennoufi, M. L. Dillmann, and H. Brussow. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417-424. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S. R., and R. W. Hendrix. 1987. Control mechanisms in dsDNA bacteriophage assembly, p. 15-91. In R. Calender (ed.), The bacteriophages, vol. 1. Plenum Publishing, New York, NY. [Google Scholar]
- 9.Casjens, S. R., and R. W. Hendrix. 1974. Locations and amounts of major structural proteins in bacteriophage lambda. J. Mol. Biol. 88:535-545. [DOI] [PubMed] [Google Scholar]
- 10.Cerritelli, M. E., and F. W. Studier. 1996. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J. Mol. Biol. 258:286-298. [DOI] [PubMed] [Google Scholar]
- 11.Chang, J., P. Weigele, J. King, W. Chiu, and W. Jiang. 2006. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073-1082. [DOI] [PubMed] [Google Scholar]
- 12.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clokie, M. R. J., et al. 2006. Transcription of a “photosynthetic” T4-type phage during infection of a marine cyanobacterium. Environ. Microbiol. 8:827-835. [DOI] [PubMed] [Google Scholar]
- 14.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Halluin, J. C., M. Milleville, P. A. Boulanger, and G. R. Martin. 1978. Temperature-sensitive mutant of adenovirus type 2 blocked in virion assembly: accumulation of light intermediate particles. J. Virol. 26:344-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dokland, T. 1999. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 56:580-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earnshaw, W. C., and S. R. Casjens. 1980. DNA packaging by the double-stranded DNA bacteriophages. Cell 21:319-331. [DOI] [PubMed] [Google Scholar]
- 18.Edvardsson, B., E. Everitt, H. Jornvall, L. Prage, and L. Philipson. 1976. Intermediates in adenovirus assembly. J. Virol. 19:533-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Effantin, G., P. Boulanger, E. Neumann, L. Letellier, and J. F. Conway. 2006. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 361:993-1002. [DOI] [PubMed] [Google Scholar]
- 20.Fane, B. A., and P. E. Prevelige, Jr. 2003. Mechanism of scaffolding-assisted viral assembly. Adv. Protein. Chem. 64:259-299. [DOI] [PubMed] [Google Scholar]
- 21.Fu, C. Y., and P. E. Prevelige, Jr. 2009. In vitro incorporation of the phage Phi29 connector complex. Virology 394:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goericke, R. 1993. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 40:2283-2294. [Google Scholar]
- 23.Hambly, E., and C. A. Suttle. 2005. The viriosphere, diversity, and genetic exchange within phage communities. Curr. Opin. Microbiol. 8:444-450. [DOI] [PubMed] [Google Scholar]
- 24.Hendrix, R. W., and S. R. Casjens. 1975. Assembly of bacteriophage lambda heads: protein processing and its genetic control in petit lambda assembly. J. Mol. Biol. 91:187-199. [DOI] [PubMed] [Google Scholar]
- 25.Hu, C. C., and S. A. Ghabrial. 1995. The conserved, hydrophilic and arginine-rich N-terminal domain of cucumovirus coat proteins contributes to their anomalous electrophoretic mobilities in sodium dodecylsulfate-polyacrylamide gels. J. Virol. Methods 55:367-379. [DOI] [PubMed] [Google Scholar]
- 26.Huiskonen, J. T., H. M. Kivela, D. H. Bamford, and S. J. Butcher. 2004. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat. Struct. Mol. Biol. 11:850-856. [DOI] [PubMed] [Google Scholar]
- 27.Iakoucheva, L. M., et al. 2001. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 10:560-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida, T., and K. Kinoshita. 2007. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35:W460-W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, W., et al. 2008. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature 451:1130-1134. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, J. E., and W. Chiu. 2007. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 17:237-243. [DOI] [PubMed] [Google Scholar]
- 31.King, J., and S. Casjens. 1974. Catalytic head assembling protein in virus morphogenesis. Nature 251:112-118. [DOI] [PubMed] [Google Scholar]
- 32.King, J., E. V. Lenk, and D. Botstein. 1973. Mechanism of head assembly and DNA encapsulation in phage P22. Part 2. Morphogenetic pathway. J. Mol. Biol. 80:697-731. [DOI] [PubMed] [Google Scholar]
- 33.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 34.Larkin, M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 35.Lindell, D., et al. 2007. Genome-wide expression dynamics of a marine virus and host reveal features of evolution. Nature 449:83-86. [DOI] [PubMed] [Google Scholar]
- 36.Lindell, D., J. D. Jaffe, Z. I. Johnson, G. M. Church, and S. W. Chisholm. 2005. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438:86-89. [DOI] [PubMed] [Google Scholar]
- 37.Lindell, D., E. Padan, and A. F. Post. 1998. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J. Bacteriol. 180:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, H. 1998. Prochlorococcus and Synechococcus growth rates and contributions to production in the Arabian Sea during the 1995 southwest and northeast monsoons. Deep Sea Res. Part II. Top. Stud. Oceanogr. 45:2327-2352. [Google Scholar]
- 39.Liu, X., et al. 2008. Genomic analysis of freshwater cyanophage Pf-WMP3 infecting cyanobacterium Phormidium foveolarum: the conserved elements for a phage. Microb. Ecol. 56:671-680. [DOI] [PubMed] [Google Scholar]
- 40.Liu, X., M. Shi, S. Kong, Y. Gao, and C. An. 2007. Cyanophage Pf-WMP4, a T7-like phage infecting the freshwater cyanobacterium Phormidium foveolarum: complete genome sequence and DNA translocation. Virology 366:28-39. [DOI] [PubMed] [Google Scholar]
- 41.Mann, N. H., et al. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187:3188-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann, N. H., A. Cook, A. Millard, S. Bailey, and M. Clokie. 2003. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424:741. [DOI] [PubMed] [Google Scholar]
- 43.McKenna, R., et al. 1992. Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications. Nature 355:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Middelboe, M. 1996. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl. Environ. Microbiol. 62:1991-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millard, A. D., K. Zwirglmaier, M. J. Downey, N. H. Mann, and D. J. Scanlan. 2009. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ. Microbiol. 11:2370-2387. [DOI] [PubMed] [Google Scholar]
- 46.Newcomb, W. W., et al. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newcomb, W. W., et al. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Padhy, R. N., and P. K. Singh. 1977. Effect of temperature on the adsorption and one-step growth of the Nostoc virus N-1. Arch. Microbiol. 115:163-167. [DOI] [PubMed] [Google Scholar]
- 49.Pope, W. H., et al. 2007. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine Synechococcus. J. Mol. Biol. 368:966-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 51.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Safferman, R. S., T. O. Diener, P. R. Desjardins, and M. E. Morris. 1972. Isolation and characterization of AS-1, a phycovirus infecting the blue-green algae, Anacystis nidulans and Synechococcus cedrorum. Virology 47:105-113. [DOI] [PubMed] [Google Scholar]
- 53.Scanlan, D. J. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40:1-12. [DOI] [PubMed] [Google Scholar]
- 54.Scanlan, D. J. 2003. Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Physiol. 47:1-64. [DOI] [PubMed] [Google Scholar]
- 55.Sharon, I., et al. 2009. Photosystem I gene cassettes are present in marine virus genomes. Nature 461:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steven, A. C., J. B. Heymann, N. Q. Cheng, B. L. Trus, and J. F. Conway. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strauss, H., and J. King. 1984. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol. 172:523-543. [DOI] [PubMed] [Google Scholar]
- 58.Studier, F. W., and J. V. Maizel, Jr. 1969. T7-directed protein synthesis. Virology 39:575-586. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan, M. B., M. L. Coleman, P. Weigele, F. Rohwer, and S. W. Chisholm. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan, M. B., et al. 2009. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial “mobilome.” Environ. Microbiol. 11:2935-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 62.Summons, R. E., L. L. Jahnke, J. M. Hope, and G. A. Logan. 1999. 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400:554-557. [DOI] [PubMed] [Google Scholar]
- 63.Suttle, C. A. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801-812. [DOI] [PubMed] [Google Scholar]
- 64.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 65.Tao, Y., et al. 1998. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 67.Tuma, R., P. E. Prevelige, Jr. and G. J. Thomas, Jr. 1998. Mechanism of capsid maturation in a double-stranded DNA virus. Proc. Natl. Acad. Sci. U. S. A. 95:9885-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 69.Wang, W. F., W. Margolin, and I. J. Molineux. 1999. Increased synthesis of an Escherichia coli membrane protein suppresses F exclusion of bacteriophage T7. J. Mol. Biol. 292:501-512. [DOI] [PubMed] [Google Scholar]
- 70.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weigele, P. R., et al. 2007. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ. Microbiol. 9:1675-1695. [DOI] [PubMed] [Google Scholar]
- 72.Wilson, W. H., N. G. Carr, and N. H. Mann. 1996. The effect of phosphate status on the kinetics of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. WH7803. J. Phycol. 32:506-516. [Google Scholar]
- 73.Wilson, W. H., I. R. Joint, N. G. Carr, and N. H. Mann. 1993. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl. Environ. Microbiol. 59:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wyman, M., R. P. Gregory, and N. G. Carr. 1985. Novel role for phycoerythrin in a marine cyanobacterium, Synechococcus strain DC2. Science 230:818-820. [DOI] [PubMed] [Google Scholar]
- 75.Zeidner, G., et al. 2005. Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ. Microbiol. 7:1505-1513. [DOI] [PubMed] [Google Scholar]
- 76.Ziegelhoffer, T., et al. 1992. The purification and properties of the scaffolding protein of bacteriophage lambda. J. Biol. Chem. 267:455-461. [PubMed] [Google Scholar]
- 77.Zwirglmaier, K., et al. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10:147-161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.