Abstract
Paramyxovirus matrix (M) proteins organize virus assembly, linking viral glycoproteins and viral ribonucleoproteins together at virus assembly sites on cellular membranes. Using a yeast two-hybrid screening approach, we identified 14-3-3 as a binding partner for the M protein of parainfluenza virus 5 (PIV5). Binding in both transfected and PIV5-infected cells was confirmed by coimmunoprecipitation and was mapped to a C-terminal region within the M protein, namely, 366-KTKSLP-371. This sequence resembles known 14-3-3 binding sites, in which the key residue for binding is a phosphorylated serine residue. Mutation of S369 within the PIV5 M protein disrupted 14-3-3 binding and improved the budding of both virus-like particles (VLPs) and recombinant viruses, suggesting that 14-3-3 binding impairs virus budding. 14-3-3 protein overexpression reduced the budding of VLPs. Using 33P labeling, phosphorylated M protein was detected in PIV5-infected cells, and this phosphorylation was nearly absent in cells infected with a recombinant virus harboring an S369A mutation within the M protein. Assembly of the M protein into clusters and filaments at infected cell surfaces was enhanced in cells infected with a recombinant virus defective in 14-3-3 binding. These findings support a model in which a portion of M protein within PIV5-infected cells is phosphorylated at residue S369, binds the 14-3-3 protein, and is held away from sites of virus budding.
Paramyxovirus infections are transmitted in the form of particles which bud from the surfaces of virus-infected cells. Viral ribonucleoproteins (RNPs), viral glycoproteins, and internal viral proteins all accumulate together at sites on plasma membranes from which budding will take place. Coordination among the different viral components during this assembly process is necessary to ensure that a reasonable fraction of particles will contain all of the components necessary for proper infectivity. Matrix (M) proteins are the key coordinators of paramyxovirus assembly (reviewed in references 6, 31, and 37). M proteins act as adapters, linking together viral glycoprotein spikes, via their cytoplasmic tails, and viral RNPs, inducing these components to coalesce at specific locations on infected cell plasma membranes.
Parainfluenza virus 5 (PIV5; formerly known as SV5) is a paramyxovirus belonging to the Rubulavirus genus, which also includes mumps virus (MuV), human parainfluenza virus types 2 and 4, Tioman virus, and Menangle virus (15). Like other paramyxoviruses, PIV5 has a genome of negative-sense single-stranded RNA that is tightly associated with viral nucleocapsid (NP) proteins to form viral RNPs. RNPs act as templates for viral RNA-dependent RNA polymerases, which are made up of viral large protein (L) and phosphoprotein (P) subunits. RNPs are packaged into membrane-enveloped particles, which are released from host cells by budding from infected cell plasma membranes. Embedded within the virion envelopes are the viral glycoproteins, densely packed to form spike layers that are visible by electron microscopy. The hemagglutinin-neuraminidase (HN) glycoproteins provide an attachment function, binding to sialic acid receptors on target cells, and also function as sialidases to facilitate the separation of newly formed particles from host cell membranes. Fusion (F) glycoproteins direct the merging together of viral and cellular membranes at neutral pH to allow virus entry. M proteins organize the assembly and budding of virus particles, and the viral V and small hydrophobic (SH) proteins disable interferon and apoptotic signaling pathways within infected cells (8, 10).
Although M proteins are the key organizers of paramyxovirus assembly and many paramyxovirus M proteins can direct the budding of virus-like particles (VLPs) when expressed alone in cells (reviewed in reference 6), the M protein of PIV5 lacks the ability to induce VLP production when it is expressed alone. Cooperation among different PIV5 structural components, including glycoproteins as well as nucleocapsid structures, is necessary for efficient release of PIV5-like particles (33). Similar requirements for particle formation have been defined for mumps virus, as efficient mumps VLP production requires coexpression of the viral M, NP, and F proteins together in cells (16).
Recruitment of host factors is a key step in the budding of many enveloped viruses. Several retroviruses use late domains within their Gag proteins to recruit and manipulate host factors that normally function to allow the formation of multivesicular bodies (reviewed in references 1, 2, 4, and 5). Some negative-strand RNA virus matrix proteins contain the same late domain sequences as those found in retroviral Gag proteins, suggesting that in some cases the fundamental mechanisms of virus budding are conserved even among distantly related viruses (7, 25). Although paramyxovirus M proteins lack classical late domains, the sequence FPIV within the PIV5 M protein was shown to be capable of functioning as a late domain, as it restored budding function to a PTAP-disrupted HIV-1 Gag protein (32).
Several efforts have been undertaken to identify and characterize host factors that bind PIV5 M protein to allow particle formation and release. Angiomotin-like 1 was recently shown to bind M protein and to facilitate virus particle production, although binding was independent of FPIV (24). M protein association with caveolin-1 (Cav-1) has been demonstrated, and this interaction may serve to facilitate the concentration of viral proteins at caveolae prior to virus budding (27).
In this study, we identified an additional host factor, 14-3-3, which binds to the PIV5 M protein. This interaction was found to negatively influence the budding function of M protein. 14-3-3 binding was dependent on M protein residue S369, which is likely the major target of M protein phosphorylation in infected cells. Using fluorescence microscopy, we found that assembly of M protein into clusters and filaments was enhanced when 14-3-3 interaction was impaired. Our results suggest that a portion of the M protein in PIV5-infected cells is phosphorylated and bound to 14-3-3, to the detriment of virus assembly.
MATERIALS AND METHODS
Plasmids.
Plasmids pCAGGS-PIV5 NP, pCAGGS-PIV5 M, and pCAGGS-PIV5 HN have been described before (33), as have plasmids pCAGGS-MuV M and pCAGGS-NiV M (16). Altered PIV5 and MuV M cDNAs were generated by PCR mutagenesis of the wild-type (wt) sequences. Plasmids pHybLex-PIV5 M-Zeo and pHyb-PIV5 M-LexZeo, used for yeast two-hybrid library screening, have been described previously (24). To generate additional bait plasmids for yeast two-hybrid pairwise tests, subfragments of PIV5 M cDNA were amplified by PCR, with the boundaries illustrated in Fig. 1, and subcloned into the plasmid pHybLexZeo (Invitrogen, Carlsbad, CA). cDNA corresponding to full-length human 14-3-3 beta was obtained from a yeast clone isolated from a premade HeLa cell-derived yeast two-hybrid cDNA library (Invitrogen). This sequence was modified by PCR to generate an N-terminal Flag tag (amino acid sequence DYKDDDDK) and was subcloned into the eukaryotic expression vector pCAGGS (20) to generate the plasmid pCAGGS-Fl.14-3-3. A cDNA corresponding to mutant 14-3-3 beta (K51E, R58E, and R62E mutations) (3) was kindly provided by Andrey Sorokin. This sequence was modified by PCR to generate an N-terminal Flag tag and was subcloned into pCAGGS to generate pCAGGS-Fl.14-3-3mut. All PCR-amplified cDNAs were sequenced to verify their identities (Macrogen, Inc., South Korea).
FIG. 1.
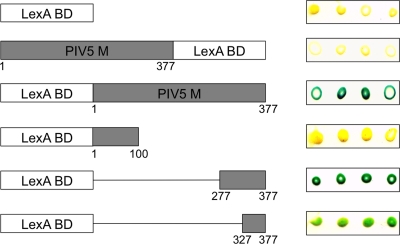
Interaction between PIV5 M protein and 14-3-3, characterized with a pairwise yeast two-hybrid assay. A prey plasmid corresponding to the YWHAB gene (encoding the full-length 14-3-3 beta protein) was used to transform yeast cells (strain L40). Yeast cells were transformed a second time with bait plasmids encoding PIV5 M protein or its derivatives, as indicated. Numbers indicate the span of M protein amino acid residues present in each construct. Transformed yeast cells were spotted onto a nitrocellulose membrane in replicates of four and then disrupted with a series of freeze-thaw cycles, and beta-galactosidase activity was detected using a colorimetric assay. BD, binding domain.
Antibodies.
Monoclonal antibodies (MAbs) M-h, HN1b, and NP-a, specific to the PIV5 M, HN, and NP proteins, respectively, have been described before (26) and were kind gifts of Richard Randall (St. Andrews University, St. Andrews, Scotland, United Kingdom). Rabbit polyclonal antibodies specific to the mumps virus and Nipah virus (NiV) M proteins have been described previously (16). A monoclonal antibody specific to the Flag tag (clone M2) was obtained from Stratagene (La Jolla, CA). A mouse monoclonal antibody specific to 14-3-3 beta, used for fluorescence microscopy experiments, was purchased from Novus Biologicals (Littleton, CO).
Yeast two-hybrid library screening and pairwise assays.
The Hybrid Hunter yeast two-hybrid system (Invitrogen) was used to identify PIV5 M-interacting host proteins from a HeLa cell-derived cDNA library by use of previously described methods (24) and based on the manufacturer's recommendations. Baits were expressed using Saccharomyces cerevisiae strain L40, which harbors two LexA-driven reporters, histidine and beta-galactosidase (Invitrogen). Transformants that tested positive for growth on plates lacking histidine were further screened for beta-galactosidase expression by use of a colony filter assay as instructed by the manufacturer (Invitrogen). For pairwise tests, prey plasmids isolated from His+ beta-galactosidase+ yeast transformants were retransformed together with bait plasmids into fresh L40 yeast cells. Colonies of transformed cells were assayed in replicates of four for beta-galactosidase expression, using the colony filter assay.
Coimmunoprecipitation.
293T cells in 6-cm-diameter dishes (70 to 80% confluent), grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), were transfected with 0.4 μg/dish of pCAGGS plasmid encoding PIV5 M protein, MuV M protein, or NiV M protein, together with the plasmid pCAGGS-Fl.14-3-3 (1.0 μg/dish). Transfections were carried out in Opti-MEM, using Lipofectamine-Plus reagents (Invitrogen). For coimmunoprecipitation from virus-infected cells, 293T cells were infected with PIV5 at a multiplicity of infection (MOI) of 1.0 PFU/cell. At 6 h postinfection (p.i.), the cells were transfected with pCAGGS-Fl.14-3-3 (1.0 μg/dish). At 16 h posttransfection (p.t.), cells were metabolically labeled with 40 μCi of [35S]Promix/ml (Perkin Elmer, Waltham, MA) for 3 h and lysed with whole-cell extraction buffer (20 mM Tris-HCl, pH 7.5, 280 mM NaCl, 10% glycerol, 0.5% Nonidet P-40, 2 mM EGTA, 0.2 mM EDTA, 100 mM phenylmethylsulfonyl fluoride), and the lysates were precleared for 0.5 h by use of protein A Sepharose beads (Invitrogen). Precleared lysates were incubated with antibody specific to PIV5 M protein, MuV M protein, NiV M protein, or Flag tag for 3 h at 4°C. Immune complexes were collected using protein A Sepharose, washed, and subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Polypeptides were visualized using a Fuji FLA-7000 laser scanner (FujiFilm Medical Systems, Stamford, CT). For immunoblotting, samples were fractionated by 10% SDS-PAGE and electrotransferred to polyvinylidene difluoride (PVDF) membranes. Immunodetection was carried out using a Flag tag-specific monoclonal antibody followed by an alkaline phosphatase-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Detection was done with a Fuji FLA-7000 laser scanner.
Measurements of VLP production and virion production.
To generate VLPs, 293T cells in 10-cm-diameter dishes were transfected with pCAGGS plasmids corresponding to PIV5 M (or derivatives) (0.8 μg/dish), PIV5 NP (300 ng/dish), and PIV5 HN (3 μg/dish). In some experiments, plasmid pCAGGS-Fl.14-3-3 or pCAGGS-Fl.14-3-3mut (1.0 μg/dish) was included as well. For measurements of virion production, 293T cells in 10-cm-diameter dishes were infected with PIV5 recombinants at an MOI of 1.0 PFU/cell. At 16 h p.t. or p.i., the culture medium was replaced with DMEM containing 1/10 the normal amounts of methionine and cysteine and 40 μCi of [35S]Promix/ml. After an additional 14 h, cell and medium fractions were harvested.
VLPs and virions were purified from medium fractions by centrifugation through 20% sucrose cushions and flotation on sucrose gradients as described previously (24). Purified VLPs/virions were loaded directly onto SDS gels. Preparation of RIPA cell lysates and immunoprecipitation of proteins were performed as described previously (33). A total of 12.5% of the cell fractions was used for immunoprecipitation analysis, whereas 100% of the medium fractions was used for VLP/virion isolation. Proteins were detected using a Fuji FLA-7000 laser scanner. Relative VLP production or virion production was calculated as the amount of M protein detected in purified VLPs/virions divided by the amount of M protein detected in the corresponding RIPA cell lysates, normalized to the value obtained with wt M protein or wt virus.
Recombinant virus generation.
PIV5 was recovered from cDNA as described previously (9), with modifications to avoid the use of vaccinia virus (38). BSR-T7 cells grown to 80% confluence in 6-cm-diameter dishes were transfected with variants of the infectious clone pSV5 M.NS (32), together with helper plasmids encoding PIV5 NP, P, and L proteins, as described previously (38). At 16 h p.t., the transfection medium was replaced with DMEM supplemented with 2% FBS, and the cells were incubated at 37°C for 3 additional days. Cell culture medium was harvested, clarified by low-speed centrifugation, and used to infect MDBK cells. At 5 days p.i., MDBK cell culture medium was harvested and used to infect BHK-21F cells for plaque purification of virus as described previously (23). Plaque-purified viruses were propagated in MDBK cells to generate virus stocks. For virus genome sequencing, total RNA was isolated from infected MDBK cells and used as a template for reverse transcription and PCR amplification as described previously (9). DNA sequencing was performed by the Penn State University Genomics Core Facility.
Virus growth curve analysis.
Growth curve analysis was performed essentially as described previously (30). MDBK cells grown in 24-well plates were infected with recombinant viruses at an MOI of 0.01 PFU/cell. After a 1-h adsorption period, cells were washed twice with phosphate-buffered saline (PBS), and 0.5 ml of DMEM supplemented with 2% FBS was added to each well. Cultures were incubated at 37°C for various times (0, 12, 24, 48, 72, or 96 h). Culture supernatants were collected, and virus titers were measured by plaque assay on BHK-21F cells as described previously (23).
Protein phosphorylation analysis.
293T cells in 10-cm-diameter dishes were infected with recombinant viruses at an MOI of 1.0 PFU/cell. At 16 h p.i., the culture medium was replaced with phosphate-free medium, and the cells were incubated at 37°C for 30 min. The culture medium was then replaced with phosphate-free medium supplemented with 100 μCi per dish of [33P]orthophosphate (Perkin Elmer, Waltham, MA). After incubation at 37°C for 24 h, cell and medium fractions were harvested and analyzed as described above.
Fluorescence microscopy.
CV-1 cells grown on glass coverslips to 50% confluence were infected with recombinant viruses at an MOI of 1.0 PFU/cell. At 16 h p.i., monolayers were fixed with 4% paraformaldehyde in PBS for 10 min. Cells were permeabilized by incubation for 30 min at room temperature in a solution containing 0.1% saponin, 5% goat serum, and 0.2% bovine serum albumin in PBS. Cells were incubated for 2 h with a mixture of M protein-specific MAb M-h (IgG3 isotype) and a MAb specific to 14-3-3 (IgG1 isotype). Cells were washed three times with PBS and incubated for 1 h with a mixture of IgG3-specific Alexa Fluor 488- and IgG1-specific Alexa Fluor 594-goat anti-mouse secondary antibodies (Invitrogen). Cells were washed an additional three times with PBS, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cell were visualized with a Zeiss Axiovert 200 M fluorescence microscope (Zeiss, Inc., Thornwood, NY). Images were captured using an Axiocam MRm charge-coupled device (CCD) camera.
Amino acid sequence comparisons.
Sequence data used for comparison of paramyxovirus M protein C-terminal regions were derived from files with the following GenBank accession numbers: PIV5, AF052755; mumps virus, AF467767; Newcastle disease virus, X04687; measles virus, AB016162; Nipah virus, AF212302; and Sendai virus, M30202.
RESULTS
Identification of 14-3-3 as a PIV5 M-interacting protein.
As part of an effort to define host factors important for paramyxovirus particle formation, we screened a yeast two-hybrid library, using the PIV5 M protein as bait. Two screens of a HeLa cell-derived cDNA library were conducted in parallel, using baits in which the LexA DNA binding domain was fused either to the N terminus or to the C terminus of the full-length M protein. In this screening system, transcription of LexA-driven reporters allows growth of yeast cells on medium lacking histidine and also results in beta-galactosidase production. His+ beta-galactosidase+ yeast colonies were selected, and the library-derived cDNAs were subjected to DNA sequence analysis. The 14-3-3 protein beta isoform (gene YWHAB; GenBank accession no. NM_003404) was the candidate identified with the greatest frequency when we used the N-terminally fused M protein as bait (accounting for 30 of 104 clones sequenced). Each of these clones encoded the full-length 14-3-3 protein. In contrast, when we used the C-terminally fused M protein as bait, the 14-3-3 protein was not identified (0 of 96 clones sequenced). To characterize the interaction further, pairwise yeast two-hybrid tests were performed. A prey plasmid encoding the 14-3-3 protein was introduced into fresh yeast cells together with bait plasmids, and beta-galactosidase activities were detected using a colony filter assay (Fig. 1). The PIV5 M protein with N-terminal fusion of the LexA DNA binding domain tested positive for interaction with 14-3-3 protein, while the PIV5 M protein with C-terminal fusion of the LexA DNA binding domain tested negative. To map the region of M protein important for binding, additional pairwise tests were conducted using bait constructs containing just the N-terminal 100 amino acid residues of M protein or just the C-terminal 50 or 100 amino acid residues of M protein, as illustrated in Fig. 1. The N-terminal fragment tested negative for 14-3-3 protein binding, while the C-terminal fragments both tested positive for the interaction. These experiments define the 14-3-3 protein (beta isoform) as a potential binding partner for the PIV5 M protein and map 14-3-3 binding activity to the C-terminal 50 amino acid residues of the M protein.
14-3-3 protein binds to PIV5 M protein in mammalian cells.
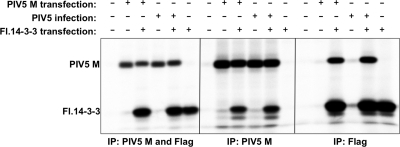
To investigate the binding of 14-3-3 protein to PIV5 M protein in mammalian cells, coimmunoprecipitation experiments were performed (Fig. 2). 293T cells were transiently transfected to produce PIV5 M protein and/or Flag-tagged 14-3-3 beta protein, as indicated. Immunoprecipitation of PIV5 M protein by use of an M protein-specific monoclonal antibody led to coprecipitation of 14-3-3 protein, and in the converse experiment, immunoprecipitation of 14-3-3 protein by use of a Flag tag-specific antibody resulted in coprecipitation of PIV5 M protein. Similar results were obtained when the M protein was supplied via virus infection instead of transient transfection (Fig. 2). These results indicate that the 14-3-3 protein binds to the PIV5 M protein in both transfected and virus-infected mammalian cells.
FIG. 2.
Interaction between PIV5 M protein and 14-3-3 detected in mammalian cells by coimmunoprecipitation. 293T cells were transfected to produce PIV5 M protein, infected with PIV5, and/or transfected to produce Flag-tagged 14-3-3 beta protein, as indicated. Proteins synthesized in the transfected/infected cells were metabolically labeled using 35S-labeled amino acids, and the cells were harvested for lysate preparation by use of 0.5% NP-40. Immunoprecipitation (IP) was carried out using the indicated antibodies, and proteins were detected using a phosphorimager. A total of 10% of cell lysates was used to generate the left panel; the remainder was used to generate the middle and right panels. The results shown are representative of 3 independent experiments.
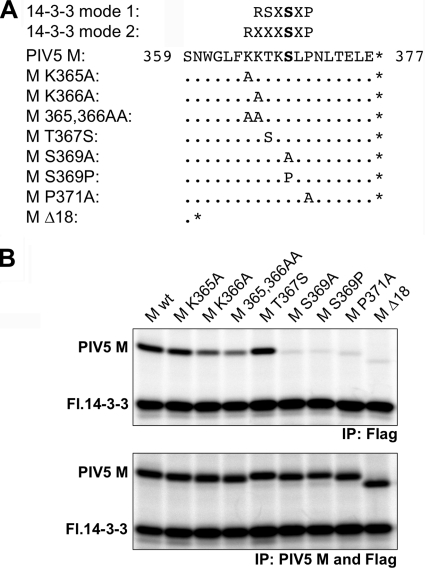
14-3-3 proteins bind to hundreds of different target proteins in eukaryotic cells. Binding usually affects target protein conformation, oligomerization, and/or intracellular localization, leading to modulation of target protein function (reviewed in references 18 and 21). Two optimal 14-3-3 binding motifs have been characterized and termed mode 1 and mode 2 sites, and these are illustrated in Fig. 3 A. A serine residue within these motifs (shown in bold) is usually phosphorylated to allow 14-3-3 binding. We scanned the C-terminal 50 amino acid residues of PIV5 M protein for potential 14-3-3 binding sites. One sequence was identified, namely, 365-KKTKSLP-371, which resembles (but does not quite match) both mode 1 and mode 2 consensus binding sites (Fig. 3A). Residue S369 within this sequence corresponds to the key phosphorylated serine residue of mode 1 and mode 2 binding sites. To investigate the importance of this region for 14-3-3 protein binding, a series of substitution mutants was generated, and the mutant proteins were subjected to coimmunoprecipitation analysis (Fig. 3B). Residues S369 and P371 of M protein were found to be critical for the interaction with 14-3-3 protein. Alanine substitutions at these positions almost completely eliminated 14-3-3 protein binding, similar to the result obtained with the M Δ18 protein, in which the C-terminal 18 amino acid residues of M protein have been removed. An additional S369P substitution was made, matching a second site mutation identified in a previous study (32). In this study, we found that S369P substitution blocked M protein interaction with 14-3-3 protein, similar to the S369A mutation (Fig. 3B).
FIG. 3.
Identification of M protein amino acid residues important for interaction with 14-3-3. (A) Well-characterized mode 1 and mode 2 consensus 14-3-3 binding sequences are illustrated, together with the C-terminal 19 amino acid residues of PIV5 M protein. Serine residues within the mode 1 and mode 2 motifs, usually phosphorylated to allow 14-3-3 binding, are indicated in bold. A series of substitution and truncation alterations affecting the PIV5 M protein sequence is also illustrated. (B) 293T cells were transfected to produce Flag-tagged 14-3-3 beta protein together with the indicated PIV5 M protein variants. Protein interactions were detected by coimmunoprecipitation as described in the legend to Fig. 2. The results shown are representative of 3 independent experiments.
Other alterations to the PIV5 M protein had less severe effects on 14-3-3 interaction. A moderate binding defect was observed upon mutation of residue K366 to alanine, while substitution affecting the adjacent K365 residue had no effect on 14-3-3 protein binding. Simultaneous replacement of both of these lysine residues had a moderate effect on 14-3-3 protein binding, similar to the effect of the K366A substitution alone. Residue T367 of M protein was changed to serine to make the M protein sequence more closely resemble a mode 1 consensus binding site. This led to a small but reproducible increase in the amount of M protein that coprecipitated with 14-3-3 protein. Overall, these mutagenesis results define a region near the C terminus of PIV5 M protein, spanning residues 365 to 371, that is necessary for efficient interaction with 14-3-3 protein.
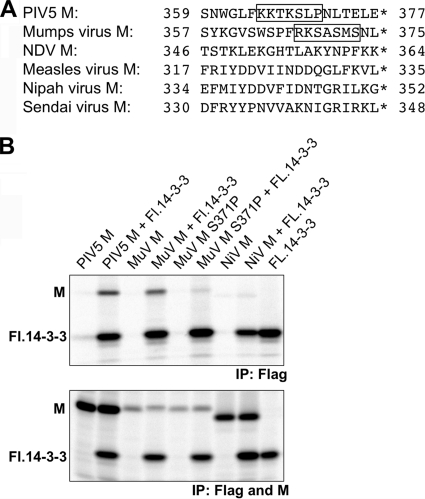
We investigated the possibility that other paramyxovirus M proteins might also have the ability to bind 14-3-3. Several paramyxovirus M proteins were scanned for potential 14-3-3 binding sites. A sequence was found within the mumps virus M protein, namely, 367-RKSASMS-373, which resembles the 365-KKTKSLP-371 region of PIV5 M protein (Fig. 4 A). Other paramyxovirus M proteins (Newcastle disease virus, measles virus, Nipah virus, and Sendai virus M proteins) do not have similar sequences, either within the C-terminal regions (Fig. 4A) or elsewhere. Coimmunoprecipitation experiments confirmed the binding of 14-3-3 protein to mumps virus M protein (Fig. 4B). Binding to mumps virus M protein was disrupted when M protein residue S371 was changed to proline. No interaction could be detected between 14-3-3 protein and Nipah virus M protein. These results indicate that only a subset of paramyxovirus M proteins have the ability to interact with 14-3-3 protein.
FIG. 4.
14-3-3 interaction with mumps virus M protein but not with Nipah virus M protein. (A) Comparison of paramyxovirus M protein C-terminal amino acid sequences. Boxed regions resemble mode 1/mode 2 14-3-3 binding sites. NDV, Newcastle disease virus. (B) 293T cells were transfected to produce the indicated paramyxovirus M proteins together with Flag-tagged 14-3-3 beta protein, as indicated. Protein interactions were detected by coimmunoprecipitation as described in the legend to Fig. 2. The mumps virus M protein-specific peptide antibody used in control experiments (lower panel) precipitates M protein with only limited efficiency. The results shown are representative of 3 independent experiments.
Enhanced VLP production directed by M protein mutants that fail to interact with 14-3-3.
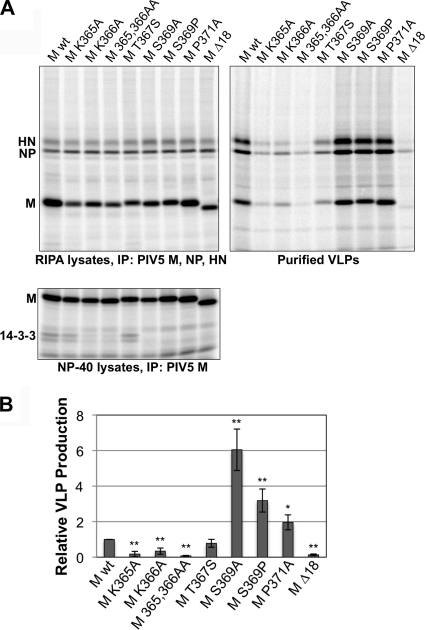
To test the effect of 14-3-3 protein binding on M protein function, VLP production experiments were carried out. Altered M proteins were expressed in 293T cells, together with PIV5 HN and NP proteins, by transient transfection. VLPs released into the culture supernatants were purified and quantified based on M protein content. Cell lysate fractions were also analyzed, and in these experiments, multiple isoforms of endogenous 14-3-3 protein could be observed coprecipitating with M protein (Fig. 5 A, lower panel), consistent with the results obtained with overexpressed 14-3-3 beta protein (Fig. 3). Interestingly, altered M proteins with substitutions at position S369 or P371 exhibited improved VLP production function compared with wt M protein (Fig. 5). VLP production efficiency increased about 6-fold after S369A substitution, 3-fold after S369P substitution, and 2-fold after P371A substitution (Fig. 5). Thus, substitutions which prevented 14-3-3 protein binding resulted in improved VLP production, suggesting that 14-3-3 protein binding is inhibitory to M protein budding function. In the case of the M Δ18 protein, which also fails to interact with 14-3-3, VLP production was reduced to nearly undetectable levels. This suggests that other parts of the M protein C-terminal region, perhaps unrelated to 14-3-3 protein binding, are important for proper M protein budding function. Consistent with this possibility, substitutions affecting K365 and/or K366 greatly impaired VLP production, even though these mutations had either moderate effects or no effect on 14-3-3 protein binding (Fig. 4 and 5). T367S substitution, which moderately increased 14-3-3 protein binding, resulted in a slight reduction in VLP production. Together, these results support a model in which PIV5 M protein is bound by 14-3-3 protein, to the detriment of virus particle formation. Lysine residues close to the 14-3-3 binding site are critical to the budding function of M protein, but for reasons that are likely unrelated to 14-3-3 protein binding.
FIG. 5.
Enhanced VLP production of M protein mutants that fail to interact with 14-3-3. (A) The indicated PIV5 M protein variants were coexpressed together with PIV5 HN and NP proteins in 293T cells for VLP production. (Top) After metabolic labeling with 35S-labeled amino acids, VLPs were purified from the culture medium by centrifugation through sucrose cushions and flotation on sucrose gradients. Viral proteins in RIPA cell lysates were immunoprecipitated using a mixture of antibodies specific to PIV5 M, NP, and HN proteins. Viral proteins from both cell lysates and VLPs were fractionated by SDS-PAGE and detected using a phosphorimager. (Bottom) A portion of the harvested cells was reserved and used to prepare NP-40 lysates. Immunoprecipitation with antibody specific to PIV5 M protein was carried out, allowing coprecipitation of endogenous 14-3-3 proteins. (B) Relative VLP production was calculated as the amount of M protein detected in VLPs divided by the amount of M protein detected in RIPA lysates, normalized to the value obtained with wt M protein. Results from three independent experiments performed as described for panel A were plotted, with error bars indicating standard deviations. Differences from the values obtained with wt M were assessed for statistical significance by Student's t test. *, P < 0.05; **, P < 0.01.
14-3-3 protein overexpression inhibits production of PIV5-like particles.
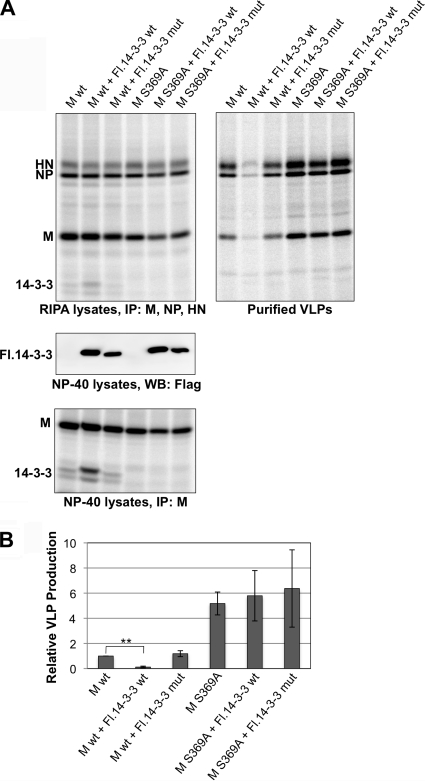
To further explore the relationship between 14-3-3 binding and M protein function, 293T cells were transfected to produce VLPs in the context of 14-3-3 protein overexpression (Fig. 6). This caused a substantial reduction in VLP release (8-fold lower than that observed in the absence of 14-3-3 protein overexpression). In contrast, overexpression of a mutant 14-3-3 protein that fails to bind target proteins (3) had no effect on VLP production. 14-3-3 protein overexpression led to increased binding to M protein, as judged by coimmunoprecipitation (Fig. 6A, bottom panel), thus linking poor VLP production to elevated 14-3-3 protein binding. Similar experiments were carried out in which VLP production was directed by S369A mutant M protein, which fails to bind 14-3-3. In this case, VLP production was elevated, consistent with earlier experiments (Fig. 5), and neither wt nor mutant 14-3-3 protein overexpression had any significant effect on VLP production efficiency. These results reinforce the conclusion that PIV5 M protein binding to 14-3-3 negatively affects particle formation.
FIG. 6.
Impaired VLP production upon 14-3-3 protein overexpression. (A) 293T cells were transfected to produce either wt PIV5 M protein or S369A mutant M protein, together with the indicated Flag-tagged 14-3-3 protein variants. (Top) Transfected cells were radiolabeled, VLPs were purified, and polypeptides were visualized as described in the legend to Fig. 5. (Middle) NP-40 lysates were prepared from duplicate, nonradiolabeled cells. Exogenous 14-3-3 protein expression levels were assessed by immunoblotting with antibody specific to Flag tag. (Bottom) A portion of the radiolabeled cells was reserved and used to prepare NP-40 lysates. Immunoprecipitation with antibody specific to PIV5 M protein was carried out, allowing coprecipitation of both endogenous and exogenous 14-3-3 proteins. (B) Relative VLP production was calculated from three independent experiments performed as described for panel A, and the results were plotted, with error bars indicating standard deviations. Differences between pairs were assessed for statistical significance by Student's t test. **, P < 0.01.
Recombinant viruses with M proteins that fail to bind 14-3-3 proteins bud particles more efficiently than does wt virus.
To investigate the consequences of M protein interaction with 14-3-3 in the context of live virus infection, we generated recombinant viruses by use of the reverse genetic system established previously for PIV5 (9). Three different recombinant viruses were recovered that encoded altered M proteins (S369A, S369P, and P371A) in place of wt M protein. Viruses were plaque purified and propagated using MDBK cells to generate virus stocks. Genome sequence analysis confirmed the presence of the engineered M protein mutations as well as the absence of any other mutations in the virus genomes. Recombinant viruses were used to infect 293T cells for assessment of 14-3-3 protein binding (Fig. 7 A). Infection with wt virus led to clear coimmunoprecipitation of endogenous 14-3-3 proteins together with the viral M protein. In contrast, infection with mutant viruses led to no detectable coimmunoprecipitation of endogenous 14-3-3 proteins, thereby confirming that these altered M proteins were defective for 14-3-3 protein interaction in the context of a productive PIV5 infection. Similar results were also obtained using infected MDBK cells as well as infected CV-1 cells (data not shown).
FIG. 7.
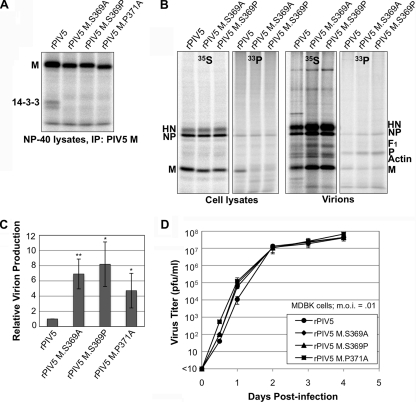
Enhanced particle production by recombinant viruses with M proteins that fail to bind 14-3-3. (A) 293T cells were infected with the indicated viruses. After metabolic labeling with 35S-labeled amino acids, NP-40 cell lysates were prepared. Immunoprecipitation with antibody specific to PIV5 M protein was carried out, allowing coprecipitation of endogenous 14-3-3 proteins. (B) 293T cells were infected with the indicated viruses, followed by metabolic labeling with 35S-labeled amino acids or [33P]orthophosphate. Cell and medium fractions were harvested, and virions were purified from the medium by centrifugation through sucrose cushions and flotation on sucrose gradients. RIPA cell lysates were prepared from cells, followed by immunoprecipitation with the indicated antibodies. Polypeptides were fractionated by SDS-PAGE and detected using a phosphorimager. (C) Relative virion production, based on 35S-labeled M protein content, was calculated from three independent experiments performed as described for panel B, and the results were plotted, with error bars indicating standard deviations. Differences from the values obtained with recombinant PIV5 were assessed for statistical significance by Student's t test. *, P < 0.05; **, P < 0.01. (D) MDBK cells were infected with recombinant viruses at an MOI of 0.01 PFU/cell. Culture medium was harvested at the indicated times, and virus titers were measured by plaque assay on BHK-21F cells. Values represent averages for three independent experiments, with error bars indicating standard deviations.
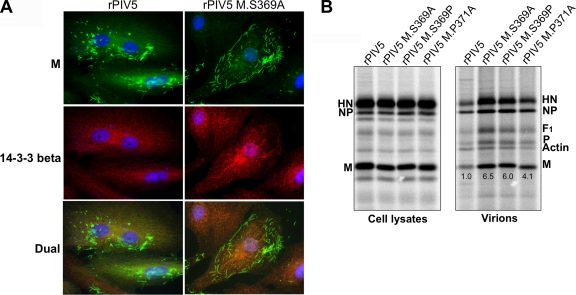
Recombinant viruses were analyzed for particle formation ability. After infection of 293T cells and metabolic labeling with 35S-labeled amino acids, medium and cell fractions were harvested. Virions were purified from the medium by use of sucrose gradients and were loaded directly onto SDS gels. Viral proteins from cell lysates were collected by immunoprecipitation. Virion production, measured based on M protein content, was substantially elevated for recombinant viruses compared with wt virus (Fig. 7B and C). Particle release efficiency increased >6-fold for the virus with S369A mutation, >8-fold for the virus with S369P mutation, and >4-fold for the virus with P371A mutation. Although these alterations increased the quantity of virion release, there was no significant change in virion polypeptide composition, as judged by the relative abundances of M, NP, and HN proteins (Fig. 7B and data not shown). Overall, the results obtained with recombinant viruses are in agreement with those obtained with VLP-producing cells and indicate that M protein interaction with 14-3-3 protein impairs virus budding.
Residue S369 of M protein is important for interaction with 14-3-3, and the 365-KKTKSLP-371 region of M protein resembles mode 1 (RSXpSXP) and mode 2 (RXXXpSXP) 14-3-3 binding sites (in which pS denotes phospho-serine) (18). This suggested to us that residue S369 of PIV5 M protein might be a target for phosphorylation, which would in turn allow 14-3-3 protein to bind. To test this possibility, cells infected with recombinant viruses were incubated with [33P]orthophosphate to label phosphorylated proteins (Fig. 7B, right panels). 33P-labeled M protein could be detected clearly in cell lysates derived from wt PIV5 infection, confirming previous observations that at least a portion of M protein is phosphorylated in infected cells (34). M protein phosphorylation was almost completely eliminated in cells infected with recombinant viruses with M protein mutations at position S369. Thus, at least a portion of M protein in PIV5-infected cells is likely phosphorylated at residue S369. 33P-labeled NP protein could also be observed in this experiment, consistent with previous observations of phosphorylated NP protein in PIV5-infected cells (34). NP protein phosphorylation in cells infected with M protein-altered recombinant viruses was not significantly different from NP protein phosphorylation observed in wt virus-infected cells. 33P-labeled NP, P, and M proteins were detected in purified virions, indicating that the phosphorylated forms of these proteins can be incorporated into budding virus particles.
Multiple-step growth curve experiments were carried out to determine the effect that disruption of 14-3-3 protein interaction has on virus multiplication in cultured cells. All of the recombinant viruses were capable of replicating to high titers in MDBK cells, similar to wt virus, with peak titers between 5 × 107 and 1 × 108 PFU/ml (Fig. 7D). Thus, failure of M protein to bind 14-3-3 protein did not impair virus multiplication in MDBK cells. At early time points postinfection (12 and 24 h p.i.), the mutant viruses reached titers that were higher than those observed for wt virus, by up to 10-fold, consistent with the enhanced particle production observed in Fig. 7B and C. However, this difference was transient. At 2 days or more postinfection, there was no significant difference between titers reached by mutant viruses and the titers reached by wt virus. Plaque size and morphology were unaffected by the M protein alterations (data not shown). Together, these results indicate that a virus with an M protein defective in 14-3-3 binding buds particles more efficiently than does wt virus and that failure to bind 14-3-3 does not prevent virus from replicating to high titers in cultured cells.
Interaction with 14-3-3 protein reduces M protein accumulation at virus assembly sites.
14-3-3 proteins usually influence the functions of target proteins which have been bound, in some cases by inducing altered intracellular localization. To test the effect of 14-3-3 binding on M protein intracellular localization, CV-1 cells infected with either wt PIV5 or rPIV5 M.S369A were examined by fluorescence microscopy (Fig. 8 A). CV-1 cells were selected for this analysis because they allow for easy visualization of PIV5 components, which assemble into clusters and filaments on infected cell surfaces (27, 30, 33, 38). We confirmed that particle production of recombinant viruses was enhanced in infected CV-1 cells, similar to the phenotype observed in 293T cells (Fig. 8B). As expected, infection of CV-1 cells with wt virus led to the concentration of M protein into clusters and filaments, some of which likely represent filamentous virions (Fig. 8A). Previous studies have shown that these clusters are plasma membrane associated and likely function as sites of active virus budding, as clustering of M protein did not occur readily in cells infected with assembly-defective recombinant viruses harboring glycoproteins with truncated cytoplasmic tails (30, 33, 38). M protein clusters and filaments were recently shown to be associated with Cav-1, and it is likely that caveolae play an important role in PIV5 assembly (27). Although much of the M protein in PIV5-infected cells was contained within clusters, a portion of M protein was found outside the clusters and dispersed throughout the cytoplasm (Fig. 8A, left panels). In dual-labeling experiments, we found that M protein that was outside the clusters and filaments colocalized with 14-3-3 beta, whereas the M protein found within clusters was for the most part not colocalized with 14-3-3 beta. In cells infected with rPIV5 M.S369A, nearly all of the M protein was found in clusters and filaments (Fig. 8A, right panels). The amount of M protein detected outside the clusters was reduced, and no colocalization with 14-3-3 protein was observed. This result is consistent with a model in which a portion of M protein in wt virus-infected cells is phosphorylated, bound to 14-3-3 protein, and held away from virus assembly sites, to the detriment of virus budding.
FIG. 8.
Intracellular localization of M protein in cells infected with recombinant viruses. (A) CV-1 cells grown on glass coverslips were infected with the indicated viruses for 16 h, fixed with paraformaldehyde, and bound with a mixture of M-specific MAb (IgG3 isotype) and 14-3-3 beta-specific MAb (IgG1 isotype). Isotype-specific secondary antibodies (IgG3-specific Alexa Fluor 488 and IgG1-specific Alexa Fluor 594) were used for detection. Fluorescence was visualized with a Zeiss Axiovert 200 M microscope. (B) CV-1 cells were infected with the indicated viruses, followed by metabolic labeling with 35S-labeled amino acids. Cell and medium fractions were harvested, and virus budding efficiency was measured as described in the legend to Fig. 7. Values indicate relative virion production efficiencies and are based on M protein content. The results shown are representative of 2 independent experiments.
DISCUSSION
Host factor recruitment contributes in a positive way to the budding of many retroviruses and negative-strand RNA viruses (reviewed in references 1, 2, and 4). Here we have identified a host factor, 14-3-3, which binds to the M protein of PIV5 and negatively affects virus budding. 14-3-3 binding was mapped to a region near the C-terminal end of the M protein, centered around residue S369, which is likely phosphorylated to allow 14-3-3 interaction. Recombinant viruses with M proteins defective in 14-3-3 interaction released particles even more efficiently than did wt virus, and fluorescence microscopy experiments suggested that 14-3-3 interaction with M protein leads to a reduction in the assembly of viral components into clusters at infected cell surfaces.
14-3-3 proteins are expressed abundantly in all eukaryotic cells. They function as either homodimers or heterodimers to bind and modulate the functions of target proteins. Over 300 different 14-3-3 target proteins have been identified so far, and 14-3-3 binding consequently affects a wide range of cellular processes, including cell cycle progression, apoptosis, cellular stress responses, cytoskeleton formation, and protein trafficking (reviewed in references 17, 18, and 21). The binding of 14-3-3 proteins by viral proteins has been observed before. For example, the HIV-1 Vpr protein binds to 14-3-3, and this interaction is important for the cell cycle-arresting activity of Vpr (13). The severe acute respiratory syndrome coronavirus nucleocapsid (N) protein binds 14-3-3, allowing the regulation of N protein nucleocytoplasmic shuttling (36). In this study, the M proteins of PIV5 and mumps virus were found to bind 14-3-3 beta, and at least in the case of PIV5, the bound M protein seemingly does not participate in virus budding. These findings support a model in which PIV5 M protein is multifunctional, with a portion of M protein free of 14-3-3 and able to contribute classical virus assembly function and the 14-3-3-bound remainder of M protein presumably providing functions that are unrelated to virus budding. A precedent for such a scenario exists, as the vesicular stomatitis virus (VSV) M protein has long been known to be multifunctional. In addition to its roles related to virus assembly and budding, VSV M protein functions to inhibit host mRNA synthesis, prevent the nuclear export of host mRNAs, and induce cytopathic effects in VSV-infected cells (reviewed in references 12 and 28). It is not clear at present what additional functions, if any, might be provided by the portion of PIV5 M protein which is bound to 14-3-3. One possibility is that this M protein simply acts as a decoy substrate, binding 14-3-3 and preventing it from binding to some of its normal cellular target proteins. If so, M protein would seemingly be well suited for such a task, as it is a major structural component of virions and accumulates to very high concentrations in virus-infected cells (likely in excess of even the relatively abundant 14-3-3 proteins). It is unclear at present what benefit would be gained by the virus through disabling 14-3-3-mediated signaling processes, although it has been suggested that 14-3-3 signaling could be important for antiviral responses (22).
14-3-3 binding to PIV5 M protein was dependent on M protein residue S369. Interestingly, this residue was identified in a previous study as a target for a second-site mutation by a budding-defective recombinant virus (32). This virus was engineered with a mutation to the FPIV sequence within the PIV5 M protein, which was shown to function as a late domain based on its ability to replace the PTAP sequence of the HIV-1 Gag protein for VLP budding. Recombinant PIV5 harboring an M protein with the FPIV sequence changed to FAIV was recovered twice independently, but the viruses were debilitated and second-site mutations arose rapidly. One virus acquired an L336P mutation within M protein, and the other virus acquired the M protein mutation S369P. Our results here provide new insight into the nature of the S369P alteration, indicating that an FPIV-defective virus partially restored its budding function through a change to its M protein that prevented 14-3-3 interaction. This may have served to partially alleviate the budding defect through an increase in the concentration of M protein at virus assembly sites. In addition, the introduction of a proline at position 369 could have been particularly beneficial to this virus, as the new proline-containing sequence was able to partially restore budding function to PTAP-defective HIV-1 Gag protein (32).
Phosphorylated M protein was detected in PIV5-infected cells, and the phosphorylation was nearly eliminated in cells infected with recombinant viruses with M proteins altered at residue S369. Phosphorylation of viral M proteins has been observed before for Sendai virus (11, 14), mumps virus (19), Newcastle disease virus (35), and PIV5 (34), although the roles and mechanisms of M protein phosphorylation have never been defined. For Sendai virus M protein, the major site of phosphorylation was identified as residue S70 (29). The amino acid sequence surrounding this residue does not resemble mode 1 or mode 2 14-3-3 target sites, however. A recombinant Sendai virus was generated in which residue S70 of M protein was changed to alanine. The virus was defective in M protein phosphorylation but did not exhibit any changes from the wt virus in terms of replication kinetics in cultured cells or pathogenicity in mice (29). Our results suggest that during PIV5 infection, the M protein is phosphorylated at residue S369 to allow 14-3-3 binding. Many target proteins regulate 14-3-3 binding through changes in phosphorylation status (17). It is possible that different functions of PIV5 M protein are also controlled through phosphorylation status, with M protein dephosphorylation triggering release from 14-3-3 and allowing trafficking to virus assembly sites. Another (not exclusive) possibility is that the quantity of phosphorylated M protein in infected cells eventually becomes so high that much of it can no longer be bound by 14-3-3. Consistent with this possibility, we observed incorporation of phosphorylated M protein into PIV5 virions. In addition, PIV5-like particle production was readily inhibited through overexpression of 14-3-3 protein, suggesting that M protein which normally would participate in virus assembly is in fact capable of binding 14-3-3. In this scenario, the timing of virus particle release might be influenced by 14-3-3, as saturation of the available 14-3-3 would have to occur prior to the participation of phosphorylated M protein in particle release. Results from growth curve experiments were consistent with this idea, as defects in 14-3-3 binding allowed recombinant viruses to reach higher titers than those of wt virus at early times postinfection but not at later time points.
Recent studies have identified multiple interacting partner proteins for PIV5 M protein, in addition to 14-3-3. For example, the host protein angiomotin-like 1 binds to M protein in a way that is beneficial to PIV5 infection, as judged by small interfering RNA (siRNA) depletion experiments (24). An association between PIV5 M protein and Cav-1 has also been described (27). Clusters at the surfaces of PIV5-infected cells were found to be concentrated not only with viral M and HN proteins but also with Cav-1. Cav-1 was found to be incorporated into budding virions, and PIV5 infectivity was reduced upon infection of cells lacking Cav-1 (27). These findings all support a model in which PIV5 budding occurs from caveolae. A potential Cav-1 binding site was identified within the PIV5 M protein amino acid sequence, and this site is conserved among related paramyxoviruses in the Rubulavirus genus (27). Interestingly, this sequence, spanning amino acid residues 355 to 363 of PIV5 M protein, is very close to the 14-3-3 binding site, which spans residues 365 to 371. We found that mutation of lysine residues adjacent to the potential Cav-1 binding site (positions 365 and 366) to either alanine or arginine severely impaired VLP production (Fig. 5 and data not shown). The proximity of the potential Cav-1 binding site to the 14-3-3 binding site raises the interesting possibility that only one of these two host proteins may be bound by M protein at any given time. This would suggest the possibility of a switching mechanism in which M protein to be held away from virus assembly sites would be bound by 14-3-3 and free of Cav-1, while M protein destined to participate in virus budding would instead be bound by Cav-1 and free of 14-3-3. Further work will be needed to fully explore the interplay between these host proteins that may occur to allow modulation of M protein function.
Acknowledgments
We thank Rick Randall and Bob Lamb for PIV5 antibody reagents and Andrey Sorokin for cDNA corresponding to binding-defective 14-3-3 beta. We are grateful to Dengyun Sun and Biao He for helpful discussions and assistance with phosphorylation experiments. We thank the laboratory of Kouacou Konan for assistance with fluorescence microscopy.
This work was supported in part by the Middle Atlantic Regional Center of Excellence (MARCE) for Biodefense and Emerging Infectious Disease Research (NIH grant AI057168) and by research grant AI070925 from the National Institute of Allergy and Infectious Diseases to A.P.S.
Footnotes
Published ahead of print on 8 December 2010.
REFERENCES
- 1.Bieniasz, P. D. 2005. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 2.Calistri, A., C. Salata, C. Parolin, and G. Palu. 2009. Role of multivesicular bodies and their components in the egress of enveloped RNA viruses. Rev. Med. Virol. 19:31-45. [DOI] [PubMed] [Google Scholar]
- 3.Chahdi, A., and A. Sorokin. 2008. Protein kinase A-dependent phosphorylation modulates beta1Pix guanine nucleotide exchange factor activity through 14-3-3beta binding. Mol. Cell. Biol. 28:1679-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, B. J., and R. A. Lamb. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison, M. S., T. Sakaguchi, and A. P. Schmitt. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int. J. Biochem. Cell Biol. 42:1416-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harty, R. N., M. E. Brown, G. Wang, J. M. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U. S. A. 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, B., G. Y. Lin, J. E. Durbin, R. K. Durbin, and R. A. Lamb. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 10.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 271:4621-4628. [DOI] [PubMed] [Google Scholar]
- 11.Hsu, C. H., and D. W. Kingsbury. 1982. Topography of phosphate residues in Sendai virus proteins. Virology 120:225-234. [DOI] [PubMed] [Google Scholar]
- 12.Jayakar, H. R., E. Jeetendra, and M. A. Whitt. 2004. Rhabdovirus assembly and budding. Virus Res. 106:117-132. [DOI] [PubMed] [Google Scholar]
- 13.Kino, T., et al. 2005. Vpr protein of human immunodeficiency virus type 1 binds to 14-3-3 proteins and facilitates complex formation with Cdc25C: implications for cell cycle arrest. J. Virol. 79:2780-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb, R. A., and P. W. Choppin. 1977. The synthesis of Sendai virus polypeptides in infected cells. III. Phosphorylation of polypeptides. Virology 81:382-397. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 16.Li, M., et al. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J. Virol. 83:7261-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackintosh, C. 2004. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 381:329-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison, D. K. 2009. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naruse, H., et al. 1981. The polypeptides of mumps virus and their synthesis in infected chick embryo cells. Virology 112:119-130. [DOI] [PubMed] [Google Scholar]
- 20.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 21.Obsilova, V., J. Silhan, E. Boura, J. Teisinger, and T. Obsil. 2008. 14-3-3 proteins: a family of versatile molecular regulators. Physiol. Res. 57(Suppl. 3):S11-S21. [DOI] [PubMed] [Google Scholar]
- 22.Ohman, T., et al. 2010. Cytosolic RNA recognition pathway activates 14-3-3 protein mediated signaling and caspase-dependent disruption of cytokeratin network in human keratinocytes. J. Proteome Res. 9:1549-1564. [DOI] [PubMed] [Google Scholar]
- 23.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. Davidson and R. M. Elliott (ed.), Molecular virology: a practical approach. IRL Oxford University Press, Oxford, United Kingdom.
- 24.Pei, Z., Y. Bai, and A. P. Schmitt. 2010. PIV5 M protein interaction with host protein angiomotin-like 1. Virology 397:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U. S. A. 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall, R. E., D. F. Young, K. K. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 27.Ravid, D., G. P. Leser, and R. A. Lamb. 2010. A role for caveolin 1 in assembly and budding of the paramyxovirus parainfluenza virus 5. J. Virol. 84:9749-9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rieder, M., and K. K. Conzelmann. 2009. Rhabdovirus evasion of the interferon system. J. Interferon Cytokine Res. 29:499-509. [DOI] [PubMed] [Google Scholar]
- 29.Sakaguchi, T., et al. 1997. Phosphorylation of the Sendai virus M protein is not essential for virus replication either in vitro or in vivo. Virology 235:360-366. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt, A. P., B. He, and R. A. Lamb. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 73:8703-8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt, A. P., G. P. Leser, E. Morita, W. I. Sundquist, and R. A. Lamb. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt, A. P., G. P. Leser, D. L. Waning, and R. A. Lamb. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheshberadaran, H., and R. A. Lamb. 1991. Simian virus 5 membrane protein maturation: expression in virus-infected cells and from a eukaryotic vector. Virology 183:803-809. [DOI] [PubMed] [Google Scholar]
- 35.Smith, G. W., and L. E. Hightower. 1981. Identification of the P proteins and other disulfide-linked and phosphorylated proteins of Newcastle disease virus. J. Virol. 37:256-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surjit, M., et al. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 79:11476-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takimoto, T., and A. Portner. 2004. Molecular mechanism of paramyxovirus budding. Virus Res. 106:133-145. [DOI] [PubMed] [Google Scholar]
- 38.Waning, D. L., A. P. Schmitt, G. P. Leser, and R. A. Lamb. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 76:9284-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]