Abstract
Enviroxime is an antienterovirus compound that targets viral protein 3A and/or 3AB and suppresses a step in enterovirus replication by unknown mechanism. To date, four antienterovirus compounds, i.e., GW5074, Flt3 inhibitor II, TTP-8307, and AN-12-H5, are known to have similar mutations in the 3A protein-encoding region causing resistance to enviroxime (a G5318A [3A-Ala70Thr] mutation in poliovirus [PV]) and are considered enviroxime-like compounds. Recently, antienterovirus activity of a phosphatidylinositol 4-kinase III beta (PI4KB) inhibitor, PIK93, was reported, suggesting that PI4KB is an important host factor targetable by antienterovirus compounds (N. Y. Hsu et al., Cell 141:799-811, 2010). In this study, we analyzed the inhibitory effects of previously identified enviroxime-like compounds (GW5074 and AN-12-H5) and a newly identified antienterovirus compound, T-00127-HEV1, on phosphoinositide (PI) kinases. We found that T-00127-HEV1 inhibited PI4KB activity with a higher specificity for than other PI kinases, in contrast to GW5074, which had a broad specificity for PI kinases. In contrast, AN-12-H5 showed no inhibitory effect on PI4KB activity and only moderate inhibitory effects on PI 3-kinase activity. Small interfering RNA (siRNA) screening targeting PI kinases identified PI4KB is a target of GW5074 and T-00127-HEV1, but not of AN-12-H5, for anti-PV activity. Interestingly, T-00127-HEV1 and GW5074 did not inhibit hepatitis C virus (HCV) replication, in contrast to a strong inhibitory effect of AN-12-H5. These results suggested that PI4KB is an enterovirus-specific host factor required for the replication process and targeted by some enviroxime-like compounds (T-00127-HEV1 and GW5074) and that enviroxime-like compounds may have targets other than PI kinases for their antiviral effect.
Poliovirus (PV) is a small nonenveloped virus with a single-strand positive genomic RNA of about 7,500 nucleotides (nt) belonging to Human enterovirus species C in the genus Enterovirus, family Picornaviridae. PV is the causative agent of poliomyelitis, which is caused by the destruction of motor neurons by direct infection by PV in the cells (12, 19). With the established live attenuated oral PV vaccine (OPV) and inactivated PV vaccine (IPV) for PV (43, 44), the global eradication program for poliomyelitis has been continued by the Global Polio Eradication Initiative (GPEI) of the World Health Organization (WHO) since 1988. Currently, indigenous wild PVs are restricted to four countries of endemicity, with a substantial reduction of the cases in the major countries of endemicity, India and Nigeria (with only 40 and 11 cases, respectively, as of November, 2010). However, at least 10 countries are suffering from reestablishment of PV transmission or active outbreaks (e.g., there were more than 450 cases in Tajikistan) (http://www.polioeradication.org/casecount.asp). In the eradication program for poliomyelitis, antivirals for PV are anticipated to have roles in the PV posteradication era, i.e., in control of a circulating vaccine-derived PV (cVDPV) along with IPV and for treatment of patients chronically infected with PV and persons exposed to PV (17, 18). However, there is currently no antiviral available for PV infection.
Compounds with anti-PV activity can be classified as capsid-binding inhibitors, replication inhibitors, and encapsidation inhibitors in terms of the target stages in PV infection. Capsid-binding inhibitors target hydrophobic pockets on the virion and inhibit the PV uncoating process by stabilizing the virion or the human rhinovirus attachment process by inducing a conformational change of the virion (27, 40). Replication inhibitors target both the viral proteins and host proteins. Viral proteins 2A, 2C, 3A, 3C, and 3D have been identified as direct or indirect targets of anti-PV compounds, including elastase inhibitors (38), guanidine hydrochloride (GuHCl) (16), enviroxime (29, 50), rupintrivir (AG7088) (23, 39), and gliotoxin (42), respectively. An elastase inhibitor, methoxy-succinyl-Ala-Ala-Pro-Val-chloromethyl ketone (MPCMK), also acted as a viral 2A protease inhibitor by forming a covalent bond with the active site of 2A protease (Cys109) and possibly the Val residue of MPCMK, and it reduced the production of infectious PV but not of infectious encephalomyocarditis virus (EMCV) (38, 49). GuHCl inhibits the initiation of negative-strand RNA synthesis by targeting viral proteins 2C and/or 2BC (8, 11, 16). GuHCl and some benzimidazole derivatives, including 2-(alpha-hydroxybenzyl)-benzimidazole (26), MRL-1237 (45), and TBZE-029 (22), belong to a group of 2C inhibitors in terms of the resistance mutations in the 2C-encoding region. Enviroxime inhibits positive-strand RNA synthesis by preventing normal formation of the replication complex, possibly by targeting viral proteins 3A and/or 3AB, although its direct interaction with the 3A protein was not detected (15, 29, 50). Recently, anti-PV compounds that have little structural similarity to enviroxime but induce common resistance mutations in the 3A-encoding region (enviroxime-like compounds) have been discovered, including TTP-8307 (24), some cellular protein kinase inhibitors (GW5074 and Flt3 inhibitor II) (4, 5), and the bifunctional antienterovirus compound AN-12-H5, which targets the replication processes of PV and enterovirus 71 (EV71) and also an early stage of EV71 infection (3). Rupintrivir was originally discovered as an irreversible inhibitor of human rhinovirus 3C protease by protein structure-based drug design methodologies (36). Inhibitory effects of the compounds on the function of viral proteins and/or evidence of direct interaction of viral proteins with the compounds suggested that these anti-PV compounds directly target viral proteins, except for enviroxime and enviroxime-like compounds (15, 36, 38, 41, 42).
Some host proteins have been identified as the targets of replication inhibitors as well as viral protein. To date, eIF4A, GBF1, and phosphatidylinositol 4-kinase III beta (PI4KB) have been identified as the targets of the replication inhibitors hippuristanol, brefeldin A, and PIK93, respectively (14, 30, 31, 37). Hippuristanol, a natural product of the coral Isis hippuis, suppresses initiation of translation by inhibiting RNA binding of eIF4A, and it delayed the expression of viral proteins in PV replication for 2 h (14). Brefeldin A blocks membrane traffic between the cis- and trans-Golgi compartments by targeting the cellular guanine nucleotide exchange factor GBF1 and inhibits PV replication but not EMCV replication (9, 21, 31, 37). Recently, a PI4KB inhibitor, PIK93, was identified as a potent anti-PV compound that targets PI4KB to suppress interaction of phosphatidylinositol 4-phosphate with viral 3D polymerase on the reorganized membrane vesicle for the formation of the viral replication complex (30). Encapsidation inhibitors target both viral and host proteins. Viral protein 2C and host protein Hsp90 have been identified as the targets of the encapsidation inhibitor hydantoin, which targets viral protein 2C and blocks polyprotein processing (46, 47), and of geldanamycin, which suppresses the encapsidation process by interfering with the folding of capsid proteins (28, 34), respectively. The emergence of mutants resistant to anti-PV compounds targeting host proteins is generally limited; no mutant resistant to geldanamycin was isolated in all attempts (28), and mutants resistant to brefeldin A were isolated after five passages in cultured cells (20). Mutants resistant to anti-PV compounds targeting host factors might have emerged by overcoming suppression of the specific target step by increasing corresponding viral protein activities as a result of the resistance mutation; brefeldin A resistance mutations conferred a resistant phenotype specific to brefeldin A treatment but not to other replication inhibitors (3).
In the present study, we have identified a novel anti-PV compound, T-00127-HEV1, by high-throughput screening with a large-scale chemical library (72,000 compounds). T-00127-HEV1 belonged to the enviroxime-like compounds based on the resistance mutation of PV. Surprisingly, we found that the PI4KB inhibitor PIK93 also belonged to the group of enviroxime-like compounds. This prompted us to analyze the inhibitory effects of enviroxime-like compounds on the activity of phosphoinositide (PI) kinases, including PI4KB. To analyze the specificity of enviroxime-like compounds to enterovirus, we examined the inhibitory effect of the compounds on the replication of another family of positive-strand RNA viruses, hepatitis C virus (HCV). The results suggested that PI4KB is a specific target of some enviroxime-like compounds, including GW5074 and T-00127-HEV1, but not AN-12-H5, for anti-PV activity. Strong anti-HCV activity of AN-12-H5, but not of GW5074 and T-00127-HEV1, suggested that enviroxime-like compounds may have targets other than PI kinases for their antiviral effect.
MATERIALS AND METHODS
Cells, viruses, and chemical library.
RD cells (a human rhabdomyosarcoma cell line) and HEK293 cells (human embryonic kidney cells) were cultured as monolayers in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). RD cells were used for titration of viruses and pseudoviruses and for screening. HEK293 cells were used for small interfering RNA (siRNA) screening to identify cellular targets of antienterovirus compounds. The Huh7.5.1 cell line was a kind gift from Frank Chisari (Scripps Research Institute). Huh7.5.1 cells were used for analysis of the inhibitory effects of anti-PV compounds on HCV replication. PV and EV71 pseudoviruses (TE-PV-Fluc mc and TE-EV71-Fluc mc), which encapsidated luciferase-encoding PV and EV71 replicons with capsid proteins derived from PV1(Mahoney) and EV71(Nagoya), respectively, were used for screening of antienterovirus compounds (1, 2, 5). PV1(Mahoney), EV71(Nagoya), and coxsackievirus B3 (CVB3) (Nancy) were used to analyze the inhibitory effects of identified compounds on enterovirus infection. PV pseudovirus mutants that have known drug resistance mutations, including G5318A (enviroxime and GW5074 resistance, 3A-Ala70Thr) (4, 29), U4614A (guanidine hydrochloride [GuHCl] resistance, 2C-Phe164Tyr) (7), and G4361A and C5190U (brefeldin A resistance, 2C-Val80Ile and 3A-Ala27Val) (20), were used for characterization of identified antienterovirus compounds (3). A diverse subset of 72,000 compounds from a chemical library at the University of Tokyo was used for screening. T-00127-HEV1 was supplied by Pharmeks Ltd. (Moscow, Russia), and the purity was checked by liquid chromatography-mass spectrometry (LC/MS). An siRNA library targeting human GBF1, ARF1, phosphatidylinositol 3-kinase-related kinase (ATM, ATR, FRAP1, PRKDC, SMG1, and TRRAP), and phosphoinositide (PI) kinase (PI4KA, PI4KB, PI4K2A, PI4K2B, PIK3C2A, PIK3C2B, PIK3C2G, PIK3C3, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PIP4K2A, PIP4K2B, PIP4K2C, PIP5K1A, PIP5K1B, PIP5K1C, PIP5KL1, and PIP5K3) genes was purchased from Thermo Fisher Scientific as a form of siGENOME SMART pools, which contain four sets of different siRNAs for each mRNA and were validated as having over 75% knockdown efficiency for target mRNAs. As control siRNAs, siGLO cyclophilin B control siRNA, siGLO lamin A/C control siRNA, siGENOME nontargeting siRNAs 1 and 2, siGENOME RISC-FREE control siRNA, and siGENOME Tox transfection control were used in each experiment.
Screening of antienterovirus compounds.
Five microliters of compound solution (60 μM; the final concentration of 10 μΜ) and 5 μl of PV pseudovirus solution (800 infectious units [IU]) were added to RD cells (5.0 × 103 cells per well in 20 μl medium) in 384-well plates (catalog no. 781080; Greiner Bio-One), and then the cells were incubated at 37°C for 7 h. Luciferase activity of the infected cells was measured at 7 h postinfection (p.i.) with the Steady-Glo luciferase assay system (Promega) using a 2030 ARVO X luminometer (Perkin-Elmer) according to the manufacturer's instructions. PV pseudovirus infection was calculated as a percentage of luciferase activity of the infected cells, where the luciferase activity in the infected cells in the absence of compounds was taken as 100%. The cutoff value of the inhibitory effect of candidate compounds was set to <10% of PV pseudovirus infection in the treated cells.
The cytotoxicity of the compounds was evaluated by viability of compound-treated cells in the screening. For candidate compounds, cytotoxicity was further evaluated by determination of the 50% cytotoxic concentration (CC50) of the compounds. RD cells (1.0 × 104 cells per well in 50 μl medium) were cultured at 37°C in 96-well plates (Becton Dickinson), followed by addition of 50 μl compound solution (final concentrations of 10 μM for screening and 16 to 500 μM for determination of CC50). The cells were incubated at 37°C for 2 days and then subjected to measurement of ATP as a marker of metabolically active cells by using a Cell Titer-Glo luminescent cell viability assay kit (Promega) according to the manufacturer's instructions. The cutoff value of screening for candidate compounds was set at >90% of viability of treated cells compared to that of mock-treated cells.
The inhibitory effect of anti-PV activity of the compounds was evaluated by 50% effective concentration (EC50). RD cells (1.4 × 104 cells per well in 100 μl medium) in a 96-well plate were inoculated with 50 μl of PV pseudovirus (400 IU) and 50 μl of compound solution (final concentrations of 0.078 to 10 μM). The cells were incubated at 37°C for 7 h. Luciferase activity in the infected cells was measured at 7 h p.i. The EC50 values of a compound were obtained by nonlinear regression analysis of the dose-response curve.
Inhibitory effect of T-00127-HEV1 on enterovirus infection.
The inhibitory effect of T-00127-HEV1 on PV, EV71, and CVB3 infection was evaluated by measurement of number of copies of the viral genome in the infected cells. RD cells (1.0 × 104 cells per well in 100 μl medium) in a 96-well plate were infected with PV1(Mahoney), EV71(Nagoya), or CVB3(Nancy) at multiplicities of infection (MOI) of 10, 1.0, and 0.1 at 37°C for 1 h in the absence of T-00127-HEV1. The cells were washed three times with 10% FCS-DMEM, followed by the addition of 100 μl of 10% FCS-DMEM containing 10 or 0 μM T-00127-HEV1. Cells were collected at 16 h p.i., and then viral RNA was extracted from the cells using a High Pure viral RNA purification kit (Roche). The number of copies of the viral genome was quantified using a real-time PCR system.
Quantification of viral RNA by real-time PCR.
Real-time PCR was performed as previously described by Dierssen et al. with modification (25). Viral RNA was assayed in a 20-μl reaction mixture containing 5 μl of viral RNA by using the one-step SYBR PrimeScript Plus reverse transcription-PCR (RT-PCR) kit (TaKaRa) with primers EQ-1 and EQ-2 (25). Viral RNA of PV1(Sabin) was used to control the quantification of the number of copies. The mixtures were subjected to real-time PCR; the PCR conditions consisted of a reverse transcription step at 42°C for 30 min and 40 cycles of thermal cycling at 95°C for 3 s and 60°C for 30 s. The fluorescence emission of SYBR green I was monitored and analyzed by using an Applied Biosystems 7500 Fast real-time PCR system (Applied Biosystems).
Inhibitory effect of antienterovirus compounds on in vitro activity of cellular protein kinases and PI kinases.
The inhibitory effect of T-00127-HEV1 at a concentration of 10 μM on in vitro cellular protein kinase activities was assessed by kinase profiling with an ATP concentration near the Km for each kinase (Carna Biosciences, Inc., Japan). Inhibitory effects of GW5074, AN-12-H5, and T-00127-HEV1 at a concentration of 10 μM on in vitro PI kinase activities were assessed by the SelectScreen kinase profiling service with an ATP concentration of 10 μM (Invitrogen). For T-00127-HEV1, the 50% inhibitory concentration (IC50) for in vitro PI4KB activity was also measured with an ATP concentration of 10 μM.
Inhibitory effects of antienterovirus compounds on HCV replication.
RNA transcripts of HCV replicons were obtained using a RiboMAX large-scale RNA production system T7 kit (Promega), using XbaI-linearized DNAs of the HCV replicon (the replicon clone pSGR-JFH-LucNeo-4 and a replication-defective clone with a disrupted polymerase motif [GDD to GND], JFHluci-GND) as the templates. The in vitro-synthesized RNA transcripts were transfected into Huh7.5.1 cells by electroporation as described previously with modifications (48). Ten micrograms of in vitro-synthesized RNA transcripts was mixed with 3.2 × 106 Huh7.5.1 cells in 400 μl Opti-MEM (Invitrogen). The mixture was transferred to an electroporation cuvette (Gene Pulser cuvette [0.4-cm electrode]; Bio-Rad) and then pulsed at 270 V and 950 μF using a Gene Pulser II apparatus (Bio-Rad). Transfected cells were immediately suspended in 10 ml of 10% FCS-DMEM and then transferred to 96-well plates (Microtest 96-well assay plate; Becton Dickinson) at 100 μl per well. The cells were incubated at 37°C for 1 h, and then 25 μl of antienterovirus compound solutions (final concentrations of 0.078 to 10 μM) or medium was added. The cells were incubated at 37°C for 23 h, and then the luciferase activity in the cells was measured at 24 h posttransfection (p.t.) with the Steady-Glo luciferase assay system (Promega) using a 2030 ARVO X luminometer (Perkin-Elmer) according to the manufacturer's instructions.
siRNA transfection.
An RNA duplex of each siRNA (final concentration of 50 nM) was transfected into HEK293 cells (5.0 × 103 cells in 100 μl medium per well) in 96-well plates (Microtest 96-well assay plate; Becton Dickinson) by using DharmaFECT1 transfection reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. The cells were incubated at 37°C for 24 h, and then the supernatant was removed and replaced with 100 μl per well of 10% FCS-DMEM. The transfection efficiency of siRNA in the cells was evaluated by the efficiency of incorporation of fluorescence-labeled siRNA (siGLO control siRNAs) in the transfected cells at 24 h p.t. and by the efficiency of cell death in the cells transfected with siGENOME Tox transfection control at 96 h p.t. (cells transfected with this control reagent die by apoptosis). siRNA-transfected cells were used for experiments at 96 h p.t.
Western blot analysis.
siRNA-treated cells in three wells of 96-well plates were collected at 96 h p.t. in 100 μl cell lysis buffer (21 mM HEPES buffer [pH 7.4], 1.8 mM disodium hydrogen phosphate, 137 mM NaCl, 4.8 mM KCl, 0.5% Nonidet P-40, and 0.5 mM EDTA) and then were subjected to 5 to 20% gradient polyacrylamide gel electrophoresis (e-PAGEL; Atto Corporation) in a Laemmli buffer system (33). The proteins in the gel were transferred to a polyvinylidene difluoride filter (Immobilon; Millipore) and blocked by using a blocking reagent (Qiagen) (for staining with anti-GBF1 and -ARF1 antibodies) or 5% nonfat dry milk (for staining with anti-PI4KB antibody). The filters were incubated with rabbit anti-GBF1, -ARF1, and -PI4KB antibodies (Abcam) (1:500, 1:250, and 1:50 dilutions, respectively) at room temperature for 1 h. The filters were washed with Tris-buffered saline (TBS) (20 mM Tris-HCl [pH 7.4], 137 mM NaCl) containing 0.1% Tween 20 (TBS-T) three times for 5 min each and then incubated with goat anti-rabbit IgG antibodies conjugated with horseradish peroxidase (Pierce) (1:1,000 dilution) at room temperature for 1 h. The filters were washed with TBS-T three times for 5 min each, and then treated with SuperSignal West Femto Maximum Sensitivity substrate (Pierce) for the detection of the signal.
Target identification by siRNA sensitization (TISS) assay.
siRNA-transfected cells were inoculated with 800 IU PV pseudovirus in the presence of suboptimal concentrations of anti-PV compounds or medium (as a mock-treated control) in a total of 200 μl per well at 96 h p.t. of siRNAs. The cells were incubated at 37°C for 7 h, and then the luciferase activity in the cells was measured with the Steady-Glo luciferase assay system (Promega) using a 2030 ARVO X luminometer (Perkin-Elmer) according to the manufacturer's instructions. The mean net relative light units (RLU) detected in mock-treated cells were 1.3 × 104 to 1.4 × 104 RLU, with standard deviations of 14% of the mean.
PV pseudovirus infection in drug-treated cells was calculated as a percentage of luciferase activity of the infected cells, where the luciferase activity in the infected cells in the absence of compounds was taken as 100% (for Fig. 1 and 5). PV pseudovirus infection in siRNA-transfected cells was calculated as a percentage of luciferase activity of the infected cells, where the luciferase activity in mock-transfected cells in the absence of compounds was taken as 100% (for Fig. 2). PV pseudovirus infection in drug-treated and siRNA-transfected cells was calculated as a percentage of luciferase activity of the infected cells, where the luciferase activity in mock-transfected cells in the presence of the compounds was taken as 100%.
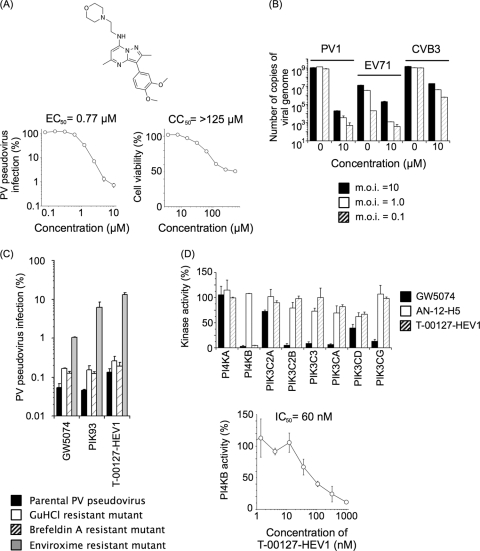
FIG. 1.
Characterization of T-00127-HEV1 and inhibitory effects on PI kinases. (A) Characterization of T-00127-HEV1. Upper panel, structure of T-00127-HEV1. Lower panels, inhibitory effect of T-00127-HEV1 on PV pseudovirus (left panel) and viability of RD cells (right panel). PV pseudovirus infection in the absence of compounds was taken as 100%. (B) Inhibitory effect of T-00127-HEV1 on enterovirus infection. RD cells were infected with PV1(Mahoney), EV71(Nagoya), or CVB3(Nancy) at MOI of 10, 1.0, and 0.1 in the absence of T-00127-HEV1 and then were treated with 0 or 10 μM T-00127-HEV1 from 1 h p.i. The total number of copies of viral genomes in the cells at 16 h p.i. is shown. (C) Specificity of mutations causing resistance to antienterovirus compounds. RD cells were infected with PV pseudovirus mutants that have mutations causing resistance to GuHCl (U4614A), brefeldin A (G4361A plus C5190U), and enviroxime (G5318A) in the presence of antienterovirus compounds GW5074 (25 μM), PIK93 (1.3 μM), and T-00127-HEV1 (4.0 μM). PV pseudovirus infection in the absence of compounds was taken as 100%. (D) Inhibitory effects of enviroxime-like compounds on in vitro activities of PI kinases. Upper panel, in vitro activities of PI kinases were analyzed in the presence of enviroxime-like compounds (GW5074, AN-12-H5, and T-00127-HEV1 at a concentration of 10 μM) with an ATP concentration of 10 μM. Lower panel, inhibitory effect of T-00127-HEV1 on the in vitro PI4KB activity measured with an ATP concentration of 10 μM.
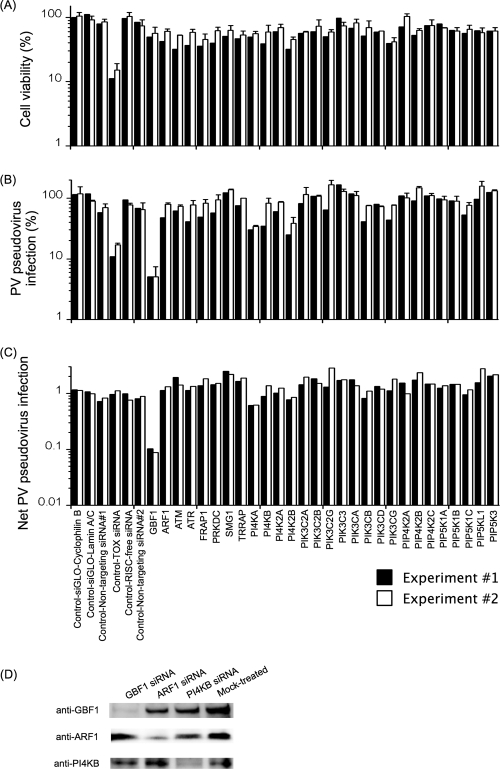
FIG. 2.
Effect of siRNA treatment targeting PI kinases on PV pseudovirus infection. (A) Effect of siRNA treatment on cell viability. The viability of mock-treated cells was taken as 100%. (B) Effect of siRNA treatment on PV pseudovirus infection. PV pseudovirus infection in mock-transfected cells was taken as 100%. (C) Net PV pseudovirus infection in siRNA-treated cells. Net PV pseudovirus infection is the ratio of PV pseudovirus infection in siRNA-transfected cells (percent) to cell viability (percent). The net PV pseudovirus infection in mock-transfected cells was 1. (D) Western blot analysis of cell lysates of the cells transfected with siRNAs targeting the GBF1, ARF1, and PI4KB genes.
To evaluate the direct effect of siRNA treatment, net PV pseudovirus infection, which is the ratio of PV pseudovirus infection in siRNA-transfected cells (percent) to cell viability (percent), was determined for each siRNA treatment (Fig. 2). The net PV pseudovirus infection in mock-transfected cells was 1. To evaluate sensitization of the cells to the compounds induced by siRNA treatment, normalized PV pseudovirus infection, which is the ratio of PV pseudovirus infection in drug-treated and siRNA-transfected cells (percent) to PV pseudovirus infection in siRNA-transfected cells in the absence of compounds (percent), was determined for each siRNA treatment. The normalized PV pseudovirus infection in mock-transfected cells was 1.
Statistical analysis.
The results of experiments are shown as the averages with standard deviations. For comparison of the effect of siRNA treatment on sensitization of the cells to enviroxime-like compounds between two different siRNAs, a paired t test was performed with normalized PV pseudovirus infection obtained in four independent experiments. P values of less than 0.05 were considered to represent significant differences and are indicated by asterisks in the figures.
RESULTS
T-00127-HEV1 is a highly specific inhibitor for PI4KB.
To identify potent anti-PV compounds that target conserved factors required for enterovirus replication, we performed a screening of 72,000 compounds by using a PV pseudovirus infection system (2, 5). We identified 488 compounds as initial hits in the screening. Candidate compounds were further selected from the initial hits according to the following criteria: (i) no apparent cytotoxicity was observed after 2 days of treatment with compounds at 10 μM (>90% viability with no morphological changes of the cells) (18 compounds were selected out of the 488 initial hits); (ii) compounds target the replication step (>90% inhibitory effect after the uncoating step) (8 compounds were selected out of the 18 noncytotoxic compounds); and (iii) compounds inhibit replication of enterovirus 71 (EV71), which belongs to Human enterovirus species A, a different species from PV (5 compounds were selected out of the 8 replication inhibitors). Thus, we finally identified 5 candidate compounds that met these criteria from the 72,000 compounds. Among these candidate compounds, T-00127-HEV1 showed more potent anti-PV activity (EC50 of 0.77 μM) than the other candidate compounds (EC50 of 1.7 to 4.7 μM), anti-EV71 activity (EC50 of 0.73 μM), a low cytotoxicity (CC50 of >125 μM), and a high selectivity index (SI) for PV (>162) (Fig. 1A and data not shown). The inhibitory effect of T-00127-HEV1 on enterovirus infection was analyzed with PV1(Mahoney), EV71(Nagoya), and CVB3(Nancy), which belongs to Human enterovirus species B (Fig. 1B). T-00127-HEV1 showed a broad inhibitory effect on infection with these viruses, with the highest activity against PV infection under the conditions examined.
To characterize the anti-PV activity of T-00127-HEV1 in terms of the resistance mutation of PV, we analyzed the inhibitory effect of T-00127-HEV1 on a panel of PV pseudovirus mutants with drug resistance mutations, including brefeldin A resistance, GuHCl resistance, and enviroxime resistance (Fig. 1C). In this characterization, we also examined the inhibitory effect of the recently identified novel antienterovirus compound PIK93 (a PI4KB inhibitor) on this panel (30), because its virus resistance determinants have not been identified. Surprisingly, we found that both of the compounds belong to the group of enviroxime-like compounds targeting the 3A/3AB proteins of PV. This prompted us to analyze the inhibitory effects of previously identified enviroxime-like compounds, including GW5074 (5), AN-12-H5 (3), and T-00127-HEV1, on PI kinases (Fig. 1D). GW5074 and T-00127-HEV1 almost completely inhibited PI4KB kinase activity at 10 μM (3% and 5% of residual activity, respectively), in contrast to AN-12-H5 (108% of activity [no inhibition]). GW5074 inhibited most of the PI3 kinases, in contrast to T-00127-HEV1, which showed a high specificity to PI4KB with a moderate inhibitory effect on PIK3CD activity (67% of activity). AN-12-H5 showed only a moderate inhibitory effect on PI kinases, and no complete inhibition was observed.
To further characterize the specificity of T-00127-HEV1 for PI4KB, we analyzed the inhibitory effect of T-00127-HEV1 on in vitro activity of cellular protein kinases, including tyrosine kinases and serine/threonine kinases (a total of 150 kinases), because one of the enviroxime-like compounds, GW5074, targets several cellular protein kinases, including Raf-1 (6) (see Table S1 in the supplemental material). The inhibitory effect of T-00127-HEV1 on the examined kinases was weak, i.e., at most 29% inhibition for TTK kinase and <10% inhibition for other kinases at a concentration of 10 μM.
These results suggested that PI4KB is a target of some enviroxime-like compounds as well as PIK93 and that T-00127-HEV1 is a highly specific inhibitor of PI4KB.
PI4KB is a cellular target of GW5074 and T-00127-HEV1 for anti-PV activity.
To analyze the cellular target of the anti-PV activities of enviroxime-like compounds, we first performed siRNA knockdown analysis targeting known PI kinases and related host factors for PV replication (GBF1 and ARF1), which are involved in membrane traffic, as well as PI4KB, on PV pseudovirus infection (Fig. 2). siRNA transfection efficiency was monitored by incorporation of fluorescence-labeled siRNAs and the Tox transfection control. Incorporation of fluorescence-labeled siRNAs was observed in virtually all the cells at 24 h p.t. (data not shown). The viability of cells treated with the siGENOME Tox transfection control was 11 to 15% of that of mock-treated cells at 96 h p.t. (Fig. 2A), suggesting a high transfection efficiency. To analyze the net inhibitory effect of siRNA treatment, net PV pseudovirus infection was determined by normalization of PV pseudovirus infection in siRNA-transfected cells to cell viability (Fig. 2B and C). siRNA targeting the GBF1 gene strongly inhibited the infection (0.089 to 0.1 of net PV pseudovirus infection), and a partial inhibition by siRNA targeting the PI4KA gene was observed (0.61 to 0.62 of net PV pseudovirus infection). For other siRNA treatments, including that targeting the PI4KB gene, significant inhibition of PV pseudovirus infection was not observed in normalized inhibition. To confirm specific reduction of the target proteins in the cells treated with the siRNAs, Western blot analysis was performed for siRNA-transfected cells targeted for the GBF1, ARF1, and PI4KB genes (Fig. 2D). The expression levels of the targeted proteins were substantially reduced, with specificity, by the corresponding siRNAs targeting the GBF1, ARF1, and PI4KB genes. These results suggested that reduction of PI kinases by siRNA treatment was not sufficient to affect PV replication except in the case of reduction of GBF1.
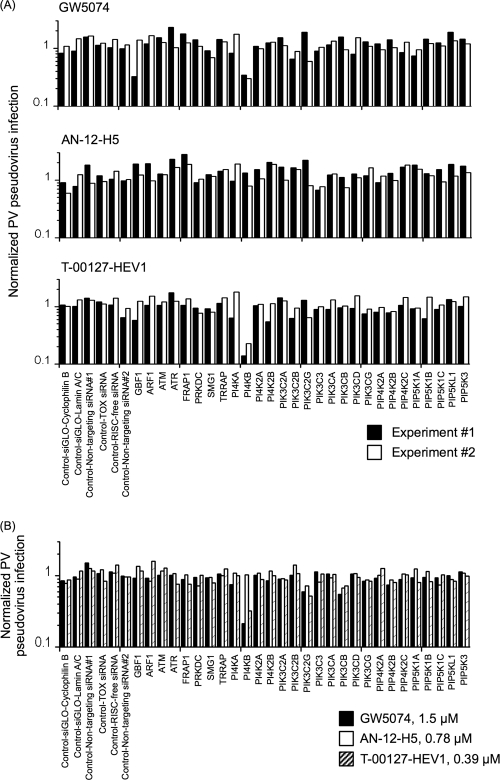
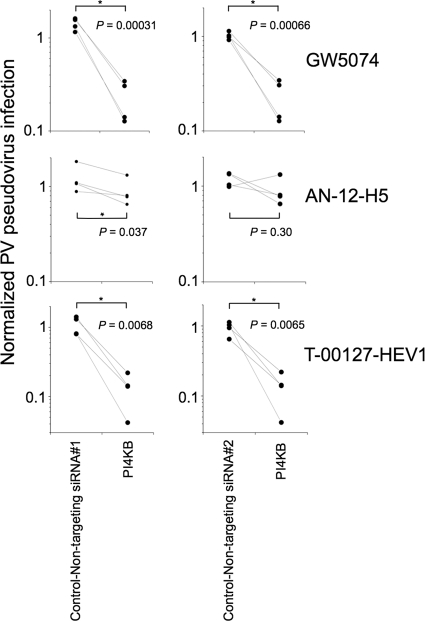
To identify the cellular targets of GW5074, AN-12-H5, and T-00127-HEV1, we developed a target identification by siRNA sensitization (TISS) assay to identify genes that affect the sensitivity of the cells to these compounds. In this assay, the effect of siRNA treatment on the sensitivity of the cells to the compounds was analyzed to identify the target proteins of the compounds for their anti-PV activity, which could be increased by reduced expression levels of the target proteins. The sensitivity of the cells to the compounds was analyzed at suboptimal concentrations of the compounds (GW5074, AN-12-H5, and T-00127-HEV1), with which only moderate inhibition of PV pseudovirus infection was observed in mock-transfected cells (22 to 47% and 57 to 83% of the infection level in mock-treated cells under the conditions used for Fig. 3A and B, respectively), but substantial inhibition could be observed in the cells transfected with siRNAs targeting the target proteins. In the two independent experiments, cells transfected with siRNA targeting the PI4KB gene showed increased sensitivity to GW5074 and T-00127-HEV1 (Fig. 3A). Increased sensitivity was also observed with lower concentrations of these compounds (Fig. 3B). However, in contrast to the case for GW5074 and T-00127-HEV1, cells transfected with the siRNAs examined did not show any increased sensitivity to AN-12-H5. The sensitization of the cells transfected with siRNA targeting the PI4KB gene to GW5074 and T-00127-HEV1 was specific and significant compared with that with nontargeting siRNA controls (Fig. 4). The increase of the sensitization of the cells to AN-12-H5 by treatment with siRNA targeting the PI4KB gene was modest (comparison with nontargeting siRNA 1) or not significant (comparison with nontargeting siRNA 2). These results suggested that GW5074 and T-00127-HEV1 target PI4KB for their anti-PV activities in the cells and that AN-12-H5 has another target(s) for its anti-PV activity.
FIG. 3.
Effects of enviroxime-like compounds on PV pseudovirus infection in siRNA-transfected cells. (A) Normalized PV pseudovirus infection in the presence of GW5074 (3.1 μM), AN-12-H5 (1.5 μM), and T-00127-HEV1 (0.78 μM). Normalized PV pseudovirus infection is the ratio of PV pseudovirus infection in drug-treated and siRNA-transfected cells (percent) to PV pseudovirus infection in siRNA-transfected cells in the absence of compounds (percent). The normalized PV pseudovirus infection in mock-transfected cells was 1. (B) Normalized PV infection in the presence of GW5074, AN-12-H5, and T-00127-HEV1 at lower concentrations (1.5, 0.78, and 0.39 μM).
FIG. 4.
Effect of enviroxime-like compounds on PV pseudovirus infection in PI4KB siRNA-transfected cells. HEK293 cells were transfected with siRNA targeting the PI4KB gene or with nontargeting siRNAs (nontargeting siRNAs 1 and 2), and then sensitization of the siRNA-transfected cells was analyzed in the presence of GW5074 (3.1 μM), AN-12-H5 (1.5 μM), and T-00127-HEV1 (0.78 μM). *, P < 0.05.
Comparison of inhibitory effects of antienterovirus compounds on PV and HCV replication.
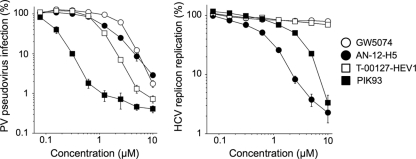
PI4KB has been reported as a host factor for enterovirus replication (30), and its role in HCV replication has also been suggested from experiments using PIK93 and siRNA (13). On the other hand, Berger et al. showed that only PI4KA among the PI4Ks was required for HCV replication (10). To clarify the role of PI4KB in enterovirus and HCV replication, we analyzed the effects of identified anti-PV compounds on the replication of the HCV replicon (Fig. 5). PIK93 showed anti-PV activity and anti-HCV activity, consistent with previous reports (13, 30), although the observed anti-HCV activity was weaker that that previously reported (EC50 of 1.9 μM). The order of potency of the compounds on PV pseudovirus infection was PIK93 > T-00127-HEV1 > AN-12-H5 > GW5074. PIK93 showed the highest anti-PV activity among the compounds, with an EC50 of 0.14 μM for PV pseudovirus infection (Table 1). In contrast to the case for PIK93, no anti-HCV activity was observed for GW5074 and T-00127-HEV1. Surprisingly, AN-12-H5 showed a strong anti-HCV activity even higher than that of PIK93. This suggested that AN-12-H5 has targets other than PI4KB for its anti-PV and anti-HCV activities.
FIG. 5.
Effects of enviroxime-like compounds on PV pseudovirus infection and HCV replicon replication. Inhibitory effects of enviroxime-like compounds on PV pseudovirus infection (left panel) and HCV replicon replication (right panel) are shown. PV pseudovirus infection or HCV replication in the absence of compounds was taken as 100%.
TABLE 1.
Properties of antienterovirus compounds in PV and HCV replicationa
| Compound | CC50 (2 days), μM | Mean (SD) EC50, μM |
SIb (PV) | |
|---|---|---|---|---|
| PV | HCV | |||
| GW5074 | 96 | 4.6 (0.85) | >10 | 21 |
| AN-12-H5 | 78 | 1.1 (0.18) | 0.69 (0.11) | 74 |
| T-00127-HEV1 | >125c | 0.77 (0.10) | >10 | >162 |
| PIK93 | 12 | 0.14 (0.0086) | 1.9 (0.5) | 85 |
Data for GW5074 and AN-12-H5 were adapted in part from reference 3.
SI, selectivity index (ratio of CC50 to EC50).
T-00127-HEV1 showed precipitation at above 125 μM.
DISCUSSION
In the present study, we performed high-throughput screening with a large-scale chemical library (72,000 compounds) and identified a novel enviroxime-like compound, T-00127-HEV1. T-00127-HEV1 was the most potent candidate compound, with broad specificity for enteroviruses (PV1, EV71, and CVB3) and low cytotoxicity. In the characterization of the anti-PV activity of T-00127-HEV1, we fortuitously found that T-00127-HEV1 and PI4KB inhibitor, PIK93, which also has a potent anti-PV activity (30), belong to the same group of anti-PV compounds, the enviroxime-like compounds, in terms of the resistance mutations (Fig. 1). PIK93 is a PI4KB inhibitor (IC50 of 19 nM) but also has an almost equal (for PIK3CA and PIK3CG) or lesser (for PIK3CB, PIK3CD, PIK3C2B, and PIK3C3) inhibitory effect on other PI kinases (32). Biological evidence (including physical complex with viral replication complex, colocalization with viral 3A protein, siRNA knockdown analysis targeting PI4KB, and binding activity of PI4P with viral polymerase) indicated the involvement of PI4KB in enterovirus replication (30). T-00127-HEV1 showed a less potent inhibitory effect on in vitro PI4KB activity (IC50 of 60 nM) than that of PIK93 but with a high specificity for PI4KB (at most 33% inhibition of PIK3CD activity and no inhibitory effect on other PI3 kinases with 10 μM T-00127-HEV1) (Fig. 1). T-00127-HEV1 did not show a significant inhibitory effect on cellular protein kinases examined, as reported for PIK93 (32) (see the supplemental material). The highly specific inhibitory effect of T-00127-HEV1 on PI4KB activity provides pharmacological evidence to support the importance of PI4KB activity in PV replication.
To date, four enviroxime-like compounds have been reported, i.e., TTP-8307 (24), the cellular protein kinase inhibitors GW5074 and Flt3 inhibitor II, and the bifunctional enterovirus inhibitor AN-12-H5 (3-5, 24). To test the hypothesis that PI4KB is the target of enviroxime-like compounds, we analyzed the inhibitory effects of these enviroxime-like compounds on PI4KB activity in vitro and in an in vivo TISS assay (Fig. 1D, 3, and 4). In the in vitro assay, GW5074, but not AN-12-H5, showed a significant inhibitory effect on PI4KB activity. In the in vivo assay, we first analyzed the effect of siRNA treatment targeting the PI4KB gene on PV infection, but we found that the treatment was not sufficient to suppress PV infection, in contrast to that targeting the GBF gene, despite the efficient knockdown of PI4KB (Fig. 2C and D). This observation was not consistent with a previous report that showed about 70% inhibition of PV replication by siRNA knockdown of PI4KB (30). This apparent discrepancy might be caused by normalization of the observed PV infection by cell viability examined in this study, because siRNA treatment targeting the PI4KB gene affected cell viability under the conditions examined (39 to 60% of the viability of mock-treated cells), which had significant weight in the evaluation of net PV infection (Fig. 2C). This suggested that residual PI4KB activity was sufficient to support PV replication. The in vivo TISS assay showed that siRNA treatment targeting the PI4KB gene, but not those targeting other genes, conferred significant sensitivity to GW5074 and T-00127-HEV1 but not to AN-12-H5, suggesting that PI4KB is a target of some enviroxime-like compounds for their anti-PV activities (Fig. 3 and 4). Interestingly, we could not observe an effect of siRNA treatment targeting the GBF1 gene on the sensitivity to the examined compounds, which is inconsistent with a proposed role of GBF1 as a tether between viral protein 3A and PI4KB in a model of PV replication (30). However, strong suppression of PV infection by the siRNA treatment, to levels was even lower than that in Tox transfection-treated cells, suggested that the signal detected in the GBF1 knockdown cells might be derived from residual untransfected cells (Fig. 2B). These findings suggested that enviroxime-like compounds have PI4KB and some targets other than PI kinases as targets for anti-PV activity and that the role of GBF1 in the inhibitory effect of enviroxime-like compounds remains to be further elucidated.
We tested the specificity of the effects of enviroxime-like compounds on PV and HCV replication because PI4KB has been suggested as a host factor of HCV replication by experiments using PIK93 and siRNA (13, 30). However, Berger et al. showed that PI4KA, but not PI4KB, is required for HCV replication by providing biological evidence (colocalization of PI4KA with the double-stranded RNA [dsRNA] and NS5A and cofractionation with NS5A) and using siRNA (10). An inhibitory effect of PIK93 on both PV and HCV replication was observed, but the effect on HCV replication was weaker than that on PV replication (Fig. 5 and Table 1), and T-00127-HEV1 did not show any anti-HCV activity. Surprisingly, AN-12-H5 showed the highest anti-HCV activity among the compounds. Our data support that PI4KB activity is not essential for HCV replication, consistent with the report by Berger et al. (10), and that PI4KB is an enterovirus-specific host factor.
Enviroxime-like compounds have been identified with a high hit rate, i.e., approximately 0.1% of analyzed compounds, in small-scale screenings (4, 5). Identification of the cellular protein kinase inhibitors GW5074 and Flt3 inhibitor II as enviroxime-like compounds, both of which were suggested to interact with ATP-binding sites of kinases (32, 35), suggested that a structural similarity to ATP is essential for the enviroxime-like activity. This common structural requirement might have contributed to the high hit rate for enviroxime-like compounds. Another factor contributing to the high hit rate would be the apparent low cytotoxicity of enviroxime-like compounds. This might be attributable to a relatively high specificity of enviroxime-like compounds for limited sets of cellular protein kinases and PI kinases (6, 32) (see the supplemental material). In our current screening, four out of five identified candidate compounds were enviroxime-like compounds (data not shown). We also identified 10 noncytotoxic compounds that target the early stage of infection, possibly capsid-binding inhibitors, in the current screening (data not shown). This suggested that enviroxime-like compounds and capsid-binding inhibitors are predominant groups of anti-PV compounds with low cytotoxicity.
In summary, we identified a novel enviroxime-like compound, T-00127-HEV1, that is a highly specific inhibitor of PI4KB. T-00127-HEV1 did not inhibit HCV replication, in contrast to PV replication. Another enviroxime-like compound, AN-12-H5, did not show any inhibitory effect on PI4KB activity, and its anti-PV activity was not affected by knockdown of PI4KB. These results suggested that PI4KB is an enterovirus-specific host factor required for the replication process and targeted by some enviroxime-like compounds (T-00127-HEV1 and GW5074) and that enviroxime-like compounds may have targets other than PI kinases for their antiviral effect.
Supplementary Material
Acknowledgments
We are grateful to Junko Wada for her excellent technical assistance. We are grateful to Koji Ishii and Takashi Shimoike for technical assistance.
This study was supported in part by Grants-in-Aid for the Promotion of Polio Eradication and Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor and Welfare, Japan, and by a grant from the World Health Organization for a collaborative research project of the Global Polio Eradication Initiative.
Footnotes
Published ahead of print on 22 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Arita, M., Y. Ami, T. Wakita, and H. Shimizu. 2008. Cooperative effect of the attenuation determinants derived from poliovirus Sabin 1 strain is essential for attenuation of enterovirus 71 in the NOD/SCID mouse infection model. J. Virol. 82:1787-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arita, M., N. Nagata, T. Sata, T. Miyamura, and H. Shimizu. 2006. Quantitative analysis of poliomyelitis-like paralysis in mice induced by a poliovirus replicon. J. Gen. Virol. 87:3317-3327. [DOI] [PubMed] [Google Scholar]
- 3.Arita, M., Y. Takebe, T. Wakita, and H. Shimizu. 2010. A bifunctional anti-enterovirus compound that inhibits replication and early stage of enterovirus 71 infection. J. Gen. Virol. 91:2734-2744. [DOI] [PubMed] [Google Scholar]
- 4.Arita, M., T. Wakita, and H. Shimizu. 2009. Cellular kinase inhibitors that suppress enterovirus replication have a conserved target in viral protein 3A similar to that of enviroxime. J. Gen. Virol. 90:1869-1879. [DOI] [PubMed] [Google Scholar]
- 5.Arita, M., T. Wakita, and H. Shimizu. 2008. Characterization of pharmacologically active compounds that inhibit poliovirus and enterovirus 71 infectivity. J. Gen. Virol. 89:2518-2530. [DOI] [PubMed] [Google Scholar]
- 6.Bain, J., et al. 2007. The selectivity of protein kinase inhibitors; a further update. Biochem. J. 408:297-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baltera, R. F., Jr., and D. R. Tershak. 1989. Guanidine-resistant mutants of poliovirus have distinct mutations in peptide 2C. J. Virol. 63:4441-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belov, G. A., Q. Feng, K. Nikovics, C. L. Jackson, and E. Ehrenfeld. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger, K. L., et al. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodian, D. 1949. Histopathologic basis of clinical findings in poliomyelitis. Am. J. Med. 6:563-578. [DOI] [PubMed] [Google Scholar]
- 13.Borawski, J., et al. 2009. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 83:10058-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bordeleau, M. E., et al. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213-220. [DOI] [PubMed] [Google Scholar]
- 15.Brown-Augsburger, P., et al. 1999. Evidence that enviroxime targets multiple components of the rhinovirus 14 replication complex. Arch. Virol. 144:1569-1585. [DOI] [PubMed] [Google Scholar]
- 16.Caliguiri, L. A., and I. Tamm. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35:408-417. [DOI] [PubMed] [Google Scholar]
- 17.Collett, M. S., J. Neyts, and J. F. Modlin. 2008. A case for developing antiviral drugs against polio. Antiviral Res. 79:179-187. [DOI] [PubMed] [Google Scholar]
- 18.Committee on Development of a Polio Antiviral and Its Potential Role in Global Poliomyelitis Eradication, NRC. 2006. Exploring the role of antiviral drugs in the eradication of polio: workshop report. The National Academies Press, Washington, DC.
- 19.Couderc, T., et al. 1989. Molecular pathogenesis of neural lesions induced by poliovirus type 1. J. Gen. Virol. 70:2907-2918. [DOI] [PubMed] [Google Scholar]
- 20.Crotty, S., M. C. Saleh, L. Gitlin, O. Beske, and R. Andino. 2004. The poliovirus replication machinery can escape inhibition by an antiviral drug that targets a host cell protein. J. Virol. 78:3378-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuconati, A., A. Molla, and E. Wimmer. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 72:6456-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Palma, A. M., et al. 2008. The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C. J. Virol. 82:4720-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Palma, A. M., et al. 2008. Potential use of antiviral agents in polio eradication. Emerg. Infect. Dis. 14:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Palma, A. M., et al. 2009. Mutations in the nonstructural protein 3A confer resistance to the novel enterovirus replication inhibitor TTP-8307. Antimicrob. Agents Chemother. 53:1850-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dierssen, U., F. Rehren, C. Henke-Gendo, G. Harste, and A. Heim. 2008. Rapid routine detection of enterovirus RNA in cerebrospinal fluid by a one-step real-time RT-PCR assay. J. Clin. Virol. 42:58-64. [DOI] [PubMed] [Google Scholar]
- 26.Eggers, H. J., and I. Tamm. 1961. Spectrum and characteristics of the virus inhibitory action of 2-(alpha-hydroxybenzyl)-benzimidazole. J. Exp. Med. 113:657-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox, M. P., M. J. Otto, and M. A. McKinlay. 1986. Prevention of rhinovirus and poliovirus uncoating by WIN 51711, a new antiviral drug. Antimicrob. Agents Chemother. 30:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geller, R., M. Vignuzzi, R. Andino, and J. Frydman. 2007. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 21:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinz, B. A., and L. M. Vance. 1995. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J. Virol. 69:4189-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu, N. Y., et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irurzun, A., L. Perez, and L. Carrasco. 1992. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191:166-175. [DOI] [PubMed] [Google Scholar]
- 32.Knight, Z. A., et al. 2006. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell 125:733-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Macejak, D. G., and P. Sarnow. 1992. Association of heat shock protein 70 with enterovirus capsid precursor P1 in infected human cells. J. Virol. 66:1520-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahboobi, S., et al. 2006. Novel bis(1H-indol-2-yl)methanones as potent inhibitors of FLT3 and platelet-derived growth factor receptor tyrosine kinase. J. Med. Chem. 49:3101-3115. [DOI] [PubMed] [Google Scholar]
- 36.Matthews, D. A., et al. 1999. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. U. S. A. 96:11000-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maynell, L. A., K. Kirkegaard, and M. W. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molla, A., C. U. Hellen, and E. Wimmer. 1993. Inhibition of proteolytic activity of poliovirus and rhinovirus 2A proteinases by elastase-specific inhibitors. J. Virol. 67:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patick, A. K., et al. 1999. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 43:2444-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pevear, D. C., et al. 1989. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez, P. L., and L. Carrasco. 1992. Gliotoxin: inhibitor of poliovirus RNA synthesis that blocks the viral RNA polymerase 3Dpol. J. Virol. 66:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabin, A. B. 1965. Oral poliovirus vaccine. History of its development and prospects for eradication of poliomyelitis. JAMA 194:872-876. [DOI] [PubMed] [Google Scholar]
- 44.Salk, J. E., et al. 1954. Studies in human subjects on active immunization against poliomyelitis. II. A practical means for inducing and maintaining antibody formation. Am. J. Public Health Nations Health 44:994-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu, H., et al. 2000. Mutations in the 2C region of poliovirus responsible for altered sensitivity to benzimidazole derivatives. J. Virol. 74:4146-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verlinden, Y., A. Cuconati, E. Wimmer, and B. Rombaut. 2000. The antiviral compound 5-(3,4-dichlorophenyl) methylhydantoin inhibits the post-synthetic cleavages and the assembly of poliovirus in a cell-free system. Antiviral Res. 48:61-69. [DOI] [PubMed] [Google Scholar]
- 48.Wakita, T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei, A. Z., I. Mayr, and W. Bode. 1988. The refined 2.3 A crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor. FEBS Lett. 234:367-373. [DOI] [PubMed] [Google Scholar]
- 50.Wikel, J. H., et al. 1980. Synthesis of syn and anti isomers of 6-[[(hydroxyimino)phenyl]methyl]-1-[(1-methylethyl)sulfonyl]-1H-benzimidaz ol-2-amine. Inhibitors of rhinovirus multiplication. J. Med. Chem. 23:368-372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.