Abstract
Evidence for an essential role of the herpes simplex virus type 1 (HSV-1) tegument protein VP1-2 originated from the analysis of the temperature-sensitive (ts) mutant tsB7. At the nonpermissive temperature (NPT), tsB7 capsids accumulate at the nuclear pore, with defective genome release and substantially reduced virus gene expression. We compared the UL36 gene of tsB7 with that of the parental strain HFEM or strain 17 and identified four amino acid substitutions, 1061D → G, 1453Y → H, 2273Y → H, and 2558T → I. We transferred the UL36 gene from tsB7, HFEM, or strain 17 into a KOS background. While KOS recombinants containing the HFEM or strain 17 UL36 gene exhibited no ts defect, recombinants containing the tsB7 UL36 VP1-2 exhibited a 5-log deficiency at the NPT. Incubation at the NPT resulted in little or no virus gene expression, though limited expression could be detected in a highly delayed fashion. Using shift-down regimes, gene expression recovered and recapitulated the time course normally observed, indicating that the initial block was in a reversible pathway. Using temperature shift-up regimes, a second defect later in the replication cycle was also observed in the KOS.ts viruses. We constructed a further series of recombinants which contained subsets of the four substitutions. A virus containing the wild-type (wt) residue at position 1453 and with the other three residues being from tsB7 VP1-2 exhibited wt plaquing efficiency. Conversely, a virus containing the three wt residues but the single Y → H change at position 1453 from tsB7 exhibited a 4- to 5-log drop in plaquing efficiency and was defective at both early and late stages of infection.
The tegument protein VP1-2, encoded by the UL36 gene of herpes simplex virus (HSV), is a key structural component. It is conserved across the herpesvirus family, is essential for virus replication, and plays multiple roles in the virus life cycle, including entry, transport, and assembly (6, 9, 11, 15, 16, 18, 28). VP1-2 is classified as an inner tegument protein and in HSV and pseudorabies virus (PrV) is tightly bound to the capsid as analyzed both biochemically during differential extraction protocols and in vivo during entry (12, 14, 17, 21, 23, 36). This tight and selective binding of VP1-2 to capsids, together with immunoelectron microscopy and fluorescence microscopy of VP1-2 on capsids at the earliest stages of infection, supports a role for VP1-2 in capsid transport within the cytoskeletal architecture and in docking and genome release at nuclear pores early in the infectious process. A key role for VP1-2 at the earliest stage in infection is strongly supported by results showing the failure of VP1-2-negative capsids to enter nuclei within polykaryocytes of infected and uninfected cells created by artificial fusion (28).
However, it was perhaps early analysis of the HSV temperature-sensitive (ts) mutant tsB7 which provided the first indication of an essential role for VP1-2 in HSV replication (3, 4, 15). Temperature-sensitive mutants generally exhibit a significant reduction in the level or activity of the corresponding gene product when the gene is expressed above a certain temperature (the restrictive temperature or nonpermissive temperature [NPT]). The analysis of ts mutants provides an extremely powerful approach for studying protein function and assembly in a wide range of fields. Many such mutants have been isolated and exploited in the field of HSV replication and gene function (5, 8, 10, 26, 27, 30). With HSV tsB7, at the NPT replication is blocked at a very early stage, virus gene expression does not take place, and capsids apparently accumulate at the nuclear pore without releasing their genomes. A second defect in tsB7 was identified by using temperature shift experiments, where initial infection was carried out at the permissive temperature, allowing entry and gene expression, followed by a shift to the NPT. This delayed shift to the NPT resulted in a shutoff of late protein synthesis and a substantial reduction in production of progeny virus (2, 3). Mapping of the genetic lesions in tsB7 was performed by marker rescue, which located both defects to a region around 0.501 map units (3). Although several candidates were proposed (15), at the time of the characterization of tsB7, neither the complete HSV transcript map nor the genome sequence were available, limiting any definitive identification of the gene or genes responsible. Additional transcript mapping (35, 38), together with genome sequencing (20), allowed the subsequent association of the tsB7 defects with the UL36 gene, encoding VP1-2.
Nevertheless, the defect in tsB7 has not been definitively shown to reside in the UL36 gene, nor have any sequence differences which might account for VP1-2 temperature-sensitive function been identified. Moreover, tsB7 was constructed by broad chemical mutagenesis of the parental strain HFEM, with a possibility that the virus may contain additional mutations potentially compounding interpretation of the ts phenotypes. Here we sequenced the UL36 genes from tsB7 and the parental virus, identifying four amino acid substitutions which may be responsible for the ts defect, and took two approaches of either constructing a tsB7 strain with wild-type (wt) UL36 or transferring the UL36 gene from tsB7 to an otherwise wt backbone in HSV strain KOS to pursue analysis of UL36 function. In particular from the latter approach, we isolated individual recombinants containing the UL36 gene from tsB7 and compared these to recombinants in which we introduced the paired UL36 gene from the parent strain HFEM. All of the recombinants contained the majority of the tsB7 UL36 gene and the associated difference at four positions compared to the HFEM gene, and all exhibited an almost 5-log difference in the ratio of plaque formation at 39°C to that at 33°C in Vero cells. We demonstrated a reversible block in virus gene expression in such mutants after infection at the NPT and an additional block in virus production later in the infection process. By construction of additional recombinants, we identified the substitution of residue 1453Y with H as being solely responsible for the reversible temperature phenotypes at entry and assembly. These results are discussed in relation to VP1-2 structure-function relationships, the roles of VP1-2, and the consequences of its dysfunction.
MATERIALS AND METHODS
Cells and viruses.
Vero, HS30, COS-1, and Hep2 cells were grown in Dulbecco's modified minimal essential medium (DMEM) (Gibco) containing 10% newborn calf serum (NCS), penicillin, and streptomycin. HS30 cells contain the HSV type 1 (HSV-1) strain KOS full-length UL36 gene and have been reported to complement the temperature-sensitive defect in tsB7 (9). These cells were maintained in DMEM containing 500 μg/ml of Geneticin (Gibco). The mutant temperature-sensitive virus tsB7 and its parental strain HFEM were kind gifts of A. Buchan (15). The UL36 deletion virus KΔUL36 was propagated on the complementing cell line HS30 (9). Plaque size was calculated using Image Pro Plus software analysis of at least 50 plaques for each virus strain.
Sequencing of the UL36 gene.
We first cloned the full-length UL36 genes into pUC19 following the same strategy as previously described for the plasmid pTD1 containing the UL36 gene from strain 17 (1). For HSV-1 tsB7 or HFEM, purified viral DNA was digested with XbaI and SpeI and the fragment corresponding to nucleotides 69247 to 80722 isolated and inserted into pUC19 digested with XbaI to construct plasmids pFA11 and pFA12. Automated sequencing was then performed for sequence determination across the complete genes for each strain. For sequencing of the UL36 gene from recombinant viruses, Vero cells were infected at 33°C, harvested after cytopathic effect (CPE), and processed for total DNA extraction using the QIAamp DNA blood kit (Qiagen). Automated sequencing was then performed with appropriate primers to ensure coverage of the entire gene. Note that the numbering of residues in tsB7 in this paper is with reference to the sequence of the parental strain HFEM. For reference, position 1061 in tsB7 corresponds to position 1066 in the published sequence of UL36 from strain 17, due to a short repeat of five residues in the strain 17 gene in a poorly conserved proline-rich region at position 375. Thus, the numbering of residues discussed here is decreased by 5 relative to the sequence of strain 17 VP1-2.
Construction of recombinant viruses rescuing tsB7 or viruses with defined mutations in UL36 in a KOS background.
To construct a recombinant virus with a rescued UL36 gene in the background of tsB7, the plasmid pTD4 carrying the UL36 gene from strain 17 was cotransfected with purified tsB7 viral DNA. pTD4 was derived from pTD3 (1) by the insertion of a short (150-bp) 5′ flanking region of homology upstream of the VP1-2-coding region. To construct recombinant viruses based on HSV-1 strain KOS but containing the potential mutations in UL36 from HSV-1 tsB7, cotransfections were performed with purified virus DNA from KΔUL36 together with plasmid DNA from either pTD1 (strain 17), pFA11 (strain tsB7), or pFA12 (strain HFEM).
To construct viruses containing subsets of the four changes in the tsB7 VP1-2, we first created chimeric genes, swapping appropriate sections of HFEM or tsB7 genes. This was achieved as follows (for reference, numbering refers to nucleotide positions in the strain 17 genome sequence). The 5′ ends of tsB7 and HFEM UL36 were first excised from pFA11 and pFA12 with HindIII (position 80708) and EcoRI (position 74916) and ligated into pcDNA3 cut with the same enzymes to create pcDNA3HFEM-5′end and pcDNA3tsB7-5′end. The region covering the 1061D → G mutation site was excised with SgrAI (positions 76683 to 80400) from both pcDNA3HFEM-5′end and pcDNA3tsB7-5′end and ligated into the corresponding swapped 5′-end plasmids to create pcDNA3HFEM(1061D → G)-5′end and pcDNA3tsB7(1061G → D)-5′end plasmids. The 3′ ends of HFEM and tsB7 UL36 were excised from pFA11 and pFA12 with EcoRI (positions 74916 to 69697) and cloned into each of the four 5′-end constructs to create eight plasmids with reconstituted UL36 genes as follows: pcDNA3-HFEM, pcDNA3-tsB7, pcDNA3-HFEM/tsB7EcoRI (named pCH1), pcDNA3-tsB7/HFEMEcoRI (named pCH2), pcDNA3-HFEM(1061D → G)/tsB7EcoRI (named pCH3), pcDNA3-tsB7(1061G → D)/HFEMEcoRI (named pCH4), pcDNA3-HFEM(1061D → G) (named pCH5), and pcDNA3-tsB7(1061G → D) (named pCH6). Based on the most informative constructs to delineate substitutions responsible for the ts phenotype, we chose the pCH1, pCH3, and pCH4 plasmids to generate the recombinant viruses expressing the UL36 genes with defined mutations (see Fig. 7).
For isolation of viruses, cotransfections of plasmid and viral DNAs were carried out on subconfluent monolayers of COS cells using the calcium phosphate precipitation technique with N,N-bis (2-hydroxyl)-2-aminoethanesulfonic acid-buffered saline (BES). Infected cells were screened 4 days later and the cells and medium harvested after extensive CPE. After three cycles of freezing and thawing, the resulting virus preparation was used to obtain pure recombinant viruses stocks by either of two methods. Recombinant viruses with a rescued UL36 gene were plaque purified by limiting dilution to obtain single plaques per well. Alternatively, viruses were isolated after infection of confluent monolayers of Vero cells with serial dilutions at 33°C in the presence of DMEM-2% NCS containing 0.4% agar (Gibco). Isolated plaques were picked and purified by a further three successive rounds of growth and single plaque isolations on Vero cells at 33°C.
Infections and temperature shift assays.
For analysis of parental or recombinant viruses, cell monolayers were incubated at 4°C for 1 h prior to adsorption and washed with cold DMEM. The virus inoculum (multiplicity of infection [MOI], 5) was then added in cold DMEM without serum. After 1 h at 4°C, monolayers were washed with cold DMEM, and DMEM containing 2% NCS, prewarmed to 39°C, was then added. The cells were then further incubated at 33°C or 39°C as indicated. For temperature shift experiments, infected cultures were transferred at different times after infection at 33°C up to 39°C (shift up) or after infection at 39°C down to 33°C (shift down) for further incubation. Cultures were transferred and submerged in a water bath at 39°C to control the temperature as synchronously and rapidly as possible. Cultures continuously incubated at 33°C or 39°C or the shifted samples were subsequently harvested at different times for analysis depending on the assay.
Immunofluorescence studies.
Immunofluorescence analysis was performed exactly as described previously (2) using the following antibodies: VP5 (East Coast Bio; 1:500) and VP1-2 (αVP1-2NT1r; 1:250) (2). Samples from mock-infected and infected cells were collected at the times indicated, washed with phosphate-buffered saline (PBS), fixed with methanol for 5 min at −20°C, and blocked with PBSB (PBS containing 10% NCS), supplemented for αVP1-2NT1r with 0.5 mg/ml human IgG (Sigma), for 1 h at room temperature. The coverslips were then incubated with the primary antibody diluted in PBSB for 45 min at room temperature, washed with PBS, and incubated with the corresponding fluorochrome (Alexa 488 or Pierce 549)-conjugated secondary antibodies for 45 min. The coverslips were washed with PBS, dried, and mounted with Mowiol containing antifade reagent. Images were collected using a Zeiss Axiovision imaging system and either a Zeiss ×10 or ×100 (Plan-Apochromat; 1.4 numerical aperture) lens. Images for each channel were captured sequentially with a Retiga 2000R camera using Image Pro plus software. Composite illustrations were prepared using Adobe software. The example images shown are representative of numerous images gathered for each virus and condition.
SDS-PAGE and Western blotting.
Mock-infected and infected cells were washed in PBS, and total lysates were prepared by adding SDS lysis buffer containing 25 mM dithiothreitol (DTT). Samples were boiled for 5 min and briefly sonicated prior to electrophoresis. Equal amounts of sample were separated by SDS-PAGE using linear 10% or 3 to 8% gradient gels and transferred onto nitrocellulose membranes (Whatman) for Western blotting. Primary antibodies for immunodetection were diluted in PBST (PBS plus 0.1% Tween 20) containing 5% nonfat dry milk. Target proteins were visualized using DyLight-conjugated secondary antibodies (Pierce) and developed using the LiCor Bioscience Odyssey infrared imaging system. Odyssey v3.0 software was used for quantification of the detected proteins, with linearity of measurement being confirmed using a standardization bioassay with serial dilutions of sample inputs. Values of specific viral protein intensities were normalized against actin values. Antibodies for Western blotting were used as follows: ICP4 (Virusys; 1:1,000), ICP8 (a kind gift from Roger Everett; 1:2,000), VP5 (East Coast Bio; 1:3,000), and monoclonal antiactin (Sigma; 1:500).
RESULTS
Identification of mutations in the UL36 gene from tsB7.
The HSV-1 ts mutant virus tsB7 was derived by chemical mutagenesis of its parental strain, HFEM, using bromodeoxyuridine (15). Two types of defect were identified in tsB7. The first was identified by continuous incubation at the NPT (39°C) and resulted in capsid accumulation at the nuclear pore, defective deencapsidation of the genome, and associated lack of gene expression. A second defect later in the replication cycle was identified by allowing infection to initially proceed at the permissive temperature (33°C), thus overcoming the very early defect, and then shifting infected cells to the NPT. This regime resulted in a block to subsequent progression of infection, indicating a second malfunction beyond the early defect in nuclear pore docking. Both defects were mapped, by intertypic marker rescue initially using HSV-2 and subsequently using HSV-1 strain F DNA overlapping SalI, XhoI, or BamHI restriction fragments, to a region located between 0.501 and 0.503 map units. The gene or genes responsible for the defect were not identified, although the close linking of a number of protein products, including ICP2, -10, -43, and -44 was noted. Subsequent completion of the HSV sequence and gene assignment (20) together with transcript mapping (35) indicate that the region conferring the defect in tsB7 is located within the UL36 gene, encoding the large tegument protein VP1-2.
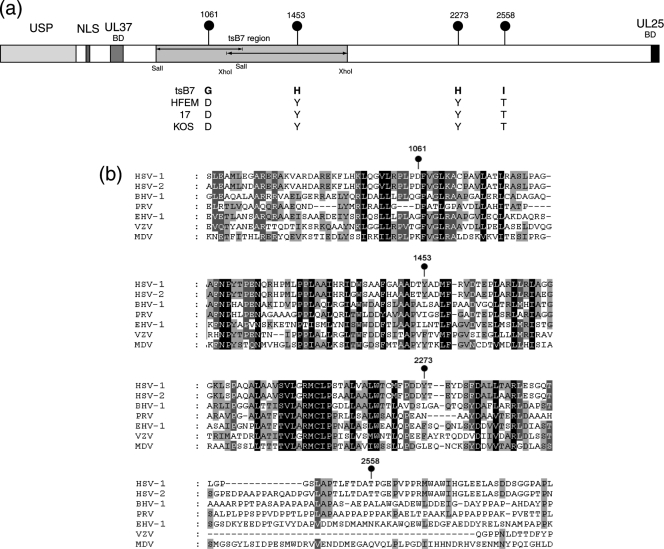
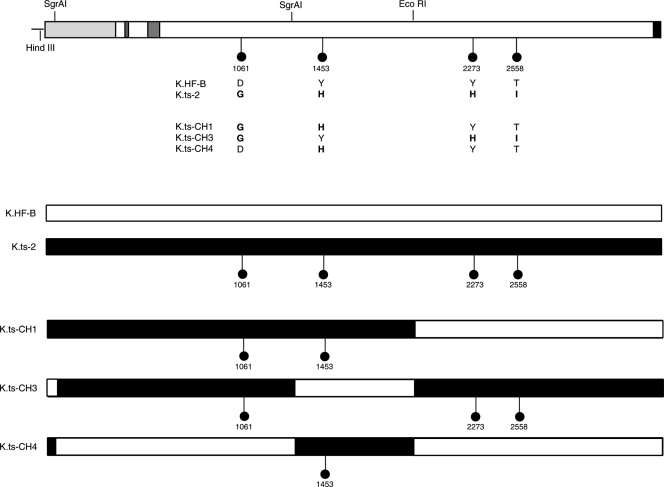
To refine analysis of the ts defect and provide information to aid our understanding of structure-function relationships within VP1-2, we sequenced the entire UL36 genes from tsB7 and its parent strain HFEM and compared these to each other and to available sequence data for wt HSV-1 strains 17 and KOS. While nucleotide substitutions which did not affect amino acid sequence were observed between strains, there were only four changes which resulted in amino acid differences between tsB7 UL36 and the parental HFEM UL36 (Fig. 1 a). In these four positions (numbered with reference to the amino acid sequence of HFEM), 1061D → G, 1453Y → H, 2273Y → H, and 2558T → I (HFEM residues are first), the HFEM residues were identical in strains 17 and KOS, increasing the significance of the changes in the tsB7 sequence. These were the only differences between tsB7 and HFEM resulting in amino acid changes in VP1-2. Interestingly, the residue at position 1061 is encoded within a SalI fragment (Fig. 1, shaded) that yielded some rescue of the ts defect in marker rescue experiments. Residue 1453 is encoded within an XhoI fragment which also yielded some rescue, although a larger SalI fragment encompassing this region and the remainder of the gene was unable to confer significant rescue (3). The residue 1061D is within a short island showing good conservation across members of the alphaherpesvirus family, although 1061D itself, while present in HSV-2, is not as well conserved (Fig. 1b). Residues 1453 and 2273, which are both tyrosines, have both been replaced by histidines. They are also present in regions of good homology and, while conserved in HSV-2 (as well as the HSV-1 strains), are not themselves conserved across the all alphaherpesvirus family members. However, at these positions, all the homologues, if not Y, have bulky hydrophobic residues (F, L, or I). The residue 2558T, while conserved in HSV-2, is toward the C terminus of the protein and in a much more highly variable region not at all conserved between homologues.
FIG. 1.
The tsB7 UL36 gene contains four amino acid substitutions. (a) The UL36 genes from tsB7, parental strain HFEM, and wt strain 17 were sequenced. Positions of amino acid substitutions (numbered with reference to the HFEM sequence [see Materials and Methods]) are indicated together with the residues at these positions in tsB7 or the other strains. Features of VP1-2 illustrated in the diagram include the ubiquitin-specific protease (USP), nuclear localization signal (NLS), UL37-binding domain (UL37BD), and conserved C-terminal UL25-binding domain (UL25BD). The positions of fragments used for marker rescue of tsB7 encompassed by SalI or XhoI sites are also indicated (3). (b) Sequence homology around the tsB7 substitutions in the VP1-2 proteins of alphaherpesvirus subfamily members. Alignments were made using ClustalX alignment software and illustrated using Genedoc. Residues with complete identity allowing for close chemical similarity are indicated in white text with a black background, over 85% by white text on dark gray, and over 50% by black text on light gray.
Construction of a rescued recombinant mutant virus based on tsB7.
Having identified these four amino acid changes in the tsB7 VP1-2, we first wished to rescue the defect in tsB7 by recombining into the tsB7 backbone a defined UL36 gene from a wt virus. Purified DNA from tsB7 was cotransfected with a linearized plasmid (pTD4) containing intact UL36 (from strain 17) with a short flanking region on the 5′ side and extending to the XbaI site on the 3′ side of the gene. Virus progeny was isolated at 33°C, and individual isolates were plaque purified, amplified, and then tested for plaquing ratio at 39°C versus 33°C.
A number of isolates showed pronounced recovery of the ts defect in plaque formation, and isolate tsB7.33a was selected for further characterization. The parental strain HFEM showed a very slight reduction in plaque formation at 39°C, while strain 17 showed a modest increase of 3- to 4-fold (Fig. 2 a). Consistent with the original characterization (15), plaque formation for tsB7 was on the order of 5 to 6 logs reduced at 39°C compared to 33°C (Fig. 2a). In contrast, for tsB7.33a, plaquing efficiency had recovered virtually all of this defect. We sequenced the UL36 gene from the rescued virus tsB7.33a, and the results indicated that virtually the entire wt UL36 had been successfully recombined and that all the four mutations were now replaced by the wt residues. The crossover at the 5′ end of the gene mapped to between residues 1018 and 1061 using nucleotide sequence polymorphisms (Fig. 2b).
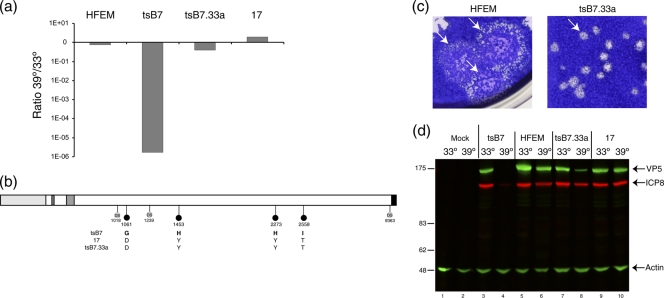
FIG. 2.
Construction of a recombinant tsB7 virus containing the wt UL36 gene. tsB7 DNA was cotransfected with a plasmid carrying the wt UL36 gene from strain 17, and the isolate tsB733a was further characterized. (a) Restoration of plaquing efficiency at 39°C. Plaque assay was performed on Vero cells infected at 33°C or 39°C with the HFEM, tsB7, tsB733a, and 17 viruses. The graph shows the relative ratio of the plaque formation of each virus. (b) The UL36 gene from tsB733a was subcloned and sequenced. Annotation is as for Fig. 1. DS, diagnostic nucleotide polymorphisms allowing sequence attribution. (c) Vero cells were infected with dilutions of HFEM and tsB733a viruses and incubated at 39°C, and plates were stained with crystal violet. Representative images of relative plaque size are shown. (d) Expression of candidate viral protein ICP8 and VP5 expression levels at 33°C and 39°C in Vero cells infected with tsB7, HFEM, tsB733a, and strain 17 at an MOI of 5. For quantitation using the LiCor Odyssey system, relative levels of each protein were normalized with actin levels.
However, while the mutations in UL36 were restored to the wt residues and plaquing efficiency was almost completely restored, plaque formation by tsB7.33a nevertheless was still different from that by the parental strain HFEM. Although both viruses exhibited a syncytial plaque morphology, tsB7.33a plaques were significantly smaller at 39°C, with plaque formation taking 5 to 6 days, compared to the large HFEM syncytial plaques, which merged together over the same time frame (Fig. 2c). This difference in plaques was still evident at 33°C (data not shown), indicating a potential difference between tsB7 and HFEM that was unrelated to the ts defect mapped to UL36.
We also examined infection during single-cycle replication after high-multiplicity infection at 33°C or 39°C, using the accumulation of candidate proteins ICP8 and VP5 as markers for early and late virus protein synthesis (Fig. 2d). As anticipated, at 39°C tsB7 exhibited a pronounced decrease in expression of both products, although very minor amounts of ICP8 were detectable. In contrast, infection with tsB7.33a at 39°C resulted in substantial levels of expression. Nevertheless, quantitative analysis indicated that levels of both proteins were reduced compared to those in infection at 33°C, with the reduction being slightly more (4-fold) for VP5 than for ICP8 (2-fold). A slight reduction in levels of these proteins was also observed at 39°C for the parental strain, though this was not observed with strain 17.
From the sequencing and recombination results, we have identified the mutations in UL36 which alone or in combination confer temperature sensitivity on the replication of tsB7. Restoration of UL36 results in a rescue in plaquing efficiency of 5 logs, presumably reflecting a combination of rescue of the early defect in capsid and the later defect attributed to UL36 function. However, tsB7.33a exhibited differences from the parental strain HFEM. It is possible that this could be due to alterations in other aspects of the genome which resulted from the nonspecific chemical mutagenesis of HFEM used to create tsB7. We next therefore adopted a different approach to exploit the utility of the ts defect in UL36 for exploring function, by introducing the UL36 gene from tsB7 into the backbone of an otherwise wt strain, KOS, and examining the transfer of temperature sensitivity.
Construction of a ts recombinant mutant virus based on KOS.
To aid selection of recombinant viruses based on HSV-1 strain KOS but containing the UL36 gene from tsB7, we employed the strain KΔUL36, which contains an internal deletion of approximately 3.6 kb in the UL36 gene (between the KpnI and EcoRV sites) (Fig. 3). This virus does not replicate in noncomplementing cells, and therefore cotransfection with the tsB7 UL36 gene and selection for growth at 33°C would facilitate isolation of KOS viruses containing defined UL36 genes for subsequent characterization. For comparison and as controls, we also recombined in the UL36 genes from the parental strain HFEM and from strain 17. Transfected cells were harvested after CPE was observed, and recombinant viruses with a rescued UL36 gene were isolated after limiting dilution and multiple rounds of plaque purification. For the UL36 genes from strains HFEM and 17, we initially isolated three independent recombinants, while for the tsB7 UL36, we isolated 15 individual recombinants with the aim of increasing the chances of obtaining different mutants. Restoration of UL36 with the tsB7 gene must in principle involve recombination upstream of the KpnI site and downstream of the EcoRV site, restoring the sequence between the KpnI and EcoRV sites, including the residues at positions 1061 and 1453. Since recombination could occur anywhere downstream of the EcoRV site, individual isolates may retain the KOS sequence or recombine in the tsB7 sequence, including the 2273H and 2558I residues. Progeny virus from each cotransfection was isolated, plaque purified, and amplified. We then sequenced across the UL36 gene for each of the isolates, in particular examining for the restoration of the intact UL36 and the provenance of the mutations at positions 1061, 1453, 2273, and 2558 (Fig. 3).
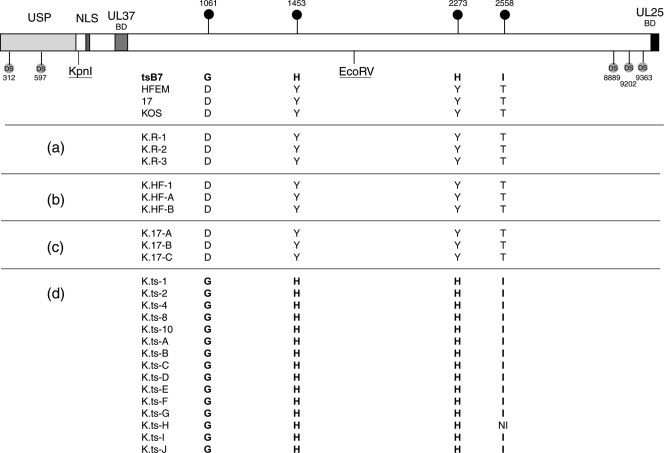
FIG. 3.
UL36 sequence analysis of the KOS recombinant viruses. COS cells were cotransfected at 33°C with KΔUL36 DNA either alone or with pUC19 plasmids carrying UL36 genes from strain HFEM, 17, or tsB7. (KΔUL36 is deleted between the indicated KpnI and EcoRV sites.) Several isolates from each transfection were purified by successive single-plaque isolations on Vero cells at 33°C. The resulting viruses were amplified and purified viral DNA sequenced. The table shows for each isolate virus the identity of the residue at the position of the four substitutions in tsB7, organized per transfection group: (a) KΔUL36 transfection alone (i.e., revertant KOS viruses K.R-x); (b) KΔUL36 plus HFEM UL36 (K.HF-x); (c) KΔUL36 plus strain 17 UL36 (K.17-x); and (d) KΔUL36 plus tsB7 UL36 (K.ts-x). Each individual virus was assigned a specific number or letter. The positions of diagnostic polymorphic nucleotide sequences (DS) are also indicated, which aided in demonstrating the extent of recombined sequences. NI, no sequence information available.
Transfection of KΔUL36 DNA alone gave rise to isolates containing KOS UL36 (Fig. 3a). This is due to a very minor population of wt KOS virus present in the KΔUL36 passaged in the complementing line, as indicated previously (9). Cotransfection with UL36 from HFEM resulted in isolates containing the HFEM UL36, including the wt residues 1061D, 1453Y, 2273Y, and 2558T. In all three isolates, the UL36 sequence was that of HFEM, which could be differentiated from KOS on the basis of nucleotide polymorphisms, at least up to the C-terminal end of UL36, the limit of our sequencing analysis (Fig. 3b). Similarly, for recombination with the UL36 from strain 17, each isolate contained the strain 17 sequence, including 1061D, 1453Y, 2273Y, and 2558T (Fig. 3c). All isolates from the cotransfections with the UL36 from tsB7 contained the full tsB7 UL36 sequence (Fig. 3d), including mutant residues 1061G, 1453H, 2273H, and 2558I (except one isolate for which sequence data toward the 3′ end was incomplete). Unfortunately, we did not obtain any reassortants in which an isolate contained the tsB7 UL36 at 1061G and 1453H together with KOS sequences toward the 3′ end (thus including the KOS 2273H and/or 2558T), which might have allowed for subsequent comparative analysis of different variants.
Characterization of growth and plaque formation.
Viruses were named for the provenance of the UL36 gene, e.g., K.ts-x, and three independent isolates were initially taken forward for analysis in comparison to parental viruses HFEM, tsB7, and KOS and to KOS isolates containing the HFEM or strain 17 UL36 gene.
First, in terms of plaque formation at 33°C versus 39°C, while wt KOS actually exhibited a modest increase in plaque numbers at 39°C, each K.tsB7 isolate exhibited a 4- to 5-log reduction in plaque formation, similar to the reduction seen with tsB7 itself (Fig. 4 a). In contrast, the KOS recombinants with the UL36 gene from HFEM, 17, or a revertant of KΔUL36 exhibited no temperature sensitivity and again if anything showed an increase in plaque number of between 4- and 8-fold. This defect was also observed in single-step growth curves. For K.HF-B at 33°C, maximum yields were obtained at approximately 25 h after infection, while at 39°C infection progressed faster, resulting in similar or modestly increased final yields. For K.ts-2, infection progressed similarly to that for K.HF-B at 33°C, (though with some reduction in final yield), while at 39°C, there was essentially no production of infectious virus, resulting in a final 39°C/33°C yield ratio of a 3- to 4-log reduction (data not shown).
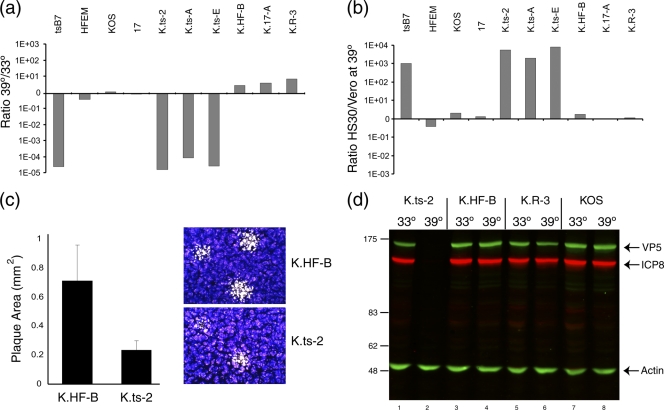
FIG. 4.
Temperature sensitivity of plaque formation by K.ts recombinant viruses. (a) Several isolates of each group of KOS recombinant viruses were analyzed for plaque formation in Vero cells at 39°C versus 33°C. Each of the K.ts isolates exhibited a reduction of approximately 5 logs in plaque formation. (b) Complementation of plaque formation in HS30 cells. Plaque formation in Vero or HS30 cells was quantitated at 39°C and the ratio determined. tsB7 plaque formation was of the order of 3 log units higher in HS30 cells, a result also obtained with each of the K.ts isolates. Comparatively minor differences between Vero and HS30 cells were observed for the other strains. (c) Plaque size for K.HF-B and K.ts-2 viruses at 33°C. The K.ts-2 plaque size was approximately one-third of the area of the corresponding isolates containing the HFEM UL36 gene. Plaque size was estimated using Image Pro Plus software from approximately 50 plaques for each virus, with representative images shown by crystal violet staining. Error bars indicate standard deviations. (d) Temperature sensitivity of gene expression. Vero cells were infected (MOI, 5) with K.ts-2, K.HF-B, K.R-3, or wt KOS at 33°C or 39°C, and extracts collected at 24 h postinfection and analyzed for the accumulation of viral proteins VP5 and ICP8. Actin levels were measured as a loading control.
The cell line HS30 is a derivative of Vero cells containing the UL36 gene and was originally used to complement the deletion of UL36. HS30 cells were also shown to complement growth of tsB7 (9). We therefore asked whether HS30 cells would complement the growth of the K.ts strains (Fig. 4b). The results demonstrate that each of the K.ts strains exhibited approximately a 3-log-greater efficiency in plaque formation on HS30 cells than Vero cells, while in comparison, the wt KOS strain or isolates containing the HFEM or strain 17 UL36 gene exhibited a modest 2- to 3-fold increase in plaquing efficiency (Fig. 4b). We note that in addition to the striking reduction in plaque formation at 39°C, plaques formed by the K.ts strains at 33°C were about 30% of the size of those formed by the corresponding strain K.HF. Results for K.ts2 are shown in Fig. 4c. The smaller plaque size indicates that the substitutions in the VP1-2 protein sequence confer some debilitation in growth even at 33°C. These results taken together provide compelling evidence that one or more of the four mutations 1061G, 1453H, 2273H, and 2558I in UL36 confer stringent temperature sensitivity of plaque formation, now in the background of the KOS strain. For further characterization, we used one isolate of each virus, thereafter named K.ts-2, K.HF-B, K.17-A, and K.R-3.
Characterization of virus yield and protein expression.
We measured the progression of infection at the level of the accumulation of representative viral proteins, ICP8 and VP5, initially at a single time point. For K.HF-B and K.R-3, ICP8 and VP5 were detected at both 33°C and 39°C, with a modest reduction at 39°C (Fig. 4d, lanes 4 to 6). In contrast, for K.ts-2, there was essentially no protein synthesis detected at 39°C (Fig. 4d, lanes 1 and 2, and Fig. 5). Additional comparative time course analysis demonstrated that for K.ts-2, expression could eventually be detected in a much reduced and much delayed fashion at 39°C. In contrast, for K.HF-B, the kinetics and relative abundance of ICP8 and VP5 synthesis were similar at the two temperatures (Fig. 4 and 5 and data not shown).
FIG. 5.
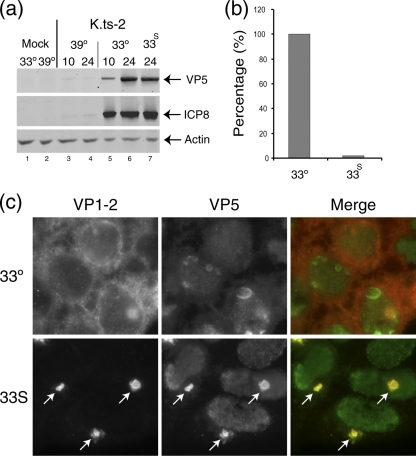
Postentry defect of K.ts-2. Monolayers of Hep2 cells were infected (MOI, 5) with K.ts-2 at 33°C, and at 10 h one set of cultures was maintained at 33°C while a duplicate set was shifted up to 39°C (33S). Incubation was continued and cultures harvested at 24 h postinfection. (a) Examination of candidate virus protein expression (ICP8 and VP5) expressed after continuous incubation at 33°C (lanes 5 and 6) or 39°C (lanes 3 and 4) or under the shift regime (33S) (lane 7). Actin levels were measured as loading control. (b) Parallel examination of total virus yield determined by titration on Vero cell monolayers at 33°C. The yield under the shift regime was reduced almost 2 log units compared to that at 33°C (set as 100%). (c) Altered localization of ts VP1-2 after temperature shift. Hep2 cells were infected with K.ts-2 at 33°C, and at 10 h postinfection were either maintained at 33°C or shifted up to 39°C. The cells were fixed at 16 h postinfection and the localization of VP1-2 and VP5 analyzed. At 39°C, pronounced relocalization of VP1-2, colocalizing with VP5 in a single juxtanuclear aggregate, is evident (arrows).
Defect in K.ts-2 after entry.
In the original characterization of tsB7, by allowing the early phase of infection to take place at the permissive temperature and then shifting temperature to 39°C, a second defect was detected. Thus, while immediate-early protein synthesis was then observed, late protein synthesis was not sustained unless infection was allowed to proceed for a period of approximately 4 h at the permissive temperature. To examine recovery of gene expression in K.ts-2 we compared expression at a single late time point (24 h) after continued incubation at 39°C (i.e., where we normally failed to see significant gene expression) with conditions where infection was first initiated at 33°C for 10 h and then shifted to 39°C until harvest (Fig. 5). Expression was then examined for representative early and late proteins, ICP8 and VP5. As shown above, expression was virtually absent after continued incubation at 39°C (Fig. 5a, compare lanes 3 and 4 to 5 and 6). However, incubation at 33°C for 10 h followed by temperature shift to 39°C (lane 7) allowed recovery to levels virtually identical to those seen after continual infection at 33°C (lane 6). Nevertheless, despite protein synthesis now accumulating to virtually normal levels in the temperature shift regime, parallel assay of yields with infectious virus showed a substantial reduction (approximately 50-fold) from those seen at the same time during continuous incubation at the permissive temperature (Fig. 5b).
We previously reported that during infection with tsB7 under the temperature shift regime, where infection is initiated at 33°C and then shifted to 39°C to allow late protein synthesis, VP1-2 is relocalized in a characteristic pattern resulting in a prominent and usually single juxtanuclear aggregate (2). Similar experiments were performed with K.ts-2, and the results demonstrated a dramatic relocalization of VP1-2, with the recruitment of additional candidate structural proteins (e.g., VP5) into a singular prominent juxtanuclear aggregate under the temperature shift regime (Fig. 5c, 33S arrows).
Examination of the precise nature of the requirement for infection at the permissive temperature to allow sustained later gene expression will be pursued in future work using K.ts-2. However, together these data demonstrate that in addition to the defect at early stages in entry, the four residue changes in VP1-2 in K.ts-2 are responsible for a defect(s) in VP1-2 function at later stages in virus assembly, presumably related to its role in the initial stages of inner tegument assembly or capsid transport functions to sites of envelopment.
Resumption of gene expression during shift down.
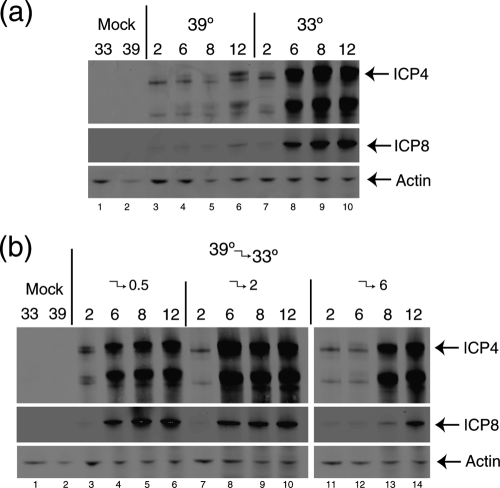
To begin to address the nature of the block to entry early in infection at the NPT, we wished to examine whether the block was reversible, i.e., whether after infection and maintenance at 39°C, infection and gene expression would ensue if infection was resumed at 33°C. To this end, we next performed shift-down experiments where infection was initiated at 39°C, held at 39°C for different lengths of time, and then shifted down to 33°C and incubation continued, examining expression levels of ICP4 and ICP8 (Fig. 6). In the control samples, robust synthesis of both proteins was detected at 33°C (Fig. 6a, lanes 7 to 10), and very substantially reduced amounts of ICP4 protein were detected in a delayed manner at 12 h postinfection after continuous incubation at 39°C (Fig. 6a, lanes 3 to 6). Initiation and maintenance of infection at 39°C for 0.5 or 2 h, followed by shift down and assay at 6 to 12 h, actually made little detectable difference in overall protein levels (Fig. 6b, lanes 3 to 10 [c.f. Fig. 6a, lanes 7 to 10]). This assay is limited by measurement of the amount of increase in protein expression between 2 and 6 h, but nevertheless the result indicates that maintenance at 39°C at least for 2 h did not disrupt subsequent progression at 33°C (though it is possible that there may have been some effect or delay). When the duration of infection and maintenance at 39°C was extended to 6 h, there was now, as expected, a substantial effect on expression levels (Fig. 6b, lane 12 [see also Fig. 6a, lane 4]). However, now upon shift down to 33°C we could clearly see that expression resumed, and within 2 h at 33°C, levels of ICP4 and ICP8 were only slightly below those seen upon continued synthesis at 33°C. At the least, during the 2 h after shift down to 33°C, expression of both ICP4 and ICP8 had accumulated to greater levels than in the initial 2 h infection at 33°C, (c.f. Fig. 6b, lanes 12 and 13, with Fig. 6a, lane 7). These results indicate not only that there is a block to the progression of infection due to the mutations in VP1-2 but that this block is at a reversible and physiologically relevant stage of infection, since shift down allows infection to proceed relatively normally, within the limits of resolution of this type of assay.
FIG. 6.
Reversible early defect in K.ts-2 upon temperature shift-down assay. Vero cells were infected with K.ts-2 (MOI, 5) continuously at 39°C or 33°C (a) or infected at 39°C and shifted down to 33°C at 0.5 h postinfection (lanes 3 to 6), 2 h postinfection (lanes 7 to 10) or 6 h postinfection (lanes 11 to 14) (b) and then harvested at the times indicated. Total times are indicated. Thus, e.g., for lanes 11 to 13, samples were at 39°C for 2 h, at 39°C for 6 h (lane 12), and at 39°C for 6 h, followed by 2 h at 33°C (lane13). Levels of representative immediate-early proteins (ICP4) and delayed-early proteins (ICP8) are shown together with actin levels as loading controls.
A single amino acid substitution is responsible for ts defects in VP1-2 function.
From these results, we conclude that K.ts2, with the four defined amino acid substitutions, exhibits reversible malfunction in VP1-2 function, resulting in defects both at the early stage of infection and later in the infectious cycle. While a priori, it may be that a combination of the four substitutions underpins the ts property, it was possible that only one mutation was responsible or that the different substitutions contributed in different ways, in particular to the possibility that the early versus the late defect may be uncoupled. To examine this, we constructed versions of VP1-2 with different combinations of mutations by swapping large restriction fragments of the wt gene from HFEM and the ts gene from tsB7 (see Materials and Methods). Having constructed these chimeric genes, they were recombined into KOS.ΔUL36 as before, and individual recombinants isolated and grown at 33°C and the sequence at the positions of the four residues determined. This created viruses with informative subsets of the four substitutions in the original tsB7 UL36. With regard to the four residues, the viruses were as follows (summarized in Fig. 7): K.ts-CH1, tsB7 at residues 1061 and 1453 and wt at positions 2273 and 2558; K.ts-CH3, tsB7 at residues 1061, 2273, and 2558 and wt at position 1453; and K.ts-CH4, tsB7 at residues 1453 and wt at positions 1061, 2273, and 2558.
FIG. 7.
Construction of the KOS recombinant viruses with selected substitutions in VP1-2. VP1-2 genes were constructed by swapping segments of parental HFEM and tsB7 genes using the indicated restriction sites (see Materials and Methods), yielding genes with various combinations of substitutions (lollipops indicate the presence of a residue from tsB7 UL36, and the absence of a lollipop indicates wt residues at those positions). Recombinant viruses were then constructed as before by cotransfecting COS cells with KΔUL36 DNA with the various plasmids. Isolates from each cotransfection were purified by successive single-plaque isolations on Vero cells at 33°C. The resulting viruses were amplified and purified viral DNA sequenced at the relevant positions of the four substitutions. Viruses were named for the provenance of the UL36 plasmids from which they were constructed, i.e., K.ts-CHx.
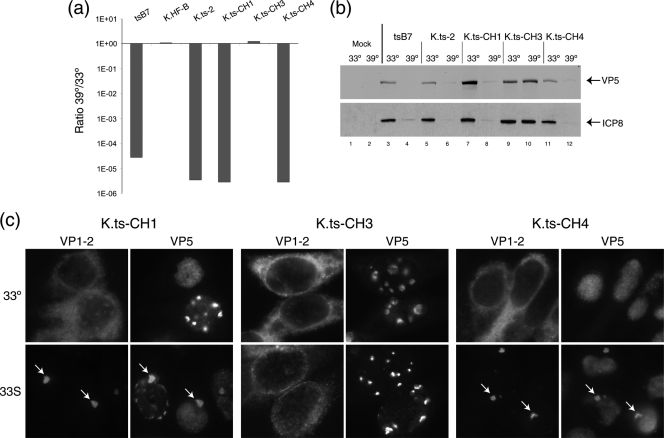
Each of these viruses was then characterized for growth at the permissive and restrictive temperatures, with consistent results which allowed definitive assignment of the substitution responsible for VP1-2 dysfunction (Fig. 8 a). Thus, K.ts-CH1 has only residues 1061G and 1453H from tsB7, with residues 2273 and 2558 being wt, and it exhibited virtually the same 5-log-unit decrease in plaque formation as K.ts2 or tsB7. This result indicates that residue 1061 or residue 1453, or both, is responsible for the defect. The variant K.ts-CH3 has residues 1061G, 2273H, and 2558I all from tsB7, but position 1453Y is wt. Despite having the tsB7 residues at three of the four positions, K.ts-CH3 exhibited completely normal plaque formation (Fig. 8a). The combination of phenotypes from these two variants provides strong evidence that residue 1453 is solely responsible for the ts defect. This conclusion is robustly confirmed with analysis of K.ts-CH4, which contains the wt residues at positions 1061, 2272, and 2558 but the single 1453Y → H change from tsB7. Plaque formation by K.ts-CH4 exhibited a reduction in efficiency at 39°C that was virtually identical to that by K.ts-CH1, K.ts2, or tsB7, on the order of 5 log units.
FIG. 8.
Identification of a single residue change responsible for the ts phenotype of tsB7 and K.ts-2. (a) tsB7, K.ts-2 (containing the four changes present in tsB7 UL36), and variants with subsets of these changes were analyzed for plaque formation in Vero cells at 39°C versus 33°C. K.ts-CH1 and K.ts-CH4 exhibited a reduction of approximately 5 log units in plaque formation, while K.ts-CH3 (containing three of the four substitutions but wt at position 1453) exhibited no defect in plaque formation. (b) Temperature sensitivity of gene expression. Vero cells were infected (MOI, 5) with viruses as indicated at 33°C or 39°C, and extracts were collected at 24 h postinfection and analyzed for the accumulation of viral proteins VP5 and ICP8. Actin levels were measured as a loading control. (c) Altered localization of ts VP1-2 after temperature shift. Hep2 cells were infected with the indicated viruses at 33°C and at 10 h postinfection either maintained at 33°C or shifted up to 39°C. Cells were fixed at 16 h postinfection and the localization of VP1-2 and VP5 analyzed. At 39°C, pronounced relocalization of VP1-2, colocalizing with VP5 in a single juxtanuclear aggregate, is evident (arrows) for K.ts-CH1 and for K.ts-CH4, which contain, respectively, 1061D → G plus 1453Y → H or just 1453Y → H. No alteration in VP1-2 localization was observed for K.ts-CH3, which contains three tsB7 substitutions, but the wt residue at position 1453.
The conclusions from these results are first that replacement of the single residue at position 1453 accounts for the defect in VP1-2 function and second that the requirements within VP1-2 for entry and for the later role in replication cannot be separated based on the effect of alteration at position 1453. The results are not consistent with a role for the other three positions in the ts defect. Thus, K.ts-CH3, with substitutions in these three positions, shows completely normal plaque formation, and K.ts-CH4 exhibits as much of a reduction in plaque formation as K.ts2 itself or K.ts-CH4. To confirm this proposition, we analyzed the defect in gene expression during high-multiplicity experiments at the NPT (Fig. 8b) and the defect under the shift-up regime as described above (Fig. 8c). The results demonstrate that K.ts-CH4 (with the single substitution 1453Y → H) (Fig. 8b, lanes 11 and 12) exhibited the same defect in gene expression as seen with K.ts2 (lanes 5 and 6) or K.ts-CH1 (lanes 7 and 8), while K.ts-CH3 (lanes 9 and 10), with substitutions at positions 1061, 2273, and 2258, exhibited normal gene expression and, as seen above, normal plaque formation.
In the shift-up experiments with K.ts-CH1 and K.ts-CH4, the results demonstrate that both viruses exhibited the same defect, where VP1-2 was synthesized but accumulated in a singular prominent juxtanuclear aggregate (Fig. 8c, 33S, arrows). Conversely, K.ts-CH3 exhibited normal cytoplasmic localization. We conclude from these combined data that the 1453Y → H substitution renders VP1-2 nonfunctional for its roles in entry at the earliest stage of infection and for its role(s) later in infection (Fig. 8c).
DISCUSSION
Temperature-sensitive mutations represent useful tools for the dissection of function in many diverse processes, and ts mutants have been isolated and exploited in analysis of HSV replication and gene function (5, 8, 10, 26, 27, 30). The conditional mutation approach is particularly useful for study of VP1-2, which has an essential role(s) late in the assembly process, as well as roles in early transport and nuclear pore docking. To approach dissection of these roles by the use of deletion mutants, and indeed even to uncouple and identify its early role in the life cycle, one would have to construct mutants which were competent to assemble virus yet defective in the early function. While this approach may be feasible in the longer term, the isolation of a defined ts virus mutant in VP1-2 affords a very useful approach to understand its role in various stages of the virus life cycle.
The first indication of an essential role of VP1-2 came from examination of the phenotype of the temperature-sensitive mutant tsB7, which exhibited defects very early in virus entry and also later in the replication cycle (3, 15). Although both defects were determined by marker rescue experiments to reside in the gene for UL36, confirmation and identification of the mutations responsible have not been established. Here we provide sequence analysis of the UL36 gene of tsB7 in comparison to that of the parental strain HFEM and identify mutations leading to four amino acid substitutions, 1061D → G, 1453Y → H, 2273Y → H, and 2558T → I. In constructing a version of tsB7 with a wt UL36 gene, we show recovery of the efficiency of plaque formation at the NPT, but a phenotype in plaque morphology and size persists. This observation, together with the fact that broad mutagenesis was used to generate tsB7, indicates that other features may contribute to the overall properties of tsB7. Therefore, we adopted a second approach, constructing a recombinant virus based on the KOS strain of HSV-1 and containing the UL36 gene from tsB7. This virus exhibited both a ts phenotype at the immediate-early phase, revealed by maintained incubation at the NPT, and a later defect revealed by temperature shift-up experiments after infection at the permissive temperature. Finally, we constructed a KOS variant containing the single 1453Y → H substitution which exhibited the same ts defect of approximately 5 logs in plaque formation, quantitatively similar to that of tsB7 or that of the KOS variant containing all of the four substitutions. Conversely a variant containing tsB7 mutant residues at positions 1061, 2272, and 2258 but the wt residue at position 1453 had normal plaquing efficiency, thus providing definitive evidence that the single Y → H substitution at position 1453 is responsible for the temperature-dependent and reversible defect in VP1-2.
The identity of this mutation in relation to certain features of the tsB7 defect and VP1-2 function warrants further discussion. Consistent with the results shown here for the KOS ts strains, we previously reported that in tsB7 the localization of VP1-2 is profoundly altered, localizing to a singular large perinuclear cluster, together with associated structural proteins. These clusters were seen by electron microscopy to represent coalesced capsids which were not progressing to the envelopment stage. Moreover, the clusters contained associated accumulation of ubiquitinated protein species. Although the target species were not identified and are more likely to be species other than VP1-2 itself, we speculated on whether the known ubiquitin-specific protease (USP) activity of VP1-2 might be compromised in tsB7. However, what is now clear is that the USP region itself contains no amino acid substitutions, and considering that the USP can fold and likely function in isolation (31), we believe it unlikely that the substitution at position 1453 (or at the other positions) affects the intrinsic USP activity. It remains possible though, that the altered intracellular localization of VP1-2 per se, or alterations in folding of other regions of the protein, affects USP activity or targeting. Indeed, other work has indicated the possibility that the defect in tsB7 affects the N-terminal region of the protein. It has been reported that VP1-2 undergoes a proteolytic cleavage event early in infection that is related to successful entry (13). The cleavage event resulted in an N-terminal product which, although not specifically mapped, migrated at approximately 55 kDa and would encompass the USP domain and adjacent N-terminal regions. Infection was blocked by an inhibitor of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-like proteases, with the inhibitor also blocking appearance of the VP1-2 cleavage product. Interestingly the VP1-2 product was observed after infection with tsB7 at the permissive temperature but not at the NPT. Thus, it is possible that presentation of the putative N-terminal cleavage site is influenced by folding determinants including residue 1453 or that other events requiring the determinant at residue 1453 precede and are necessary for the cleavage event. Future work will explore the production of the N-terminal product in the KOS.ts virus with mutant position 1453 and examine any effect on USP function. This will require the establishment of in vivo and in vitro assays for relevant USP function of the intact protein to explore the possibility that the distal amino acid substitutions in VP1-2, specifically at position 1453, affect USP function intrinsically or, e.g., by formation of a dominant negative effect, contributing to the defect in overall VP1-2 function.
It seems unlikely to us that the residue at position 1453 is required for global folding of the entire protein. As indicated above, the USP domain folds in isolation and in vivo would be translated and fold much earlier than distal parts of the protein. Large sections of the protein toward the C terminus can be deleted and are obviously dispensable for folding, while determinants within the last 60 residues are critical (6, 7, 16, 22). Isolated domains containing the C terminus interact with UL25 and are recruited to assembly sites (16). While possible, it would seem unlikely that residue 1453 is involved in folding the essential distal C-terminal region beyond the dispensable region. Notwithstanding the reported loss of N-terminal cleavage in tsB7, the region at position 1453 might be more likely to be involved in formation of a local determinant or potentially in intermolecular interactions. The residue is within a block of conservation from approximately position 1320 to 1910 whose alignment within the alphaherpesvirus VP1-2 species allows no gaps. Although residue 1453 is not perfectly conserved, in HSV-1, HSV-2, and Marek's disease virus (MDV) this residue is a Y and in the other members it is bulky and hydrophobic, within the sequence FGAAADTYADMF (where bold indicates very strong conservation and Y at position 1453 is underlined). Unfortunately, there is no simple pattern in the nature of the amino acid substitutions that cause temperature sensitivity, and substitutions that destabilize a candidate protein can be chemically varied (19). Nevertheless, in those proteins for which there is structural information, the locations of temperature-sensitive mutations correlate with low side chain mobility and low surface accessibility (24, 25, 34, 37). This mutation may underpin subtle conformational changes which are revealed at the elevated temperature.
In relation to defining the ts nature of mutant proteins, early work defined two general classes, termed TL for thermolabile mutants and TSS (or TSF) for mutants displaying temperature-sensitive synthesis (29, 32, 33). In TL mutants, an active protein produced at the permissive temperature is inactivated upon exposure to the restrictive temperature. For TSS proteins, the substitution appears to affect properties of folding intermediates rather than of the native state. TSS proteins are not defective if expression takes place at the permissive temperature and the proteins are then exposed to the restrictive temperature. Although it is beyond the scope of the current work, we are now in a better position to examine these features in the ts VP1-2 with the defined mutation. Having constructed a defined version of the mutation in the background of KOS and having a paired strain for comparison will enable a more robust examination of the likely TL properties of the mutant VP1-2 in virions (i.e., those properties underpinning the very early defect, where virus produced at the permissive temperature displays temperature sensitivity in entry, before new protein synthesis). The defect early in infection has been attributed to a defect after entry, at the stage of nuclear pore docking. However, the TL nature of virions themselves requires further analysis. Infectivity of isolated virions may be differentially temperature sensitive outside the cell compared to the situation upon detachment from the envelope, and other tegument proteins, after infection, situations clearly that are likely to have differing conformations and interactions for VP1-2. Having constructed viruses with defined mutations in a well-characterized background, we can also undertake a more quantitative approach in attributing the profound 5-log decrease in overall plaque formation to different requirements for VP1-2 at the early versus the late phases of infection.
Finally, since we now indentify a single mutation in a reasonably well-conserved region sufficient for the ts block, it may be possible to generate ts versions of VP1-2 in other HSV strains or in other alphaherpesviruses, which will facilitate comparative studies of early entry pathways, assembly pathways, and the role of VP1-2 in these other contexts. These results and viruses will provide useful tools for further analysis of the structure-function relationships in VP1-2 and its roles at various stages of entry and assembly and in determining the outcome of infection.
Acknowledgments
We thank A. Buchan for the tsB7 and parental HFEM viruses, Prashant Desai for HSV-1 KΔUL36, antibody, and the complementing cell line HS30, and Roger Everett for anti-ICP8 antibody.
C.M.C. was supported by a University Research Fellowship from the Royal Society. F.A. was supported by a fellowship from the Spanish Ministerio de Educacion y Ciencia. This work was funded by Marie Curé Cancer Care.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1.Abaitua, F., and P. O'Hare. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J. Virol. 82:5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abaitua, F., R. N. Souto, H. Browne, T. Daikoku, and P. O'Hare. 2009. Characterization of the herpes simplex virus (HSV)-1 tegument protein VP1-2 during infection with the HSV temperature-sensitive mutant tsB7. J. Gen. Virol. 90:2353-2363. [DOI] [PubMed] [Google Scholar]
- 3.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benyesh-Melnick, M., P. A. Schaffer, R. J. Courtney, J. Esparza, and S. Kimura. 1975. Viral gene functions expressed and detected by temperature-sensitive mutants of herpes simplex virus. Cold Spring Harbor Symp. Quant. Biol. 39:731-746. [DOI] [PubMed] [Google Scholar]
- 6.Bottcher, S., et al. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottcher, S., et al. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 80:9910-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLuca, N. A., M. A. Courtney, and P. A. Schaffer. 1984. Temperature-sensitive mutants in herpes simplex virus type 1 ICP4 permissive for early gene expression. J. Virol. 52:767-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-116018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon, R. A., and P. A. Schaffer. 1980. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J. Virol. 36:189-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luxton, G. W., et al. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luxton, G. W., J. I. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews, B. W. 1987. Genetic and structural analysis of the protein stability problem. Biochemistry 26:6885-6888. [DOI] [PubMed] [Google Scholar]
- 20.McGeoch, D. J., et al. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 21.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohl, B. S., et al. 2010. Random transposon-mediated mutagenesis of the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 84:8153-8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb, W. W., and J. C. Brown. 14 July 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed]
- 24.Pakula, A. A., and R. T. Sauer. 1989. Genetic analysis of protein stability and function. Annu. Rev. Genet. 23:289-310. [DOI] [PubMed] [Google Scholar]
- 25.Parsell, D. A., and R. T. Sauer. 1989. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J. Biol. Chem. 264:7590-7595. [PubMed] [Google Scholar]
- 26.Preston, C. M. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J. Virol. 29:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preston, V. G. 1981. Fine-structure mapping of herpes simplex virus type 1 temperature-sensitive mutations within the short repeat region of the genome. J. Virol. 39:150-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, A. P., et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadler, J. R., and A. Novick. 1965. The properties of repressor and the kinetics of its action. J. Mol. Biol. 12:305-327. [DOI] [PubMed] [Google Scholar]
- 30.Schaffer, P. A., V. C. Carter, and M. C. Timbury. 1978. Collaborative complementation study of temperature-sensitive mutants of herpes simplex virus types 1 and 2. J. Virol. 27:490-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlieker, C., et al. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturtevant, J. M., M. H. Yu, C. Haase-Pettingell, and J. King. 1989. Thermostability of temperature-sensitive folding mutants of the P22 tailspike protein. J. Biol. Chem. 264:10693-10698. [PubMed] [Google Scholar]
- 33.Thomas, G. J., Jr., R. Becka, D. Sargent, M. H. Yu, and J. King. 1990. Conformational stability of P22 tailspike proteins carrying temperature-sensitive folding mutations. Biochemistry 29:4181-4187. [DOI] [PubMed] [Google Scholar]
- 34.Varadarajan, R., H. A. Nagarajaram, and C. Ramakrishnan. 1996. A procedure for the prediction of temperature-sensitive mutants of a globular protein based solely on the amino acid sequence. Proc. Natl. Acad. Sci. U. S. A. 93:13908-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, E. 1985. Individual HSV transcripts: characterisation of specific genes. Herpesviruses 3:45-91. [Google Scholar]
- 36.Wolfstein, A., et al. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227-237. [DOI] [PubMed] [Google Scholar]
- 37.Yu, M. H., and J. King. 1988. Surface amino acids as sites of temperature-sensitive folding mutations in the P22 tailspike protein. J. Biol. Chem. 263:1424-1431. [PubMed] [Google Scholar]
- 38.Zhang, Y. F., and E. K. Wagner. 1987. The kinetics of expression of individual herpes simplex virus type 1 transcripts. Virus Genes 1:49-60. [DOI] [PubMed] [Google Scholar]