Abstract
Native hepatitis B surface antigen (HBsAg) spontaneously assembles into 22-nm subviral particles. The particles are lipoprotein micelles, in which HBsAg is believed to span the lipid layer four times. The first two transmembrane domains, TM1 and TM2, are required for particle assembly. We have probed the requirements for particle assembly by replacing the entire first or third TM domain of HBsAg with the transmembrane domain of HIV gp41. We found that either TM domain of HBsAg could be replaced, resulting in HBsAg-gp41 chimeras that formed particles efficiently. HBsAg formed particles even when both TM1 and TM3 were replaced with the gp41 domain. The results indicate remarkable flexibility in HBsAg particle formation and provide a novel way to express heterologous membrane proteins that are anchored to a lipid surface by their own membrane-spanning domain. The membrane-proximal exposed region (MPER) of gp41 is an important target of broadly reactive neutralizing antibodies against HIV-1, and HBsAg-MPER particles may provide a good platform for future vaccine development.
HBsAg is the surface antigen of hepatitis B virus. In the virus, translation of the same open reading frame can begin at one of three in-frame start codons, resulting in proteins of large (42 kDa), medium (33 kDa), and small size (27 kDa), which share the identical 226-amino-acid sequence at the carboxyl end. The 27-kDa form of HBsAg is fully capable of spontaneous assembly into virus-like particles, so we have focused on this protein to study the requirements for particle assembly. In nature, small HBsAg participates in forming at least four different kinds of particles: 42-nm infectious hepatitis B virions (Dane particles), 22-nm spherical subviral particles, 22-nm-diameter rods, or as the borrowed envelope protein in 36-nm hepatitis D virus (39, 45).
Recombinant-derived small HBsAg forms subviral particles when expressed in a number of different cell types, including mammalian (39), yeast (25), and insect cells (19), but not Escherichia coli (25, 26). The particles are lipoprotein micelles, which contain 25% lipid. In these particles, HBsAg is believed to span the lipid four times, with four transmembrane domains (TM1 to TM4). This conformation exposes three external domains on the lipid surface at the amino end, middle, and carboxyl end of the protein (7, 42). Prior studies, based on the deletion of membrane-spanning domains, showed that transmembrane domains TM1 and TM2 were required for particle formation (5, 6). However, subsequent studies have shown that, at least for TM1, the replacement of amino acids is permitted (21, 33). In this paper, we have substituted an entire TM domain of another transmembrane protein (HIV gp41) for TM1 or TM3 of HBsAg and probed the requirements for particle assembly.
When other proteins were expressed in tandem with HBsAg, the surface antigen acted as a carrier protein and incorporated proteins as large as gp120 into the particles (2). This method allowed us to express membrane proteins and transmembrane proteins like gp41 on a lipid surface (34). The membrane-proximal exposed region of gp41 (MPER) is an important target of broadly reactive neutralizing antibodies against HIV (27). Although they are detected occasionally in HIV-infected people, it has been difficult or impossible to elicit comparable antibodies by immunization with conventional MPER immunogens (14, 20, 24). The expression of MPER in HBsAg particles could improve antibody quality by combining the potency of HBsAg particles with the antigenicity of MPER displayed on a lipid surface. We expressed MPER linked to the amino end of HBsAg or within the exposed middle domain of HBsAg. At each site, MPER was linked directly to the nearest TM domain of HBsAg, or it was attached via the TM domain of gp41. In these constructs, HBsAg showed remarkable flexibility in accommodating the insertion of an entire foreign TM domain in place of its own TM1 or TM3 domain. The resulting particles expressed MPER in its natural milieu on a lipid surface, where it was anchored via the TM domain of gp41. Many viral antigens are membrane or transmembrane proteins, and HBsAg particles may be a good platform for expressing these antigens for future vaccine development.
MATERIALS AND METHODS
Antibodies and antigens.
Antibodies were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. Monoclonal antibodies 2F5 (37) and 4E10 (41) were from Hermann Katinger, and monoclonal z13e was from Michael Zwick (50). Aldrithiol-2-inactivated HIV-1 virions of the IIIB, MN, or ADA strain or SHIV virions of the 89.6 type were a kind gift of Larry Arthur and Jeffrey Lifson at the AIDS Vaccine Program, NCI (22). Recombinant HBsAg (adw) was purchased from Aldevron, LLC (Fargo, ND).
Construction of TM12, TM14, and TM16.
Codon-optimized HBsAg (2) was PCR amplified using primers HC-1B and HIL-14B (see Table 1) and cloned between EcoRI and SpeI sites downstream of the baculovirus polyhedrin promoter and an influenza hemagglutinin leader in a pFastbac vector (3, 23) to produce pFB-HALsAg. A plasmid coding for gp160 of the 89.6 strain (12) was provided by Dan Littman (New York University, New York, NY). The MPER and TM domains of gp41 were PCR amplified using primers HC-3B and KS-7. The PCR product and plasmid pFB-HALsAg were ligated to produce plasmid TM18, in which the transmembrane domain of g41 was contiguous with the first membrane-spanning domain of HBsAg. Subsequent constructs were produced by removing one TM domain or the other, or a part of each.
TABLE 1.
Oligonucleotides
| Oligonucleotide | Sequence | Product |
|---|---|---|
| ClaI-L1 | CTGGCTGTGGTATATAAATCGATTATTCATAATGATAG | TM12 |
| ClaI-L2 | CTATCATTATGAATAATCGATTTATATACCACAGCCAG | TM12 |
| ClaI-M1 | GATTATTCATAATCGATAGTAGGAGGC | TM14 |
| ClaI-M2 | GCCTCCTACTATCGATTATGAATAATC | TM14 |
| HC49 | ATCGATAGTACTGGTGCTGCAGGCCGGCTTC | TM14 |
| HC47 | ATCGATACCCCAGTCCCTGGACTCCTGGTGGACC | TM16 |
| HC48 | GAATATGCATCACTAGTCAGTACCGATG | TM16 |
| HC3B | GGGAATTCAATGAAAAAGAATTATTGGAATTGG | TM18 |
| KS-7 | GCCGGAGGTGATCGATAGTACAGCAAAAACTATTCTTAAACC | TM18 |
| HC-1B | GCGAATTCGAATCGATCACCTCCGGCTTCCTGGGCCCCCTGC | TM18 |
| HIL-14B | GCCCATCTTCTTCTGCCTGTGGGTGTACATCGGTACCTGAACTAGTCC | TM18 |
| DA31 | P-CGATCAATGAAAAAGAATTATTGGAATTGGATAAATGGGCTAGCTTGTGGG | TM32 |
| DA32 | P-CGCCCACAAGCTAGCCCATTTATCCAATTCCAATAATTCTTT TTCATTGAT | TM32 |
| DA33 | P-CTAGCTTGTGGAATTGGTTTGACATAACAAACTGGG | TM34 |
| DA34 | P-CTAGCCCAGTTTGTTATGTCAAACCAATTCCACAAG | TM34 |
QuickChange mutagenesis (8) with oligonucleotides ClaI-L1 and ClaI-L2 inserted a second ClaI site between MPER and TM sequences. Digestion with ClaI and ligation, followed by site-directed mutagenesis to add a base and restore the open reading frame, formed plasmid TM12. QuickChange mutagenesis of TM18 with oligonucleotides ClaI M1 and ClaI M2 inserted a second ClaI site within the TM domain of gp41. The plasmid was cleaved with ClaI and SpeI and ligated to a PCR product made of HBsAg amplified with the primers HC48 and HC49, followed by deleting a base C to restore the open reading frame, resulting in plasmid TM14. Plasmid TM18 then was cut with ClaI and SpeI and ligated to a PCR product of HBsAg amplified with the primers HC47 and HC48. The product, plasmid TM16, retained the TM domain of gp41 but lacked the first membrane-spanning domain of HBsAg. Each product was confirmed by DNA sequencing.
Construction of TM32F, TM34, and TM20 and double MPER insertions.
A ClaI site was inserted into the exposed middle domain of HBsAg by site-directed mutagenesis of plasmid pFB-HALsAg. This resulted in the deletion of amino acids Pro154 to Ala159 of HBsAg and the insertion of Ser in place of Pro152. 5′-Phosphorylated oligonucleotides DA31 and DA32 (Table 1) were ligated to the vector cut with ClaI to produce plasmid TM32. In one clone, four correctly oriented copies of this sequence in tandem were detected by DNA sequencing, giving rise to plasmid TM32F. The insert contained an NheI site. It was cut with NheI and ligated with 5′-phosphorylated DA33/DA34 to make the complete MPER determinant, called plasmid TM34. Plasmid TM20, containing MPER linked to its own TM domain instead of the third domain of HBsAg, was made as synthetic DNA by GeneArt (Regensburg, Germany) and was spliced into pFastbac between the SalI and SpeI restriction sites.
To make double MPER-inserted forms of HBsAg, plasmid TM16 was mutated to remove SphI from the vector. It then was cut at an internal SphI site between the second and third membrane-spanning domains of HbsAg and ligated to the corresponding SphI to SpeI fragment from TM20 to produce TM16 + 20. Alternatively, cut TM16 was ligated to TM32F to make TM16 + 32F or to TM34 to make TM16 + 34. All constructs were confirmed by DNA sequencing.
Protein expression, purification, and quantification.
pFastbac DNA was transformed into cells bearing full-length bacmid DNA coding for infectious baculovirus (23); the insert recombined with bacmid DNA via a transposon. Bacmids were screened for the uptake of the insert by PCR using primers spanning the transposon. Bacmids containing the insert were used to transform Sf-9 cells (Protein Expression Lab, Frederick, MD) as follows: 10 μg of bacmid DNA was diluted into 200 μl saline and mixed with 150 μl of expressNOW transfection reagent (Lonza, Eastport, Prague, Czech Republic). After gentle mixing, followed by 15 min of equilibration, the mixture was added to 100 ml of Sf-9 cells. These were cultured for 4 days in SFX medium, and baculovirus was harvested from the culture supernatants.
For protein expression, Hi 5 cells were cultured overnight at 0.8 × 106 per ml and then infected with baculovirus recombinants at a multiplicity of infection (MOI) of 3:1 to 5:1. After 29 h on a shaker at 27°C, the infected cells were harvested by centrifugation at 1,000 rpm for 10 min in a Sorvall RT6000 centrifuge. Preliminary experiments showed that 28 to 30 h of incubation gave the greatest yield, and that most of the protein remained as intracellular virus-like particles. The cell pellet from 200 ml of culture was suspended in 10 ml phosphate-buffered saline (PBS) and stored frozen at −80°C. For use, the cells were thawed and diluted 1:1 with PBS in 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) plus protease inhibitor cocktail (BD Pharmingen). After 30 min at 4°C, they were sonicated for 40 s in a Vibra cell sonicator with an external probe, followed by centrifugation for 10 min at 2,000 rpm in a TJ-6 desktop centrifuge to remove cell debris.
For initial characterization and partial purification, 10 ml of clarified cell lysate was layered onto a discontinuous sucrose gradient, consisting of 8-ml layers of 10, 20, and 40% sucrose, and sedimented in a Beckman SW28 rotor for 140 min at 27,000 rpm. Fractions of about 0.7 ml each were collected from the bottom of the tube and assayed for MPER content by enzyme-linked immunosorbent assay (ELISA) 2F5 as the primary antibody and goat anti-human IgG conjugated to alkaline phosphatase (MP Biomedicals, Aurora, OH) as the detecting reagent. Peak fractions were pooled and characterized for MPER content and antigenicity. Each purified TM protein was quantified by electrophoresis on an SDS gel, followed by staining with Coomassie blue and measuring the fluorescence of the HBsAg band with excitation at 700 nm in a Li-Cor Odyssey infrared imager. The concentration of HBsAg-TM was determined by comparison to a standard curve of HBsAg, which was linear over the range of 1 to 12 μg per lane.
Antibody binding ELISA.
Each TM construct was diluted to 1 μg/ml and coated onto a soft plastic ELISA plate (Falcon 3912) overnight at 4°C. The plates were blocked with 1% bovine serum albumin (BSA), and monoclonal antibodies were serially diluted 3-fold, starting at a concentration of 3 μg/ml. After 2 h at room temperature, the plates were washed, and goat anti-human IgG conjugated alkaline phosphatase was added as a second antibody. After another 90 min at room temperature, the plates were washed, phosphatase substrate was added, and the optical density at 410 nm (OD410) was measured in an ELISA plate reader. For competitive ELISA, a constant amount of monoclonal antibody (1 μg/ml) was added to serial dilutions of each antigen, starting at 100 μg/ml (4 μM). After 40 to 60 min of preincubation at room temperature, the mixture was transferred to ELISA plates coated with AT-2-inactivated virions of the MN strain (1). Antibody binding was measured as described above. Differences in apparent affinity were determined as the antibody concentration giving equal binding for each HBsAg-MPER construct.
RESULTS
Requirements for HBsAg particle assembly.
A series of HBsAg hybrids expressing the MPER of HIV are shown in Fig. 1 and 2. The MPER determinants recognized by monoclonal antibodies 2F5 and 4E10 are indicated, as well as the TM domain of gp41 and the TM1 and TM3 domains of HBsAg. MPER was expressed at the amino end (Fig. 1) or in the exposed middle domain of HBsAg (Fig. 2). We varied the MPER-HBsAg junction to create an HBsAg-MPER particle that more closely resembled MPER on the viral surface while fulfilling the minimal requirements for HBsAg particle assembly.
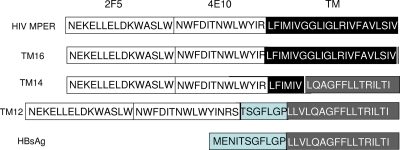
FIG. 1.
Insertion of gp41 MPER at the amino end of HBsAg. Top row, membrane-proximal exposed region (MPER) determinants bound by monoclonal antibodies 2F5 and 4E10 and the transmembrane (TM) domain of gp41. Bottom row, the amino end of HBsAg and the first transmembrane domain TM1. In each construct, the MPER-HBsAg junction was external to the membrane (construct TM12), within the TM domain (construct TM14), and inside the particles (construct TM16).
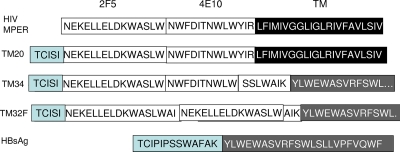
FIG. 2.
Insertion of gp41 MPER into the middle domain of HBsAg. Top row, MPER determinants and the TM domain of gp41. Bottom row, partial sequence of the exposed middle domain of HBsAg and the third transmembrane domain, TM3. In each construct, the MPER-HBsAg junction was either external to the membrane (constructs TM34 and TM32F) or inside the particles (construct TM20). Construct TM32F had four 2F5 determinants in tandem, of which two are shown.
We initially compared three MPER inserts at the amino end of HBsAg to determine the effect of the partial or complete substitution of the TM1 domain on particle assembly (Fig. 1). In construct TM12, MPER was linked through a 7-amino-acid linker to the intact TM1 domain of HBsAg. In construct TM14, the MPER-to-HBsAg junction was located within the TM domain: the first six amino acids came from the TM domain of gp41, and the next 14 amino acids came from TM1 of HBsAg. In construct TM16, the entire TM domain (21 amino acids) came from gp41 and completely replaced the TM1 domain of HBsAg. These constructs differ at the MPER-HBsAg junction: the junction moved by 10 amino acids between constructs TM12 and TM14 and by another 13 amino acids between constructs TM14 and TM16. We assumed that the partial replacement of the TM1 domain would be less disruptive of particle assembly than its complete replacement by the foreign TM domain from gp41.
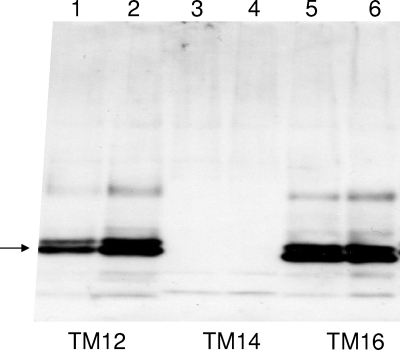
The expression of the MPER determinant was measured by Western blotting for two plasmids of each type (Fig. 3). Construct TM12, with an intact transmembrane domain 1, was expressed well, as shown by a band at 27 kDa. The expression of construct TM14 was not detected for either plasmid. The TM16 construct was expressed as well as TM12, even though the entire TM1 domain of HBsAg was replaced by the TM domain of gp41. A strong band on the Western blot generally indicates particle assembly, since unassembled HBsAg is quickly degraded (6). To test particle assembly, each particle was tested by velocity sedimentation in sucrose gradients to determine particle size. MPER expression was detected with monoclonal antibody 2F5. Construct TM12 (Fig. 4, left) sedimented at large size, comparable to the size of native HBsAg particles. Construct TM16 also assembled into virus-like particles, as shown by sedimentation at large size on a preparative scale and by electron microscopy (Fig. 5), despite the substitution of the TM sequence of gp41. The doublet bands observed for both TM12 and TM16 are typical of the two glycoforms normally observed for small HBsAg at 27 and 24 kDa.
FIG. 3.
MPER expression after transmembrane domain swapping. MPER was linked to HBsAg in three ways: to the amino end of HBsAg (TM12 in lanes 1 and 2), to a junction within the first membrane-spanning domain of HBsAg (TM14 in lanes 3 and 4), or by replacing the entire transmembrane domain TM1 of HBsAg with the TM domain of gp41 (TM16 in lanes 5 and 6). Sf-9 cells were infected with two recombinant baculoviruses of each type, and MPER expression was detected by Western blotting with monoclonal antibody 2F5. The replacement of the entire TM1 domain was allowed, but partial replacement led to no protein expression.
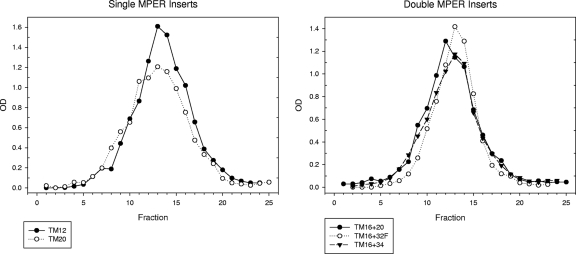
FIG. 4.
Analytical velocity sedimentation of MPER particles. Each MPER construct was expressed by recombinant baculovirus, partially purified, and analyzed by sedimentation through a 10 to 50% sucrose gradient for 100 min at 39,000 rpm in an SW41 Ti rotor. Each fraction was assayed for MPER content with monoclonal 2F5. Single MPER substitutions, TM12 and TM20, are shown on the left, and double MPER substitutions, TM16 + 20, TM16 + 32F, and TM16 + 34, are shown on the right. TM12 also was assayed for HBsAg content, and the HBsAg peak coincided with the MPER peak. Each HBsAg-MPER hybrid sedimented at large size, indicating efficient particle assembly.
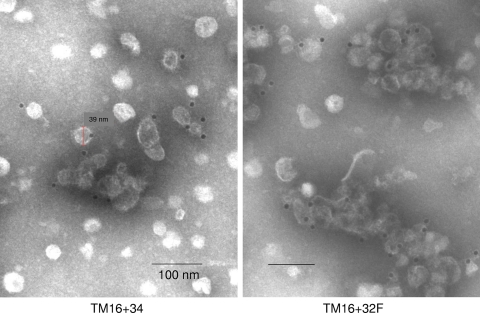
FIG. 5.
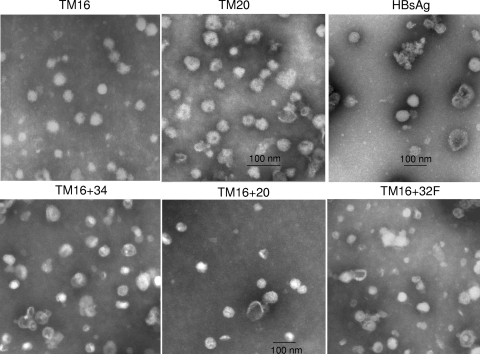
Electron microscopy of HBsAg-MPER particles with single MPER-TM substitution for the TM1 (TM16) or TM3 (TM20) domain of HBsAg, control HBsAg (top row), or double MPER substitutions (bottom row). TM16 and control HBsAg particles were purified by hydrophobic interaction chromatography, and all other particles were purified by banding in CsCl. Negatively stained samples were examined at a magnification of ×31,500 (top row) or ×25,000 (bottom row). Bar, 100 nm.
We then expressed MPER as part of the exposed middle loop of HBsAg (Fig. 2), which is considered immunodominant (39). These constructs expressed the 2F5 epitope alone (TM32), both the 2F5 and 4E10 epitopes (TM20 and TM34), or four copies of the 2F5 epitope in tandem (TM32F). In two constructs (TM32F and TM34), MPER was linked to the intact TM3 domain of HBsAg. In construct TM20, the entire TM3 domain of HBsAg was replaced by the TM domain of gp41. The assembly of TM20 was demonstrated by velocity sedimentation in sucrose gradients (Fig. 4, left). TM20 sedimented at the size of virus-like particles despite the substitution of one TM domain for the other. Assembly was an efficient process, since none of the TM20 protein sedimented at the size of monomers. TM20 expressed MPER near a major antigenic determinant of HBsAg and anchored it to lipid via the TM domain of gp41.
The particles were stable on further purification by banding in CsCl (TM20) or by hydrophobic interaction chromatography (TM16). Electron microscopy of purified TM16 showed spherical particles ranging from 35 to 47 nm in diameter (Fig. 5). The heterogeneity is typical, but the size is a little larger than that reported for native HBsAg particles (19, 25, 39). When we prepared native HBsAg particles by the same method, however, they ranged in size from 52 to 64 nm (Fig. 5). This is very similar to the size of TM20 particles, which ranged from 52 to 60 nm. The formation of TM16 and TM20 hybrid particles (but not TM14) is consistent with a two-stage model of membrane protein folding (15, 36). In the first stage, the TM domains partition independently into the lipid membrane, followed by association with other subunits to form the native protein. The results follow the prediction of the model: each TM domain is a functional unit in particle assembly, so an entire TM domain could replace another, but part of a TM domain could not. The TM2 domain may be necessary for HBsAg particle formation, but it was not sufficient by itself to form particles when the TM1 domain was partially replaced by a TM fragment, as in construct TM14.
Double MPER and TM substitutions in HBsAg.
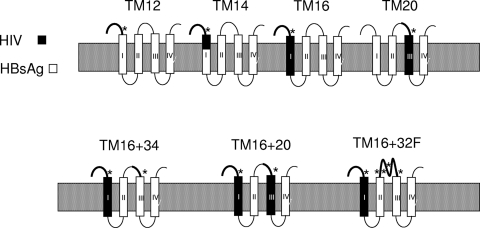
We examined particle formation when two MPER substitutions were combined in the same HBsAg construct. Constructs TM16 + 32F and TM16 + 34 combined an MPER-TM substitution for the TM1 domain (as in TM16) with another MPER substitution in the exposed middle loop of HBsAg (Fig. 6). They both formed particles, as shown by sedimentation at large size in sucrose gradients (Fig. 4, right) and by electron microscopy (Fig. 5). The particles were nearly identical to or slightly larger than TM16, with diameters of 35 to 46 nm for TM16 + 34 and 42 to 55 nm for TM16 + 32F (Fig. 5). Construct TM16 + 20 combined two MPER-TM substitutions in the same hybrid protein. Despite replacing both transmembrane domains TM1 and TM3 of HBsAg, this hybrid assembled into virus-like particles, sedimented at large size (Fig. 4, right), and formed 42- to 50-nm particles (Fig. 5). Of the four transmembrane domains of HBsAg, TM2 and TM4 were sufficient for particle formation when combined with two copies of the gp41 TM domain per molecule.
FIG. 6.
Topology of HBsAg chimeras. The four presumed transmembrane domains of HBsAg are shown for constructs with a single MPER substitution (top row) or double MPER substitutions (bottom row). The gp41 TM insert is shaded black, and the MPER determinant is indicated by an asterisk.
Antigenicity.
By comparing different HBsAg-MPER hybrids, we could determine the effect of MPER location, anchoring, and valency on antibody binding. All MPER constructs bound monoclonal antibodies 2F5 and 4E10 in a conventional ELISA, with the MPER antigen immobilized on the plate (not shown). This type of ELISA was insensitive to different forms of TM anchoring. In contrast, a competitive binding assay, with each antigen tested in solution, showed clear differences among constructs TM12, TM16, and TM20 (Fig. 7). In this assay, antibody was preincubated with the antigen, and the residual unbound antibody then was measured by ELISA. Antibody binding to the ELISA plate decreased in proportion to the amount of competitor added.
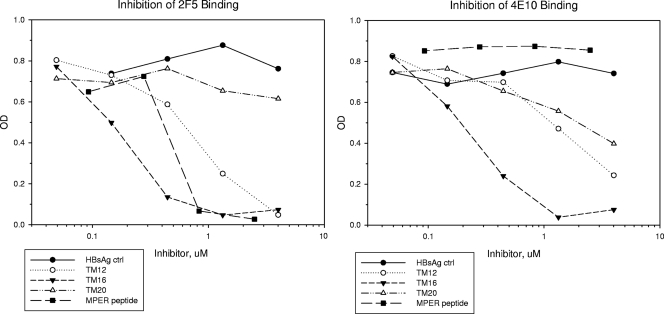
FIG. 7.
Competitive inhibition of 2F5 and 4E10 binding to AT-2-inactivated HIV virions. Monoclonal antibodies were preincubated with each HBsAg-MPER hybrid for 1 h and then transferred to ELISA wells coated with inactivated HIV virions and assayed for residual binding. Binding decreased in proportion to the amount of inhibitor added. For both monoclonal antibodies, TM16 was a more potent inhibitor than TM12, and TM20 was the least potent inhibitor. MPER located at the amino end of HBsAg and anchored via its own TM domain gave the most potent antibody binding.
The effect of TM anchoring on MPER antigenicity was shown by comparing TM12 and TM16 (Fig. 7). These two constructs share the same exposed MPER sequence at the amino end of HBsAg. However, MPER anchored via the TM domain of gp41 (TM16) gave consistently greater competitive inhibition than MPER anchored to the TM1 domain of HBsAg (TM12). Monoclonal 2F5 was almost completely inhibited by TM16 at 0.4 μM, while TM12 required five to six times more antigen (in two experiments) to achieve the same degree of inhibition. Similarly, monoclonal 4E10 was inhibited 75% by TM16 at 0.4 μM, while TM12 required nine times more antigen to inhibit to the same extent. Specificity of competition was demonstrated by using a short peptide that expressed only the 2F5 epitope. This peptide completely inhibited monoclonal 2F5 binding at 0.8 μM, but it failed to compete for monoclonal 4E10 binding at any concentration tested.
The effect of MPER location on antibody binding was shown by comparing construct TM16 to TM20 (Fig. 7). Both constructs have the same MPER sequence, and both are anchored by the TM domain of gp41. However, one was displayed at the amino end of HBsAg, and the other was in the exposed middle domain. Monoclonal 2F5 was inhibited by 0.4 μM TM16, but it was insensitive to any concentration of TM20 tested, up to 30 times the 50% inhibitory concentration (IC50) of TM16. Similarly, the inhibition of monoclonal 4E10 by TM20 required 14 times more antigen to inhibit to the same degree as TM16. The results indicate that MPER located at the amino end of HBsAg, as in TM16, was a more potent competitor than MPER located in the exposed middle domain of HBsAg, as in TM20.
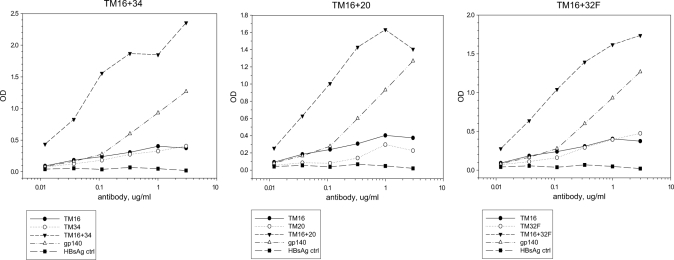
MPER valency had a consistent effect on antibody binding (Fig. 8). 2F5 binding improved considerably when we combined two MPER determinants in the same HBsAg construct, as measured in a direct ELISA binding assay. For example, divalent TM16 + 34 bound 2F5 better than either monovalent parent, TM16 or TM34, or the sum of the parental constructs (Fig. 8, left) or the gp140 control. Apparent binding affinity for the divalent form (measured on the x axis) increased about 80-fold relative to the monovalent forms, suggesting cooperative binding. Similarly, 2F5 binding to divalent TM16 + 20 was increased relative to the parental forms or the gp140 control (Fig. 8, middle), and 2F5 binding to pentavalent TM16 + 32F was much greater than that to the parental forms TM16 and TM32F (Fig. 8, right).
FIG. 8.
Effect of MPER valency on antibody binding. Monoclonal 2F5 binding was measured by direct ELISA for divalent or multivalent MPER particles and compared to the monovalent MPER parental forms. Left panel, divalent TM16 + 34 versus monovalent TM16 or TM34. Center panel, divalent TM16 + 20 versus monovalent TM16 or TM20. Right panel, pentavalent TM16 + 32F versus monovalent TM16 or tetravalent TM32F. In each case, binding affinity to divalent or multivalent MPER was increased about 80-fold relative to the monovalent form of the same MPER construct.
HBsAg-MPER constructs display MPER on the surface of virus-like particles. This was demonstrated by immune electron microscopy of TM16 + 34 and TM16 + 32F particles (Fig. 9). Using 12-nm immunogold particles, we detected specific 2F5 binding to each HBsAg-MPER construct. No immunogold labeling was observed for antibody controls that omitted monoclonal 2F5 or for HBsAg controls that lacked MPER (not shown).
FIG. 9.
Immunoelectron microscopy of MPER displayed on the surface of virus-like particles. Divalent TM16 + 34 or pentavalent TM16 + 32F particles were labeled with monoclonal antibody 2F5, followed by detection with 12-nm gold particles coated with goat anti-human IgG. No immunogold labeling was detected in the absence of 2F5 or in HBsAg controls lacking MPER.
DISCUSSION
We have examined the role of transmembrane domains TM1 and TM3 in HBsAg particle formation. The assembly of native HBsAg virus-like particles depends on the first two TM domains of HBsAg (5, 6). However, using a strategy of domain swapping, we found that the entire TM1 domain of HBsAg could be replaced by the TM domain of HIV gp41 without disrupting particle assembly. Similarly, particles formed when the TM3 domain of HBsAg was replaced by the TM domain of gp41, or when both TM1 and TM3 domains were replaced by TM of gp41. Despite little or no direct sequence homology, one functional TM domain could replace another, suggesting that TM function was more important than sequence homology. HBsAg hybrid particles provide a novel way to express membrane-associated viral proteins, including transmembrane proteins, on a lipid surface. For antigenic determinants like the MPER of HIV gp41, this approach could enhance antibody binding by increasing valency and by presenting MPER in its natural milieu on a lipid surface.
The topology of particle formation is shown in Fig. 6, based on the membrane-spanning model of Stirk et al. (42). The constructs can be divided into those where MPER was expressed at the exposed domains of HBsAg (TM12, TM32F, and TM34) and those where the transmembrane domain of the insert replaced a TM domain of HBsAg (constructs TM16 and TM20 and the double inserts). The first group was expected to form particles, since the TM domains were unaffected by the insert. However, the ability of the second group to form particles despite substitution for one or both of the TM1 and TM3 domains of HBsAg suggests that the inserted TM domain of gp41 also contributes to particle formation. The fact that the gp41 TM shared little or no sequence homology with HBsAg TM1 or TM3 suggests that other functional transmembrane domains could also permit particle assembly.
The assembly and folding of HBsAg-MPER chimeric particles is consistent with the two-stage model proposed for membrane proteins (15, 36). In the first stage, each transmembrane domain functions independently as it partitions into the lipid bilayer and forms a hydrophobic alpha helix. In the second stage, the TM domains organize through side-to-side interactions to generate functional proteins, and these can form disulfide bonds to neighboring HBsAg proteins to form dimers. The dimers oligomerize and eventually assemble into 22-nm particles. The ability to swap transmembrane domains as a complete unit (as in TM16), but not partial domains (TM14), agrees with the two-stage model, since an entire TM domain is the functional unit. Gp41 is an evolved transmembrane protein whose TM domain can partition into lipid (step 1) and self associate to form trimers (step 2). In addition, the current model of HBsAg particle formation suggests that two steps in subviral particle morphogenesis occur in distinct cellular compartments (17, 32).
In the second stage, the inserted TM domain of gp41 must interact favorably with the three remaining TM domains of HBsAg to allow protein folding and particle assembly. Unlike some enveloped viruses, such as HIV, which readily accept membrane proteins of the host cell (1) or other unrelated viruses (47), HBsAg particles rarely take up foreign proteins. HBsAg can form mixed particles when coexpressed with surface antigen of the closely related woodchuck hepatitis virus (WCsAg) but not duck hepatitis virus (16). In addition, an HBsAg/WCsAg protein chimera formed particles when transmembrane domains TM1 and TM2 of WCsAg were replaced with the corresponding domains of HBsAg (38). In these chimeras, the TM1 domain of HBsAg shared 12/21 amino acids with the WCsAg sequence it replaced (Table 2 and 3). The TM2 domains were even more conserved, sharing 19/22 amino acids between the two proteins, and this may allow both TM2 domains to function interchangeably.
TABLE 2.
TM1 domains of HBsAg chimeras capable of assemblya
| Antigen | TM1 domain sequence | Reference or source | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBsAg | F | L | G | P | L | L | V | L | Q | A | G | F | F | L | L | T | R | I | L | T | I | |||||||||
| WCsAg | L | - | - | L | - | A | G | - | - | V | V | Y | - | - | W | - | K | - | - | - | - | 16, 38 | ||||||||
| DsAg | I | - | A | G | - | I | G | - | L | V | S | - | - | - | - | I | K | - | - | E | - | 16, 38 | ||||||||
| TM16 | R | L | F | I | M | I | V | G | G | L | I | - | L | R | I | V | F | T | A | - | S | - | V | This study | ||||||
| TM20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | This study | ||||||||
| TM16 + 20 | R | L | F | I | M | I | V | G | G | L | I | - | L | R | I | V | F | T | A | - | S | - | V | This study | ||||||
| HCV E1 | W | G | V | L | A | G | I | A | Y | F | S | M | V | G | N | W | A | K | V | - | V | V | L | L | L | F | A | G | V | 11, 33 |
| HCV E2 | Y | V | L | L | - | F | L | - | L | - | D | A | R | V | C | A | C | L | W | M | M | L | L | I | A | Q | A | 11, 33 | ||
-, no change.
TABLE 3.
TM3 domains of HBsAg chimeras capable of assemblya
| Antigen | TM3 domain sequence | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBsAg | K | F | L | W | E | W | A | S | A | R | F | S | W | L | S | L | L | V | P | F | V | Q | W | F | V |
| WCsAg | N | Y | - | - | - | - | - | L | - | - | - | - | - | - | N | - | - | - | - | L | L | - | - | L | G |
| DsAg | |||||||||||||||||||||||||
| TM16 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TM20 | R | L | F | I | M | I | V | G | G | L | I | G | L | R | I | V | F | T | A | L | S | I | V | ||
| TM16 + 20 | R | L | F | I | M | I | V | G | G | L | I | G | L | R | I | V | F | T | A | L | S | I | V | ||
| HCV E1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| HCV E2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
-, no change.
In the current study, we held membrane-spanning domains 2 and 4 of HBsAg constant while replacing domain TM1 or TM3 of HBsAg with the TM domain of gp41. Although the gp41 TM domain shared only 2/21 amino acids with TM1 of HBsAg (Table 2), it could replace TM1 and still form particles, as in construct TM16. HIV envelope gp41 readily forms pseudotypes with other membrane proteins, and this may explain its favorable interactions with the remaining HBsAg domains TM2, TM3, and TM4 during particle formation. Similarly, although the transmembrane domain of gp41 shared only 2/21 amino acids with the TM3 domain of HBsAg (Table 3), construct TM20 formed particles, suggesting cooperative interactions with the remaining HBsAg domains TM1, TM2, and TM4 in particle formation. Once the first TM domain of gp41 is substituted into HBsAg particles (TM16 or TM20), it may be easier to accept the second TM substitution, as in TM16 + 20 particles. In this hybrid, two TM domains of gp41 may interact with each other, as they do in HIV envelope trimers. We have not substituted for domains TM2 or TM4, and TM2 may be essential for particle assembly.
Similar results have been reported recently for HBsAg chimeras containing the ectodomains and TM domains of envelope proteins E1 and E2 of hepatitis C virus (11, 33). These authors replaced TM1 of HBsAg with the TM domains of the HCV envelope proteins E1 and E2. Despite differences in nearly all amino acids of the TM domain (Tables 2 and 3), chimeric HBsAg-HCV proteins could participate in particle assembly. Unlike our TM constructs, particle formation depended on coexpressing HBsAg-E1 or HBsAg-E2 with native HBsAg. Difficulty in assembling HBsAg-HCV particles by themselves may reflect the size of the inserts, which included the entire E1 and E2 ectodomains and were much larger (54 and 85 kDa) than our HBsAg-MPER constructs (27 kDa).
The MPER determinant is an important target of HIV neutralizing antibodies (4, 10, 13, 27-29, 41). Its sequence is conserved among diverse HIV isolates, and human monoclonal antibodies to MPER can neutralize a broad spectrum of HIV isolates from North America and western Europe (4). MPER-specific neutralizing antibodies have been detected occasionally in infected humans, but they have never been elicited by immunization (27). Similarly, in this study, despite immunizing rabbits with monovalent, divalent, or pentavalent HBsAg-MPER particles, we have not reliably elicited antibodies to MPER. Although an MPER immunogen is highly desirable, it presents a number of challenges. First, MPER is weakly immunogenic (34). Many successful vaccines depend on particle formation to enhance vaccine potency. These include inactivated viral particles, such as inactivated polio vaccine (48) and rabies (35), and recombinant proteins that form virus-like particles, such as HBsAg (9, 40) and human papillomavirus (HPV) (18). A number of experimental vaccines have gained potency by linking antigens with HBsAg to form virus-like particles, including vaccines against malaria (43) and hepatitis C virus (30). Similarly, we have made HBsAg-MPER particles to take advantage of the effect of multivalency on vaccine potency.
Second, the MPER determinant may require expression on a lipid surface (31, 44, 49). By providing the lipid component, HBsAg-MPER particles could improve antibody quality by eliciting antibodies specific for MPER, as found on the viral surface. Third, it is located in a dynamic part of gp41, so neutralizing antibodies may need to bind a conformation of MPER that appears only transiently during viral entry (27). This form of MPER might not be found on particles, and further modifications to the MPER insert would be required to produce this epitope. Finally, MPER may cross-react with certain host proteins (46), and the immune response would need to be targeted selectively to avoid these potentially self-reactive epitopes.
HBsAg is a self-assembling membrane protein that presents viral antigens in the form of virus-like particles. We have probed its particle-forming activity by substituting an entire heterologous TM sequence for domains TM1 and TM3 of HBsAg. Particle formation depended on a functional TM domain rather than a conserved TM sequence. The particles displayed foreign envelope or transmembrane antigens, such as the HIV MPER determinant, anchored to a lipid surface. Antigenicity was enhanced by linking MPER to the amino end of the carrier, anchoring it through its own TM domain, and increasing its valency. Similarly modified HBsAg particles could provide a flexible platform for vaccine development against a variety of enveloped viruses.
Acknowledgments
We thank Carol Weiss for the critical reading of the manuscript.
This research was supported in part by the NIH Intramural AIDS Targeted Research Program.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1.Arthur, L. O., et al. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 258:1935-1938. [DOI] [PubMed] [Google Scholar]
- 2.Berkower, I., M. Raymond, J. Muller, A. Spadaccini, and A. Aberdeen. 2004. Assembly, structure, and antigenic properties of virus-like particles rich in HIV-1 envelope gp120. Virology 321:75-86. [DOI] [PubMed] [Google Scholar]
- 3.Berkower, I., et al. 2008. Targeted deletion in the beta 20-21 loop of HIV envelope gp120 exposes the CD4 binding site. Virology 377:330-338. [DOI] [PubMed] [Google Scholar]
- 4.Binley, J. M., et al. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. U. S. A. 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss, V., and D. Ganem. 1991. Mutational analysis of hepatitis B surface antigen particle assembly and secretion. J. Virol. 65:3813-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss, V. 2007. Hepatitis B virus morphogenesis. World J. Gastroenterol. 13:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke, T. F., et al. 1998. Identification of a BRCA1-associated kinase with potential biological relevance. Oncogene 16:1031-1040. [DOI] [PubMed] [Google Scholar]
- 9.Cabral, G. A., et al. 1978. Cellular and humoral immunity in guinea pigs to two major polypeptides derived from hepatitis B surface antigen. J. Gen. Virol. 38:339-350. [DOI] [PubMed] [Google Scholar]
- 10.Cardoso, R. M., et al. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion associated motif in gp41. Immunity 22:163-173. [DOI] [PubMed] [Google Scholar]
- 11.Cocquerel, L., et al. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collman, R., et al. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 14.Eckhart, L., et al. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 15.Engelman, D. M., et al. 2003. Membrane protein folding: beyond the two stage model. FEBS Lett. 555:122-125. [DOI] [PubMed] [Google Scholar]
- 16.Gerhardt, E., and V. Bruss. 1995. Phenotypic mixing of rodent but not avian hepadnavirus surface proteins into human hepatitis B virus particles. J. Virol. 69:1201-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huovila, A. P., A. M. Eder, and S. D. Fuller. 1992. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 118:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. U. S. A. 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford, R. E., et al. 1989. Expression and characterization of hepatitis B virus surface antigen polypeptides in insect cells with a baculovirus expression system. J. Virol. 63:1549-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenz, O., et al. 2005. Trimeric membrane-anchored gp41 inhibits HIV membrane fusion. J. Biol. Chem. 280:4095-4101. [DOI] [PubMed] [Google Scholar]
- 21.Lepère-Douard, C., M. Trotard, J. Le Seyec, and P. Gripon. 2009. The first transmembrane domain of the hepatitis B virus large envelope protein is crucial for infectivity. J. Virol. 83:11819-11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lifson, J. D., et al. 2004. Evaluation of the safety, immunogenicity, and protective efficacy of whole inactivated simian immunodeficiency virus (SIV) vaccines with conformationally and functionally intact envelope glycoproteins. AIDS Res. 20:772-787. [DOI] [PubMed] [Google Scholar]
- 23.Luckow, V. A., C. S. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in E. coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo, M., et al. 2006. Induction of neutralizing antibody against human immunodeficiency virus type 1 (HIV-1) by immunization with gp41 membrane proximal external region (MPER) fused with porcine endogenous retrovirus (PERV) p15E fragment. Vaccine 24:435-442. [DOI] [PubMed] [Google Scholar]
- 25.McAleer, W. J., et al. 1984. Human hepatitis B vaccine from recombinant yeast. Nature 307:178-180. [DOI] [PubMed] [Google Scholar]
- 26.Miyanohara, A., A. Toh-e, C. Nozaki, F. Hamada, N. Ohtomo, and K. Matsubara. 1983. Expression of hepatitis B surface antigen gene in yeast. Proc. Natl. Acad. Sci. U. S. A. 80:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montero, M., N. E. van Houten, X. Wang, and J. K. Scott. 2008. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 72:54-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muster, T., et al. 1993. A conserved neutralizing epitope on gp41 of human immuno-deficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, J. D., et al. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J. Virol. 81:4033-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Netter, H. J., T. B. Macnaughton, W. P. Woo, R. Tindle, and E. J. Gowans. 2001. Antigenicity and immunogenicity of novel chimeric hepatitis B surface antigen particles with exposed hepatitis C virus epitopes. J. Virol. 75:2130-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ofek, G., et al. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patient, R., et al. 2007. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 81:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patient, R., C. Hourioux, P. Vaudin, J. C. Page, and P. Roingeard. 2009. Chimeric hepatitis B and C viruses envelope proteins can form subviral particles: implications for the design of new vaccine strategies. New Biotechnol. 25:226-234. [DOI] [PubMed] [Google Scholar]
- 34.Phogat, S., et al. 2008. Analysis of the human immunodeficiency virus type 1 gp41 membrane proximal external region arrayed on hepatitis B surface antigen particles. Virology 373:72-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piza, A., et al. 2002. Effect of the contents and form of rabies glycoprotein on the potency of rabies vaccination in cattle. Mem. Inst. Oswaldo Cruz 97:265-268. [DOI] [PubMed] [Google Scholar]
- 36.Popot, J. L., and D. M. Engelman. 2000. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69:881-922. [DOI] [PubMed] [Google Scholar]
- 37.Purtscher, M., et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 38.Schirmbeck, R., et al. 2001. Targeting murine immune responses to selected T cell- or antibody-defined determinants of the hepatitis B surface antigen by plasmid DNA vaccines encoding chimeric antigen. J. Immunol. 166:1405-1413. [DOI] [PubMed] [Google Scholar]
- 39.Seeger, C., F. Zoulim, and W. S. Mason. 2007. Hepadnaviruses, p. 2977. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 40.Stevens, C. E., et al. 1987. Yeast-recombinant hepatitis B vaccine. Efficacy with hepatitis B immune globulin in prevention of perinatal hepatitis B virus transmission. JAMA 257:2612-2616. [DOI] [PubMed] [Google Scholar]
- 41.Stiegler, G., et al. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 42.Stirk, H. J., J. M. Thornton, and C. R. Howard. 1992. A topological model for hepatitis B surface antigen. Intervirology 33:148-158. [DOI] [PubMed] [Google Scholar]
- 43.Stoute, J. A., et al. 1997. A preliminary evaluation of a recombinant circumsporozoite protein vaccine against Plasmodium falciparum malaria. RTS, S Malaria Vaccine Evaluation Group. N. Engl. J. Med. 336:86-91. [DOI] [PubMed] [Google Scholar]
- 44.Sun, Z. Y. J., et al. 2008. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28:52-63. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, J. M., P. Farci, and R. H. Purcell. 2007. Hepatitis D (delta) virus, p. 3031. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 46.Verkoczy, L., et al. 2010. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc. Natl. Acad. Sci. U. S. A. 107:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, W., et al. 2008. Establishment of retroviral pseudotypes with influenza hemagglutinins from H1, H3, and H5 subtypes for sensitive and specific detection of neutralizing antibodies. J. Virol. Methods 153:111-119. [DOI] [PubMed] [Google Scholar]
- 48.Wood, D. J., et al. 1997. A new WHO international reference reagent for use in potency assays of inactivated poliomyelitis vaccine. Biologicals 25:59-64. [DOI] [PubMed] [Google Scholar]
- 49.Zhu, P., et al. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847-852. [DOI] [PubMed] [Google Scholar]
- 50.Zwick, M. B., et al. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of Human Immunodeficiency Virus Type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]