Abstract
Segment 8 of the influenza A virus codes for two proteins (NS1 and NS2/NEP) via splicing. Here, we developed a viral vector expressing a cytokine or chemokine instead of the interferon antagonist NS1. To achieve both the desired genetic stability and high transgene expression levels, NS2/NEP mRNA splicing efficacy had to be fine-tuned by modification of splicing elements. Expression levels of secreted foreign proteins could be further enhanced by fusing the N-terminal 13 amino acids of NS1 with an IgK-derived secretion signal peptide. Thus, the first start codon was used for translation initiation of both NS2/NEP and the foreign protein.
Genetic engineering allowed the rational design of influenza A virus vaccine prototypes based on partial or complete deletions (4, 7, 30) of the viral interferon (IFN) antagonist nonstructural protein 1 (NS1) (7).
A recently completed clinical phase I trial of a vaccine using the complete NS1 deletion (delNS1) virus confirmed its excellent safety profile for humans after a single intranasal application (31). These clinical data, as well as the fact that influenza viruses lack a DNA intermediate in their replication cycle, render the delNS1 virus attractive for further development as a viral expression vector.
Several bicistronic strategies have been explored for the expression of foreign genes from the segmented genome of the influenza A virus. Those include internal promoters (13), minigenes (24), internal ribosomal entry sites (8), protease cleavage sites (11, 14, 22, 28), and a stop-start translation reinitiation sequence (UAAUG) (10; for a review, see reference 16).
Here, we developed a strategy to express cytokines or chemokines from the genome of the delNS1 virus. In our initial construct (pdelNS1-IL-2-spl-wt), the human interleukin-2 (IL-2) open reading frame (ORF) including the signal peptide preceded by the stop-start pentamer UAAUG was inserted between the splice donor and the splice acceptor site of the nonstructural protein (NS) segment (Fig. 1b). The NS1 protein is thereby truncated to the N-terminal 21 amino acids, rendering it nonfunctional. Mutations were introduced by standard molecular biology techniques (e.g., restriction enzyme digests, PCR mutagenesis, overlapping PCR) into a PolI promoter-driven plasmid coding for influenza virus A/PuertoRico/8/34 (PR8) segment 8 and confirmed by sequencing. We rescued the corresponding virus, delNS1-IL-2-spl-wt, by transfecting Vero cells with the plasmid pdelNS1-IL-2-spl-wt together with seven bidirectional plasmids (9) coding for the remaining segments of a Vero cell-adapted IVR-116 (20) strain.
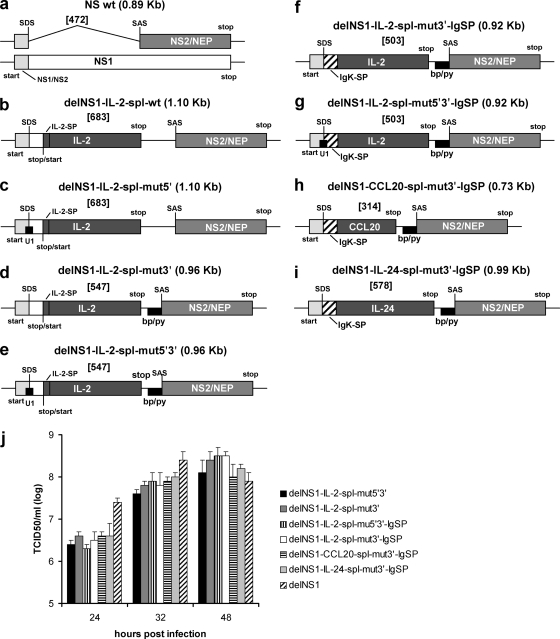
FIG. 1.
(a to i) Schematic representation of chimeric NS segments and the wild-type NS segment of the PR8 virus. For the wild-type NS segment, both the NS1 and the NS2/NEP transcript are shown. The names and segment sizes (in parentheses) are given above each construct. Intron sizes are given in brackets. Start and stop codons and the start/stop pentamer are indicated. wt, wild type; NS1, nonstructural protein 1; NS2/NEP, nonstructural protein 2/nuclear export protein; IgK-SP, partial mouse Ig kappa signal peptide; IL-2-SP, human IL-2 signal peptide; IL-2/CCL20/IL-24, ORF of either human IL-2, human CCL20, or human IL-24; U1, the viral CAG/GUAGAUUG sequence was changed to achieve high complementarity to the human U1 snRNA, CAG/GUAAGUAU for delNS1-IL-2-spl-mut5′ virus and delNS1-IL-2-spl-mut5′3′ virus or CAG/GUAAGUCU for delNS1-IL-2-spl-mut5′3′-IgSP virus (the 5′ splice junction is indicated with /, and nucleotides complementary to the U1 snRNA are underlined); bp/py, branch point sequence plus 20-nucleotide pyrimidine stretch; SDS, splice donor site; SAS, splice acceptor site. Nucleotide sequences are available upon request. (j) Growth curves of delNS1 virus and chimeric viruses in Vero cells. Vero cells were infected at a multiplicity of infection (MOI) of 0.001, and supernatants harvested at the indicated times were analyzed for infectious titers by a 50% tissue culture infective dose (TCID50) assay on Vero cells.
Surprisingly, reverse transcription-PCR (RT-PCR) (Fig. 2) and sequence analysis (data not shown) demonstrated that a major part of the introduced IL-2 sequence was already deleted from the virus after one passage in Vero cells, despite the fact that IL-2 had been stably expressed previously from the partial NS1 deletion mutant NS1-125 (10).
FIG. 2.
RT-PCR analysis of passaged stop-start delNS1-IL-2 viruses demonstrating the genetic stability of splice sequence mutants. Chimeric viruses obtained from transfection were serially passaged in Vero cells. RT-PCR was carried out from viral RNA isolated from cell culture supernatants by using oligonucleotides homologous to the NS segment. PCR controls were included by using the respective chimeric delNS1 virus plasmid DNAs as templates (p). PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Viruses and the number of passages are indicated on the top. The sizes of the marker bands are given in bp. wt, delNS1-IL-2-spl-wt virus; mut5′, delNS1-IL-2-spl-mut5′ virus; mut3′, delNS1-IL-2-spl-mut3′ virus; mut5′3′, delNS1-IL-2-spl-mut5′3′ virus; M, marker; 1, 1st passage; 5, 5th passage; p, plasmid control.
We hypothesized that the introduction of a foreign sequence at this position of the NS segment might compromise NS mRNA splicing due to an increased intron size (19), an interfering RNA secondary structure, or both. We therefore constructed three additional IL-2-expressing chimeric segments aiming to enhance NS mRNA splicing efficacy (Fig. 1c to e); we exchanged the sequence surrounding the splice donor site toward full complementarity to the 5′ end of the human U1 snRNA (5) in plasmid p-delNS1-IL-2-spl-mut5′, replaced 168 nucleotides upstream of the splice acceptor site (including the natural branch point [23] and the pyrimidine-rich region) with a synthetic sequence comprising a 20-nucleotide pyrimidine stretch directly preceded by branch point sequence (26) in plasmid p-delNS1-IL-2-spl-mut3′, or combined both of these modifications in plasmid p-delNS1-IL-2-spl-mut5′3′.
Following virus rescue, we again analyzed the genetic stability of the chimeric NS segments by RT-PCR and sequence analysis. For the delNS1-IL-2-spl-mut5′ virus, progeny virus deletions within the IL-2 ORF were observed after five passages (Fig. 2; sequencing data not shown). In contrast, the delNS1-IL-2-spl-mut3′ and delNS1-IL-2-spl-mut5′3′ viruses appeared genetically stable for up to 10 passages (Fig. 2 and data not shown). These two viruses replicated in Vero cells to titers similar to those of the IVR-116 delNS1 virus (Fig. 1j).
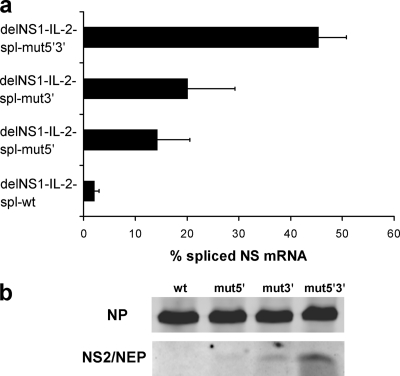
To analyze the effect of splice site modifications on splicing efficacy, we used a transient replication/transcription system (6). A PolI promoter-driven plasmid coding for the respective delNS1-IL-2 virus segment was cotransfected into Vero cells together with four bidirectional plasmids (9) coding for the viral ribonucleoprotein (RNP) complex proteins PA, PB1, PB2, and NP. Steady-state levels of spliced NS mRNA as assessed by real-time PCR for the delNS1-IL-2-spl-wt virus construct were only 3% (Fig. 3a). This was below the range of 5 to 15% described for wild-type NS (12, 25) and thus likely too low to enable sufficient NS2/nuclear export protein (NEP) expression for efficient viral growth. Modifying the 5′ splice site or the 3′ splice elements clearly increased the steady-state levels of spliced NS mRNA, with the splicing efficacy being the highest for the construct that contains both modifications (Fig. 3a). NS2/NEP protein levels as analyzed by immunoblotting paralleled NS mRNA splicing efficacies for the four constructs (Fig. 3b).
FIG. 3.
(a) Real-time PCR analysis indicating the percentage of spliced NS mRNA in an in vitro replication/transcription system. Vero cells were transfected with bidirectional plasmids (9) coding for the viral RNP proteins and a plasmid coding for the indicated chimeric delNS1-IL-2 virus segment containing either wild-type or mutated splice sequences. After 24 h, RNA was extracted from transfected cells, reverse transcribed, and analyzed for spliced and unspliced NS mRNA steady-state levels by real-time PCR using a SYBR green detection system (Applied Biosystems). For absolute quantitation, suitable serial dilutions of pcDNA-NEP (for spliced mRNA) and pHW-PR8-NS (for unspliced mRNA) were used as standards. All PCRs were run in triplicate. The amplification of single bands was verified by agarose gel electrophoresis and melting curve analysis. The values were calculated as the percentage of spliced NS mRNA and are means ± standard deviations from three independent experiments. (b) NS2/NEP protein expression parallels NS mRNA splicing efficacy. Vero cells were transfected as described for panel a and harvested 24 h posttransfection. Whole-cell extracts were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis and analyzed by immunoblotting using a polyclonal NS2/NEP-specific antiserum (unpublished data) and, as a control, a blend of two monoclonal NP-specific antibodies (Millipore). Splice site mutations are indicated on the top. wt, wild type.
Corresponding to the higher percentage of unspliced NS mRNA, IL-2 protein levels in supernatants of infected Vero cells as measured by an enzyme-linked immunosorbent assay (ELISA) were found to be about 10 times higher for the delNS1-IL-2-spl-mut3′ virus than for the delNS1-IL-2-spl-mut5′3′ virus (Fig. 4). No IL-2 was found in supernatants of Vero cells infected with the delNS1 virus. The biological activity of IL-2, as assessed by a CTLL-2 cell proliferation assay (3), was found to be in the range of 5 to 8 U/ng IL-2 for both genetically stable viruses, delNS1-IL-2-spl-mut3′ and delNS1-IL-2-spl-mut5′3′.
FIG. 4.
Absolute IL-2 levels in the supernatants of virus-infected Vero and MDCK cells as determined by ELISA. (a) Vero cells were infected with a genetically stable stop-start or an NS1-IgK signal peptide delNS1-IL-2 virus or with delNS1 virus as a control (MOI, ∼0.01), as indicated on the left, and incubated at 37°C for 12 to 16 h in the presence of 1 μg/ml trypsin to permit limited viral replication. Fetal calf serum (FCS) was added to a final concentration of 10% (vol/vol), and incubation was continued for another 24 h. IL-2 levels are given on a logarithmic scale as the means ± standard deviations from three independent experiments. (b) Vero cells or MDCK cells were infected with the indicated virus (MOI, ∼1) and incubated at 37°C for 2 h, followed by the addition of 10% (vol/vol) FCS to prevent viral replication. Supernatants were collected 24 h later and analyzed for IL-2.
The exchangeability of more- and less-effective splice sequences enables the fine-tuning of splicing efficacy, which eases viral genetic engineering. While the mutagenesis of the 3′ splice elements was sufficient to achieve genetic stability of chimeric delNS1-IL-2 segments, larger inserts may require the additional modification of the 5′ splice site.
In contrast to our delNS1-IL-2 virus constructs, Kittel and coworkers could stably express foreign sequences from a partial NS1 deletion mutant virus without the modification of splice sequences (10). This might be due to (i) differences in polymerase proteins, which have been shown to be involved in splicing (29), as Kittel et al. used PB1 derived from the PR8 virus strain, whereas we used PB1 from influenza virus A/Texas/ 1/77 (20), (ii) the fact that the partial deletion protein NS1-125 (10) may enhance the translation of NS2/NEP (1, 6, 15, 27) and thereby compensate for the reduced splicing efficacy caused by the insertion of the foreign ORF, or (iii) an mRNA secondary structure of the delNS1-IL-2 virus construct that interferes with splicing.
We next intended to fuse IL-2 to the truncated NS1 protein to allow for the usage of the first start codon, which should further enhance IL-2 expression compared to that seen with translation reinitiation via the stop-start pentamer (10). Classical secretion signal peptides are characterized by a positively charged region at the N terminus followed by a stretch of hydrophobic amino acids and a more polar region at the C terminus. In eukaryotes, the positive charge is often provided by the free N terminus. Since NS1 is an intracellular protein, its N-terminal part does not meet the requirements of a signal peptide. Accordingly, a viral vector that contained the 3′ splice modification and mature IL-2 (i.e., without its signal peptide) directly fused to the N-terminal 13 amino acids of NS1 was genetically stable but did not express IL-2 (data not shown). Thus, to allow IL-2 secretion, the truncated NS1 protein needed to be functionally converted into a signal peptide.
To achieve this, we fused the N-terminal 13 amino acids of NS1 to the hydrophobic core plus polar C terminus of the murine IgK signal peptide, followed by mature IL-2, resulting in the delNS1-IL-2-spl-mut3′-IgSP virus construct (Fig. 1f). The truncated NS1 thereby effectively replaces the N-terminal six amino acids of the murine IgK signal peptide. We evaluated the construct design with the SignalP 3.0 signal peptide prediction server (Technical University of Denmark) (2) in order to test whether this NS1-IgK hybrid amino acid sequence would be recognized as a signal peptide. Both models (2) predicted a high probability (neural network D score [2], 0.931 [cutoff, 0.43]; hidden Markov model P value, 0.999) that the NS1-IgK chimera would act as a signal peptide for IL-2 and also predicted the correct cleavage site of the signal peptide. Changing NS1 amino acid positions 12 and 13 to achieve higher complementarity to human U1 snRNA (delNS1-IL-2-spl-mut5′3′-IgSP virus construct) (Fig. 1g) did not significantly alter the outcome of the SignalP 3.0 predictions (data not shown).
Upon rescue, both viruses, delNS1-IL-2-spl-mut3′-IgSP and delNS1-IL-2-spl-mut5′3′-IgSP, proved to be genetically stable for at least 10 passages, as assessed by RT-PCR and sequencing (data not shown). Replication in Vero cells was similar to that observed for the progenitor viruses (Fig. 1j). For both viruses, the IL-2 levels in the supernatants of infected Vero cells were about 10-fold higher than those for their respective stop-start counterparts (Fig. 4) and more than 100 times higher than those measured for the previously described IL-2-expressing partial NS1 deletion mutant (10) (data not shown). The biological activity of IL-2 for both viruses as assessed by a CTLL-2 assay was found to be in the range of 5 to 6 U/ng IL-2 (data not shown).
Thus, the chimeric NS1-IgK signal peptide allows for high-level secretion of biologically active IL-2. To the best of our knowledge, this is the first time that the N-terminal part of an intracellular protein has been fused to a signal peptide to permit the high-level expression of a secretory protein.
To assess whether IL-2 secretion occurs via the classical endoplasmic reticulum (ER)-Golgi pathway, Tu-16 suspension hepatoma cells (18) were infected with the delNS1-IL-2-spl-mut3′-IgSP virus and incubated in the presence or absence of the trans-Golgi inhibitor monensin (17, 21). Following intracellular staining with an antibody specific for human IL-2, cells were analyzed by flow cytometry. The presence of monensin in the medium clearly enhanced the mean IL-2-specific fluorescence (data not shown), indicating that IL-2 is indeed secreted via the ER-Golgi pathway.
To test whether other secretory proteins can be expressed via the NS1-IgK fusion signal peptide and modified splice sites, the IL-2 ORF in the delNS1-IL-2-spl-mut3′-IgSP virus was replaced with cDNAs coding for either the human cytokine IL-24 or the human chemokine CCL20. Signal peptide sequences were optimized via SignalP 3.0 server prediction models. Both delNS1-CCL20-spl-mut3′-IgSP and delNS1-IL-24-spl-mut3′-IgSP viruses (Fig. 1h and i) were found to be genetically stable for up to 10 passages (data not shown). Substituting CCL20 or IL-24 for IL-2 did not alter the viral growth properties in Vero cells (Fig. 1j). Analysis of the supernatants of infected Vero cells by ELISA demonstrated that CCL20 was expressed at 27.3 ± 6.5 ng/ml and IL-24 at 7.7 ± 1.2 ng/ml. Neither CCL20 nor IL-24 was detected in the supernatants of cells infected with the delNS1 virus.
To assess transgene expression in an IFN-competent host, MDCK cells were infected with all four genetically stable delNS1-IL-2 viruses. IL-2 expression levels in MDCK cells were approximately 10-fold lower than those in Vero cells infected under the same conditions (Fig. 4b). This is in accordance with a previous report demonstrating lower levels of viral protein synthesis for delNS1 virus in MDCK cells than in Vero cells (7).
The influenza virus vector newly described herein combines a clinically acceptable vaccine virus profile with an optimized level of transgene expression. Site-directed mutagenesis of signal peptides and the careful adjustment of splicing efficacy allowed an optimal balance between high transgene expression levels and the genetic stability of the chimeric NS segments. Practical applications for delNS1 virus-based expression vectors encompass: (i) expression of proinflammatory cytokines to enhance the immunogenicity of a delNS1 virus-based viral vaccine, (ii) expression of a proinflammatory cytokine for the use of delNS1 virus as an armed oncolytic virus, and (iii) expression of foreign antigens to allow the use of the delNS1 virus as a vaccine vector for pathogens other than influenza virus.
Acknowledgments
This work was supported by EU grant 512864, “Chimeric Vaccines.”
We thank Erich Hoffmann for kindly providing plasmid pHW2000.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1.Aragon, T., et al. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Davis, L. S., P. E. Lipsky, and K. Bottomly. 2001. Measurement of human and murine interleukin 2 and interleukin 4. Curr. Protoc. Immunol. 2001:Unit 6.3. [DOI] [PubMed] [Google Scholar]
- 4.Egorov, A., et al. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freund, M., et al. 2005. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 33:5112-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garaigorta, U., and J. Ortin. 2007. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 35:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Sastre, A., et al. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 8.García-Sastre, A., T. Muster, W. S. Barclay, N. Percy, and P. Palese. 1994. Use of a mammalian internal ribosomal entry site element for expression of a foreign protein by a transfectant influenza virus. J. Virol. 68:6254-6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kittel, C., et al. 2005. Generation of an influenza A virus vector expressing biologically active human interleukin-2 from the NS gene segment. J. Virol. 79:10672-10677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kittel, C., et al. 2004. Rescue of influenza virus expressing GFP from the NS1 reading frame. Virology 324:67-73. [DOI] [PubMed] [Google Scholar]
- 12.Lamb, R. A., P. W. Choppin, R. M. Chanock, and C. J. Lai. 1980. Mapping of the two overlapping genes for polypeptides NS1 and NS2 on RNA segment 8 of influenza virus genome. Proc. Natl. Acad. Sci. U. S. A. 77:1857-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machado, A. V., N. Naffakh, S. van der Werf, and N. Escriou. 2003. Expression of a foreign gene by stable recombinant influenza viruses harboring a dicistronic genomic segment with an internal promoter. Virology 313:235-249. [DOI] [PubMed] [Google Scholar]
- 14.Manicassamy, B., et al. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U. S. A. 107:11531-11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marion, R. M., T. Aragon, A. Beloso, A. Nieto, and J. Ortin. 1997. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 25:4271-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Sobrido, L., and A. García-Sastre. 2007. Recombinant influenza virus vectors. Future Virol. 2:401-416. [Google Scholar]
- 17.Mollenhauer, H. H., D. J. Morre, and L. D. Rowe. 1990. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim. Biophys. Acta 1031:225-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muster, T., et al. 2004. Interferon resistance promotes oncolysis by influenza virus NS1-deletion mutants. Int. J. Cancer 110:15-21. [DOI] [PubMed] [Google Scholar]
- 19.Nemeroff, M. E., U. Utans, A. Kramer, and R. M. Krug. 1992. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol. Cell. Biol. 12:962-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolson, C., D. Major, J. M. Wood, and J. S. Robertson. 2005. Generation of influenza vaccine viruses on Vero cells by reverse genetics: an H5N1 candidate vaccine strain produced under a quality system. Vaccine 23:2943-2952. [DOI] [PubMed] [Google Scholar]
- 21.Nylander, S., and I. Kalies. 1999. Brefeldin A, but not monensin, completely blocks CD69 expression on mouse lymphocytes: efficacy of inhibitors of protein secretion in protocols for intracellular cytokine staining by flow cytometry. J. Immunol. Methods 224:69-76. [DOI] [PubMed] [Google Scholar]
- 22.Percy, N., W. S. Barclay, A. García-Sastre, and P. Palese. 1994. Expression of a foreign protein by influenza A virus. J. Virol. 68:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plotch, S. J., and R. M. Krug. 1986. In vitro splicing of influenza viral NS1 mRNA and NS1-beta-globin chimeras: possible mechanisms for the control of viral mRNA splicing. Proc. Natl. Acad. Sci. U. S. A. 83:5444-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Restifo, N. P., et al. 1998. Transfectant influenza A viruses are effective recombinant immunogens in the treatment of experimental cancer. Virology 249:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robb, N. C., D. Jackson, F. T. Vreede, and E. Fodor. 2010. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J. Gen. Virol. 91:2331-2340. [DOI] [PubMed] [Google Scholar]
- 26.Ruskin, B., A. R. Krainer, T. Maniatis, and M. R. Green. 1984. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38:317-331. [DOI] [PubMed] [Google Scholar]
- 27.Salvatore, M., et al. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sereinig, S., et al. 2006. Influenza virus NS vectors expressing the mycobacterium tuberculosis ESAT-6 protein induce CD4+ Th1 immune response and protect animals against tuberculosis challenge. Clin. Vaccine Immunol. 13:898-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih, S. R., M. E. Nemeroff, and R. M. Krug. 1995. The choice of alternative 5′ splice sites in influenza virus M1 mRNA is regulated by the viral polymerase complex. Proc. Natl. Acad. Sci. U. S. A. 92:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talon, J., et al. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wacheck, V., et al. 2010. A novel type of influenza vaccine: safety and immunogenicity of replication-deficient influenza virus created by deletion of the interferon antagonist NS1. J. Infect. Dis. 201:354-362. [DOI] [PubMed] [Google Scholar]