Abstract
Yellow dwarf viruses in the family Luteoviridae, which are the causal agents of yellow dwarf disease in cereal crops, are each transmitted most efficiently by different species of aphids in a circulative manner that requires the virus to interact with a multitude of aphid proteins. Aphid proteins differentially expressed in F2 Schizaphis graminum genotypes segregating for the ability to transmit Cereal yellow dwarf virus-RPV (CYDV-RPV) were identified using two-dimensional difference gel electrophoresis (DIGE) coupled to either matrix-assisted laser desorption ionization-tandem mass spectrometry or online nanoscale liquid chromatography coupled to electrospray tandem mass spectrometry. A total of 50 protein spots, containing aphid proteins and proteins from the aphid's obligate and maternally inherited bacterial endosymbiont, Buchnera, were identified as differentially expressed between transmission-competent and refractive aphids. Surprisingly, in virus transmission-competent F2 genotypes, the isoelectric points of the Buchnera proteins did not match those in the maternal Buchnera proteome as expected, but instead they aligned with the Buchnera proteome of the transmission-competent paternal parent. Among the aphid proteins identified, many were involved in energy metabolism, membrane trafficking, lipid signaling, and the cytoskeleton. At least eight aphid proteins were expressed as heritable, isoelectric point isoform pairs, one derived from each parental lineage. In the F2 genotypes, the expression of aphid protein isoforms derived from the competent parental lineage aligned with the virus transmission phenotype with high precision. Thus, these isoforms are candidate biomarkers for CYDV-RPV transmission in S. graminum. Our combined genetic and DIGE approach also made it possible to predict where several of the proteins may be expressed in refractive aphids with different barriers to transmission. Twelve proteins were predicted to act in the hindgut of the aphid, while six proteins were predicted to be associated with the accessory salivary glands or hemolymph. Knowledge of the proteins that regulate virus transmission and their predicted locations will aid in understanding the biochemical mechanisms regulating circulative virus transmission in aphids, as well as in identifying new targets to block transmission.
Plant viruses belonging to the family Luteoviridae, collectively referred to as luteovirids, are phloem-restricted RNA viruses that cause disease in many staple food crops (18, 32, 36, 37, 48). One of the most fascinating features of these viruses is their stealthy journey through their aphid vectors, a mode of virus transmission known as circulative-nonpropagative transmission (33, 37). Once a virion is acquired from an infected plant by an aphid, the virion circulates through the aphid digestive tract, hemocoel, and salivary tissues, presumably without successful replication in the aphid. The virus particle, which contains at least two virus-encoded proteins (32, 33, 37) and perhaps some associated plant host proteins (6), will likely interact with proteins in the aphid to facilitate virus movement through the aphid. These interactions define the vector species-specific transmission that is a hallmark of luteovirids (29, 31-34, 37, 79).
The circulative transmission pathway through the aphid begins during ingestion of phloem sap. The virus must pass along the chitin-lined foregut opening, then into the midgut, and ultimately into the thin-walled hindgut (31, 32, 37). Different luteovirid species have the capacity to pass through either the midgut or hindgut epithelium into the hemolymph (31, 37, 59). The gut epithelial cell lining is the first site where the virus enters into aphid cells and is the first potential barrier to virus transmission. Plant viruses rarely traverse insect cell membranes directly but rather appear to rely on receptor-mediated endocytosis to enter into existing endocytic trafficking pathways for movement within individual cells (28, 29, 31, 32). Indeed, microscopic evidence supports the hypothesis that luteovirids enter into aphid cells via clathrin-mediated endocytosis and are retained in membrane vesicles for transport across the cytoplasm (29, 31). Once inside gut cells, virions have been observed only in membrane-bound vesicles, and thus they appear to avoid the cytoplasm, where replication may occur. The vesicle membrane eventually fuses with the basement membrane and releases virions into the aphid hemolymph, where they must avoid components of the aphid immune system. Aphids harbor bacterial endosymbionts of the genus Buchnera, which produce an abundance of the Buchnera chaperone protein symbionin, a homologue of GroEL (21). GroEL functions in protein folding and translocation across membranes (21). Symbionin binds in vitro to luteoviruses (23, 73) as well as to circulative plant viruses transmitted by whiteflies (30, 49). It may afford protection to viruses transmitted via the circulative route from the insect immune system during their journey to the salivary tissues (30), by as yet undetermined mechanisms.
In the final leg of its journey, the virus must pass through the salivary tissues and be released into the salivary canal, where it can be deposited into a potential host plant during salivation. Luteovirids move exclusively through the paired accessory salivary glands (ASG), which also function as the principal excretory organs in aphids (31). The basal lamina encasing each ASG and the basal plasmalemma of each of the four ASG cells can both function as barriers to transmission (8, 29). The vector specificity of luteovirids lies primarily at the ASG (29, 31). Serologically related luteovirids compete for transmission through the gut and ASG, lending further support to the idea that receptor-mediated endocytosis is the mechanism that regulates the vector-specific transmission of each luteovirid species. These data indicate that virus-specific receptors may be expressed at the gut and at the ASG basal lamina and plasmalemma (32, 37). In addition, the circulative pathway described above indicates the potential involvement of multiple unknown aphid and endosymbiont proteins in virus trafficking through the endomembrane system and/or in immune evasion.
Understanding aphid components regulating the transmission of luteovirids by use of molecular biology and transgenic analyses has been difficult because the tools to generate a transgenic aphid have not yet been developed, and the genome sequence of an aphid, Acyrthosiphon pisum, was only recently published (38). However, the advent of aphid microarrays has provided transcript-level analyses of aphid-virus interactions. Genes involved in intercellular trafficking, endocytosis, and signal transduction were among the few genes differentially expressed in A. pisum intestinal epithelium following feeding on plants infected with Pea enation mosaic virus (7). The identification of a relatively small number of genes differentially expressed following virus acquisition and the functional classification of these genes support the hypotheses that viruses use a preexisting protein trafficking pathway for the circulative transmission pathway in the insect and that regulation of this pathway might be controlled primarily at the protein level.
Taking advantage of the reproductive biology of aphids, we performed sexual crosses between transmission-competent and refractive genotypes of the greenbug aphid, Schizaphis graminum. An F2 population segregating for yellow dwarf virus (YDV) transmission efficiency has been maintained as parthenogenetically reproducing genotypes, which allows repeated phenotyping of heritable traits. Genetic analysis of the F2 genotypes showed that the transmission of each YDV species is controlled by distinct but overlapping sets of loci (8, 9, 34). The virus movement across potential transmission barriers also segregates independently in the F2 genotypes, indicating that the movement of virions across the gut and ASG is also controlled by different genes (8). Furthermore, genetic analyses of the F2 population indicated that virus transmission in this population is controlled by a few major genes and a multitude of minor genes acting in an additive manner (9, 32).
We combined genetics and proteomics to identify several proteins in S. graminum that interact with and/or direct the movement of Cereal yellow dwarf virus-RPV (CYDV-RPV) along the circulative transmission pathway (77). That study demonstrated the discovery potential of the F2 population for revealing proteins involved in virus circulative transmission. At the time, limited genome sequence data for aphids allowed the identification of only two of these proteins by mass spectrometry. In the present study, we coupled two-dimensional (2-D) fluorescence difference gel electrophoresis (DIGE), one of the most widely used platforms for quantitative proteomics (68), and tandem mass spectrometry with genetic analyses to quantitatively compare the proteomes of S. graminum genotypes that differ in the ability to transmit CYDV-RPV. The recently published genome sequence of the aphid species A. pisum has provided additional resources for protein discovery in our S. graminum genotypes and has propelled proteomics to the forefront of biological discovery technologies for S. graminum and other aphid species (10, 14, 25, 26, 50, 51, 74).
MATERIALS AND METHODS
Aphids and virus transmission.
Parthenogenetically reproducing aphid colonies were maintained on caged barley (Hordeum vulgare) at 20°C with an 18-h photo period as described previously (35). The origins and CYDV-RPV transmission efficiencies of the parental genotypes of S. graminum, Sg-SC and Sg-F, as well as the F2 genotypes A3, C2, K2, and K3 were described previously (8, 9, 32, 77). Additional F2 genotypes, MM1, BB1, CC1, and CC6, are described in Table 1.
TABLE 1.
Schizaphis graminum genotypes examined using 2-D DIGE for proteins associated with CYDV-RPV transmission competency, their CYDV-RPV transmission types, and their transmission barriers
| Genotypea | Sourceb | RPV transmission phenotype (% transmission efficiency)c | RPV transmission barrierd | Refractive typee |
|---|---|---|---|---|
| Sg-F | Parent | Competent (63) | None | |
| A3 | F2 | Competent (100) | None | |
| CC6 | F2 | Competent (75) | None | |
| Sg-SC | Parent | Refractory (1) | ASG, HG | RT-3 |
| K2 | F2 | Refractory (<1) | ASG, HG | RT-3 |
| MM1 | F2 | Refractory (<1) | ASG, HG | RT-3 |
| C2 | F2 | Refractory (3.6) | HG | RT-1 |
| CC1 | F2 | Refractory (<1) | HG | RT-1 |
| K3 | F2 | Refractory (4.3) | ND | RT-ND |
| BB1 | F2 | Refractory (<1) | ASG | RT-2 |
The name of the S. graminum genotype as it is maintained in the USDA-ARS Virology and Nematology Facility on the campus of Cornell University, Ithaca, NY.
The sources of the S. graminum genotypes used in this study were the parental genotypes Sg-F (transmission competent) and Sg-SC (refractive) and selected F2 lines from random matings of F1 hybrids.
Characterization of the CYDV-RPV transmission phenotype, used for the purpose of grouping in the statistical analysis of the 2-D DIGE experiments. Transmission-competent genotypes all had >60% CYDV-RPV transmission efficiencies, and refractory genotypes had <5% CYDV-RPV transmission efficiencies.
For the refractory genotypes, the barriers to transmission, i.e., the hindgut (HG) and the accessory salivary gland (ASG), have been characterized for the genotypes as indicated. ND, the barriers have not been determined.
RT-1 genotypes possess a strong gut barrier and can transmit viruses across the ASG, RT-2 genotypes possess a strong ASG barrier and can acquire viruses into and across the gut epithelium, and RT-3 genotypes possess a dual barrier to transmission.
Experimental design and Cy dye labeling.
To determine the repeatability of the DIGE analysis, we performed a pilot experiment comparing six biological replicates of the parental Sg-F and Sg-SC genotypes. A power analysis (15, 43) conducted on these data determined that three biological replicates were sufficient to detect at least a 1.3-fold change with high statistical confidence (power of >0.8 for 100% of the data). Thus, to quantitatively compare the aphid genotypes in subsequent experiments, at least three biological replicates were analyzed. Each biological replicate consisted of protein extracts from independent pools of a single genotype collected from multiple colonies. For each sample, 3 g of aphids was ground to a fine powder in liquid nitrogen, using a prechilled mortar and pestle. The proteins were prepared and quantified for 2-D gel electrophoresis by precipitation in trichloroacetic acid and acetone (TCA-A) (13) and by the Quick Start Bradford protein assay (Bio-Rad, Hercules, CA), respectively. Cy dye-labeled samples were grouped during electrophoresis so that no two Cy3- and Cy5-labeled sample pairs were run on duplicate gels. A dye swap design was incorporated to control for labeling biases (44). A combined Cy2-labeled internal standard containing equal amounts of all protein extracts was included in every gel to normalize across the multiple gels and to perform relative quantification. The added benefit of the Cy2 internal standard lies in its ability to minimize variation in sample quantification due to differences in electrophoretic conditions and loading and to provide a template for spot matching so that there are no missing values. Proteins were labeled with the cyanine dye Cy2, Cy3, or Cy5 according to the manufacturer's instructions (GE Healthcare, Piscataway, NJ). Briefly, protein samples were adjusted to equal volumes, pH adjusted to 8.5, and allowed to react with the Cy dyes for 45 min on ice in the dark. The reactions were quenched by the addition of a molar excess of l-lysine for 10 min, and then the Cy3- and Cy5-labeled sample pairs were mixed together with the Cy2-labeled internal standard before 2-D electrophoresis. The dye/protein ratio for the experiments was 200 pmol dye to 50 μg protein.
2-D DIGE.
The following eight F2 genotypes, in addition to the two parental genotypes, were compared using DIGE: Sg-F, A3, and CC6 (transmission competent), Sg-SC, K2, and MM1 (refractive due to a dual gut-ASG barrier), BB1 (refractive due to an ASG barrier), and C2 and CC1 (refractive due to a gut barrier) (Table 1). The barrier in the refractive K3 genotype has not been determined. Analytical 2-D gels containing Cy dye-labeled samples were used for quantitative analysis, and preparative gels containing nonlabeled samples were used for spot picking and mass spectrometry. A total of 150 μg protein (50 μg of each Cy2-, Cy3-, and Cy5-labeled sample) or 500 μg of nonlabeled protein was loaded onto immobilized pH gradient (IPG) strips (pH 3 to 10, nonlinear, 24 cm; GE Healthcare) during an overnight passive rehydration of the strips according to the manufacturer's specifications for analytical or preparative gels, respectively. The first- and second-dimension conditions were run as described previously (13), with the addition of Destreak rehydration solution (GE Healthcare) in the cathodic wick during isoelectric focusing. The preparative gels were fixed in a solution of 10% methanol and 7% acetic acid for 1 h, stained overnight in colloidal Coomassie blue (Invitrogen, San Diego, CA), and destained in water for 12 h prior to scanning and spot picking.
Gel analysis.
Gels were scanned on a Typhoon variable-mode imager (GE Healthcare) at 100 dpi according to the manufacturer's specifications for Cy dyes (GE Healthcare), and colloidal Coomassie blue (Invitrogen)-stained gels were visualized with a 632.8-nm helium-neon laser with no emission filter. DIGE gel images were analyzed using Nonlinear Progenesis Samespots v. 3.2 (Nonlinear Dynamics, Newcastle Upon Tyne, United Kingdom). All images passed quality control checks for saturation and dynamic range and were cropped to adjust for positional differences in scanning. The alignment procedure was semiautomated. Fifty manual alignment seeds were added per gel (∼12 landmark spots per quadrant), and the gels were then autoaligned and grouped according to genotype for analysis. The SameSpots v. 3.2 default settings for detection, background subtraction (lowest on boundary), normalization, and matching were used (63). Spots were selected as being expressed differentially if they showed a ≥1.3-fold change in spot density and a one-way analysis of variance (ANOVA) P value of <0.05. Spots were selected based on their expression profiles (see Fig. S1 in the supplemental material). To identify proteins that are potentially involved with virus transmission, we selected spots according to whether they were upregulated in transmission-competent genotypes and downregulated in multiple refractive genotypes. To identify proteins that may potentially block virus transmission, we selected spots according to whether they were downregulated in multiple competent genotypes and upregulated in multiple refractive genotypes. To identify proteins specifically acting at either the gut or the ASG, we selected spots according to whether they were either up- or downregulated in genotypes with a single barrier to transmission (either gut or ASG).
Principal component analysis (PCA) (41) was used to identify outliers and to determine the sources of variation in the data. PCA reduces the variation in the expression of the spots to two-dimensional space to identify the sources of variation in the gel replicates and in gel spot expression. The PCA biplot was generated with Samespots v 3.2 and contains both gel and spot data. Correlation analysis was performed to identify proteins that might be acting at the same barrier to transmission. A power analysis was repeated on the complete DIGE data to confirm that the data were at sufficient power, and it was determined that 99.1% of the DIGE data were at a power of >0.8. Q values (false discovery rate-adjusted P values) were calculated to determine the number of false-positive spots in the data set (44). A Q value cutoff of 0.002 was selected (which equals 1.3 false positive results for the entire data set with a change of >1.3-fold). We used the Progenesis Samespots v 3.2 statistical modules to conduct these analyses.
Mass spectrometry.
Protein spots were picked manually from the preparative gels of genotypes Sg-F, Sg-SC, A3, K2, K3, and C2 by use of a 1.5-mm picking pen (The Gel Company, San Francisco, CA) and were then digested with trypsin (Promega, Madison, WI) as previously described (13).
For matrix-assisted laser desorption ionization-tandem mass spectrometry (MALDI MS-MS/MS) analysis, peptides were resuspended in 10 μl 0.1% trifluoroacetic acid (TFA), desalted using a 0.2-μl C18 Ziptip (Millipore, Billerica, MA), and freeze-dried in a vacuum concentrator. The samples were reconstituted in 3 μl of 0.1% TFA in 50% acetonitrile (ACN) prior to analysis by mass spectrometry. MALDI-time of flight (MALDI-TOF)-MS/MS was carried out as described previously (13, 77).
For nano-liquid chromatography-electrospray ionization-MS/MS (nano-LC-ESI-MS/MS) analysis on an LTQ-Orbitrap Velos instrument (Thermo-Fisher Scientific, San Jose, CA), the tryptic digest was reconstituted in 10 μl of 2% ACN with 0.5% formic acid (FA). The mass spectrometer was equipped with a “plug-and-play” nano ion source device (CorSolutions LLC, Ithaca, NY). The nano-LC was performed using a Dionex UltiMate3000 MDLC system (Dionex, Sunnyvale, CA). The gel-extracted peptides (5 to 10 μl) were injected using a user-defined program onto a PepMap C18 trap column (5 μm, 300 μm by 5 mm; Dionex) at a 20-μl/min flow rate for online desalting and then separated on a PepMap C18 reverse-phase (RP) nano column (3 μm, 75 μm by 15 cm; Dionex) which was installed in the “plug-and-play” device with a 10-μm spray emitter (New Objective, Woburn, MA) mounted in front of the Orbitrap ion transfer tube. The peptides were then eluted in a 20-min gradient of 10% to 40% ACN in 0.1% FA at 300 nl/min. The Orbitrap Velos instrument was operated in positive ion mode with the nano spray voltage set at 1.5 kV and the source temperature at 225°C. The instrument was either internally calibrated, using the background ion signal at m/z 445.120025 as a lock mass, or externally calibrated, using Ultramark 1621 for the Fourier transform (FT) mass analyzer. The instrument was operated in parallel data-dependent acquisition (DDA) mode, using the FT mass analyzer for survey MS scans. This was followed by MS/MS scans on the top seven most intense peaks with multiply charged ions above a threshold ion count of 5,000 in the LTQ mass analyzer. MS survey scans were done at a resolution of 60,000 full width at half max (FWHM) at m/z 400 for the mass range of m/z 375 to 1,400. Dynamic exclusion parameters were set at repeat count 1, with a 20-s repeat duration, exclusion list size of 500, 30-s exclusion duration, and ±10 ppm exclusion mass width. Collision-induced dissociation parameters were set at the following values: isolation width, 2.0 m/z; normalized collision energy, 35%; activation Q, 0.25; and activation time, 10 ms. All data were acquired using Xcalibur 2.1 software (Thermo-Fisher Scientific).
For nano-LC-ESI-MS/MS analysis on a 4000QTrap (ABI/MDS Sciex, Framingham, MA), peptides were reconstituted in 10 μl of 2% ACN with 0.5% FA. Nano-LC was carried out using an LC Packings Ultimate integrated capillary high-performance liquid chromatography (HPLC) system equipped with a Switchos valve (Dionex). The gel-extracted peptides (6.4 μl) were injected using a Famos autosampler onto a C18 PepMap trap column (5 μm, 300 μm by 5 mm; Dionex) for online desalting, separated on a PepMap C18 RP nano column, and then eluted in a 20-min gradient of 10% to 40% ACN in 0.1% FA at a 275-nl/min flow rate. The nano-LC was connected inline to a hybrid triple-quadrupole linear ion trap mass spectrometer (4000QTrap) equipped with a Micro Ion Spray Head II ion source.
MS data acquisition was performed using Analyst v 1.4.2 software (ABI/MDS Sciex) in the positive ion mode, using DDA. The nano spray voltage was 1.85 kV for all experiments in positive ion mode. Nitrogen was used as the curtain (value of 10) and collision (set to high) gas, with a heated interface at 150°C. The declustering potential was set at 50 eV, and gas 1 was 20 (arbitrary units). For DDA, after each survey scan (m/z 375 to m/z 1,550) and an enhanced-resolution scan, the three ions with the highest intensity and multiple charge states were selected for MS/MS with rolling collision energy based on the different charge states of the selected ions and their m/z values. Calibration was performed using MS/MS spectra of 100 fmol/μl of [Glu1]-fibrinopeptide B by infusion analysis for all three scan speeds (250 atomic mass units [amu]/s, 1,000 amu/s, and 4,000 amu/s). All calibrations were applied for survey scans, enhanced-resolution scans to determine the charge states, and MS/MS scans in DDA.
For nano-LC-ESI MS-MS/MS analysis on a Synapt HDMS instrument (Waters, Manchester, United Kingdom), dried peptides were reconstituted in 12 μl of 2% ACN and 0.5% FA. Five microliters of each sample was injected. Nano-LC separation of tryptic peptides was performed with a nanoACQUITY system (Waters) equipped with a Symmetry C18 (5 μm, 20 mm by 180 μm) trapping column and an ultrahigh-pressure liquid chromatography (UPLC) BEH C18 RP (1.7 μm, 15 cm by 75 μm) analytical column (Waters). The samples (5-μl partial loop injections) were transferred to the trapping column with a 0.1% solution of FA in water at a flow rate of 7 μl/min for 3 min. Mobile phase A consisted of 0.1% FA in water, and mobile phase B consisted of 0.1% FA in ACN. Following desalting and concentration, the trapping column was subjected to a reverse flush to the analytical column and separated with a gradient of 2 to 40% mobile phase B over 30 min at a flow rate of 300 nl/min, followed by a 5-min rinse with 95% mobile phase B. The column was reequilibrated under the initial conditions for 20 min. Column temperature was maintained at 35°C. A concentration of 100 fmol/μl [Glu1]-fibrinopeptide B in 25% ACN with 0.1% FA was used as the lock mass compound and was delivered via the auxiliary pump of the LC system at a flow rate of 300 nl/min to the reference sprayer of the NanoLockSpray source of the mass spectrometer. The eluent from the analytical column was delivered to the analytical sprayer of the same source through a PicoTip emitter (New Objective, Woburn, MA) with a 10-μm tip diameter.
The Synapt HDMS instrument was operated in Q-TOF V mode, with a typical resolution of at least 10,000 FWHM. Analysis was conducted using positive polarity. The TOF analyzer of the mass spectrometer was calibrated externally using fragmentation of the doubly protonated monoisotopic ion of [Glu1]-fibrinopeptide B delivered via the lock mass reference sprayer. Calibration was performed over the m/z range from 50 to 2,000. Collected data were postacquisition lock mass corrected using the same [Glu1]-fibrinopeptide B ion. The reference sprayer was sampled for 1 s every 30 s. Accurate mass LC-MS/MS DDA data were obtained as follows. MS survey scans of 1 s in duration, with an interscan delay of 0.02 s, were acquired for the m/z range from 300 to 1,500. Charge state selection was enabled such that MS/MS data were obtained for up to three ions of charge 2+, 3+, or 4+ detected in the survey scans. MS/MS spectra were acquired for the m/z range from 50 to 2,000, at a scan rate of 1 s, with an interscan delay of 0.02 s. Charge state-dependent collision energy ramps were employed to improve the quality of MS/MS spectra. A real-time dynamic exclusion window of 40 s was applied to each precursor selected for fragmentation. The acquisition mode was switched from MS to MS/MS when the abundance of an individual ion exceeded 25 cps and was returned to MS mode when the total ion current for the MS/MS acquisition exceeded 10,000 cps or after three scans had been completed.
Protein identification.
The MS and MS/MS data collected were submitted to MASCOT v 2.2 (Matrix Science, Boston, MA) (53, 55) by use of GPS Explorer software, v 3.5 (ABI/MDS Sciex), for MALDI identifications and to our in-house MASCOT v 2.2 server for LC-MS/MS identifications. The experimental data were searched against the entire NCBI nonredundant (nr) database containing the A. pisum gene models (download dates, 28 July 2008 and 12 January 2010). The following search parameters were used for identifications from data generated by the 4700 and Synapt instruments: carbamidomethyl-cysteine as a fixed modification and methionine oxidation as a variable modification; one missed tryptic cleavage was allowed. The searches were done with a mass error tolerance of 25 ppm in the MS mode and 0.1 Da in the MS/MS mode. For identifications made from data generated by the 4000QTrap instrument, carbamidomethyl-cysteine and methionine oxidations were allowed as variable modifications, one missed tryptic cleavage was allowed, the peptide mass error tolerance was 1.5 Da, and the MS/MS mass error tolerance was 0.6 Da. For the Orbitrap data, all MS and MS/MS raw spectra were processed using Proteome Discoverer 1.1 (PD1.1; Thermo-Fisher Scientific). The peptide mass tolerance was set to 10 ppm, and the MS/MS tolerance was set to 0.8 Da. Carbamidomethyl modification of cysteine was selected as a fixed modification, and variable modifications included methionine oxidation and deamidation of asparagine and glutamine. The preliminary protein identifications obtained automatically from the software were inspected manually by examining the spectra for the completeness of the Y and B ion series prior to acceptance. For predicted or hypothetical proteins, homology to known proteins was determined by searching against protein databases in NCBI with BLAST (1), and conserved domains were identified by searching the Conserved Domain Database (CDD) (46).
RESULTS
Heritable differences in expression levels of S. graminum proteins correlate with CYDV-RPV vectoring capacity.
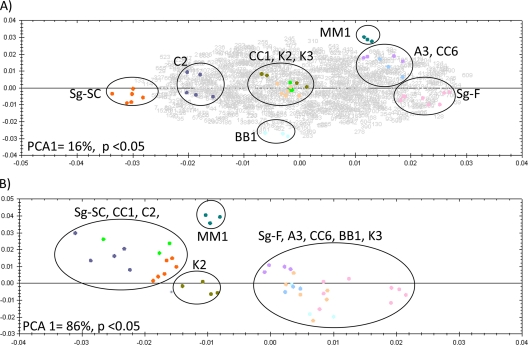
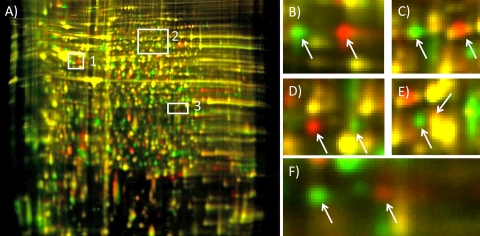
The proteomes of 10 S. graminum genotypes differing in CYDV-RPV transmission capacity (Table 1) were analyzed by quantitative DIGE. A total of 1,189 spots were visualized on the Cy2 reference gel and matched across all gels in the experiment (Fig. 1). Transmission-competent and refractive aphid proteomes had substantial levels of nonoverlap, as seen by the Cy3- and Cy5-specific signals on the 2-D gel (Fig. 1B); therefore, spot matching to a Cy2 internal standard gel image containing all of the proteins in the entire experiment was critical for alignment and quantitative comparison of spots that were completely absent in some genotypes. Of the 1,189 total spots, 687 were expressed differentially among genotypes by at least a 1.3-fold change, with a P value of <0.05 by one-way ANOVA and a Q value cutoff of 0.002. For these 687 spots, PCA revealed that the first principle component (x axis in Fig. 2A) accounted for 16% of the variation in the experiment; variation was attributed to protein expression differences between the transmission-competent parent, Sg-F, and the refractive parent, Sg-SC. The variation in the second principle component (14%) was ascribed to protein expression differences between the refractory F2 genotypes BB1 and MM1 (y axis of Fig. 2A). The protein expression data clustered each F2 genotype sample in the PCA according to genotype, not gel replicate or Cy dye label, both of which are common sources of technical variation in 2-D electrophoresis experiments (44). The F2 genotypes were interspersed between the parental genotypes in the PCA biplot (Fig. 2A), proving that the F2 genotypes overall express the same proteins as the parental genotypes but differ in their protein expression levels for many of these proteins. The transmission-competent F2 genotypes (A3 and CC6) clustered near the competent parent Sg-F, indicating that there is less biological variation between these genotypes. However, MM1, a refractory F2 genotype, also clustered near the competent genotypes and may express many proteins regulating virus transmission, but not enough to confer vectoring capacity. Taken together, these data indicate that, as expected, not all 687 proteins differentially expressed between the parental genotypes influence the virus transmission phenotype. Thus, spots were selected as described below, based on their expression patterns.
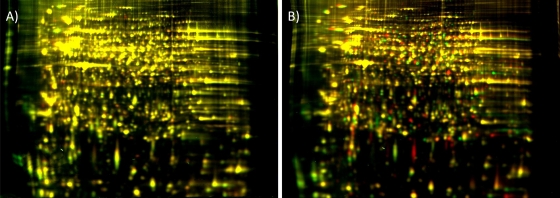
FIG. 1.
Comparison of S. graminum proteins extracted from the two biological replicates of one genotype (A) or from a sample combining two parent genotypes, Sg-F and Sg-SC (B), using 2-D DIGE. The 2-D gel format consisted of pH 3 to 10 nonlinear IPG strips for the first-dimension separation and 24-cm 12% PAGE gels for the second dimension. (A) Protein extracts from two independent Sg-F colonies, labeled with Cy3 (green) and Cy5 (red). (B) Protein extracts from the Sg-F competent parental genotype, labeled with Cy3, and the Sg-SC refractive parental genotype, labeled with Cy5. The gels were visualized using a Typhoon variable-mode imager (GE Healthcare) according to the manufacturer's instructions on imaging cyanine dyes. A majority of the spots are present for both replicate Sg-F samples, as evidenced by a general lack of distinct red and green spots and a preponderance of yellow spots in panel A. Numerous red and green spots in panel B represent proteins that are unique to either the Sg-F (Cy3) or the Sg-SC (Cy5) proteome.
FIG. 2.
(A) Graph of PCA biplot showing the first and second principle components on the x and y axes, respectively, for DIGE data comparing all 10 S. graminum genotypes. The biplot displays expression variation in all spots for all genotypes (displayed as gray numbers), determined as a ≥1.3-fold change with an ANOVA P value of <0.05, and variation in the replicate gel images for each genotype (displayed as colored spots). Gel replicates for each genotype that show similar variation in the first and second principle components are grouped in black circles. Sixteen percent of the total variance along the first principle component can be explained by the differences between the two parental genotypes, Sg-SC and Sg-F, used to generate the F2 population. (B) PCA plot displaying gel replicates based on three protein spots (spots 112, 181, and 208) (Table 4) predicted to act at the gut, which can clearly distinguish between genotypes that either have or do not have an acquisition barrier to virus transmission in the gut. PCA1 = 86% (P < 0.05 for the three selected spots).
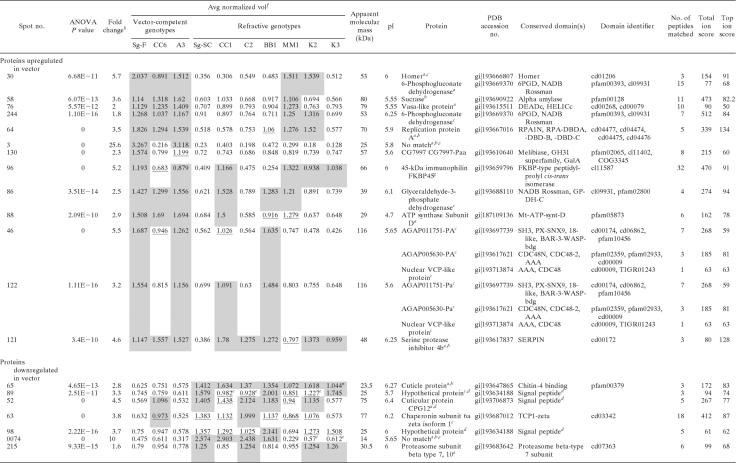
The DIGE expression data were first compared between the three transmission-competent genotypes (Sg-F, A3, and CC6) and the refractive parent genotype Sg-SC. These spots were selected without regard to their expression in the refractive F2 genotypes, since the genes controlling virus transmission are additive in effect (8, 9, 34), and as we expected, the refractive F2 genotypes expressed some of the proteins in a manner similar to that of the competent genotypes. Thirty-two such spots were identified (Tables 2 and 3), 13 of which were aphid proteins upregulated in the competent genotypes and 7 of which were downregulated in the competent genotypes (Table 2). Three of the spots (3, 130, and 122) and two of the spots (52 and 63) were not significantly upregulated and downregulated, respectively, in CC6 relative to the refractive parent, although the trend was apparent for most (Table 2). The spot volumes for the transmission-competent F2 genotypes were similar to those for the transmission-competent parent Sg-F, demonstrating that the level of expression for the proteins in these spots is heritable from the male transmission-competent parent Sg-F. The 12 other spots were later identified as endosymbiont proteins (Table 3): 6 were upregulated and 6 were downregulated in the competent vectors. Among these 32 spots, the expression levels of 25 spots in the refractive genotype MM1 matched those for the competent genotypes (Tables 2 and 3), which is consistent with the variation in all differentially expressed spots in MM1 observed in the PCA biplot (Fig. 2A).
TABLE 2.
Schizaphis graminum proteins differentially expressed between transmission-competent and refractive genotypes
a Method of protein identification, MALDI MS-MS/MS.
b Method of protein identification, ESI MS-MS/MS on 4000QTrap instrument.
c Method of protein identification, ESI MS-MS/MS on Synapt HDMS instrument.
dInterpro Scan SignalP hidden Markov model.
e Average normalized values were elevated but not statistically significant.
f Mean separations of log-normalized volumes are shown via shading and underlining, with shading indicating higher expression levels on a spot-by-spot basis (ANOVA P value, <0.05). Sample spot volumes were normalized using a background-adjusted abundance ratio to the Cy2 internal standard and were adjusted by a gain factor calculated for each sample.
g Method of protein identification, ESI MS-MS/MS on LTQ-Orbitrap Velos instrument.
hThe fold change value reported is the maximum change between the mean normalized volumes between genotypes.
TABLE 3.
Buchnera proteins differentially expressed between transmission-competent and refractive Schizaphis graminum genotypes
a Method of protein identification, MALDI MS-MS/MS.
b Average normalized values were elevated but not statistically significant.
c Mean separations of log-normalized volumes are shown via shading and underlining, with shading indicating higher expression levels on a spot-by-spot basis (ANOVA P values, <0.05). Sample spot volumes were normalized using a background-adjusted abundance ratio to the Cy2 internal standard and were adjusted by a gain factor calculated for each sample.
d The fold change value reported is the maximum change between the mean normalized volumes between genotypes.
e Isoform pairs are spots 6/12 and 106/147, spots 12 and 28, spots 131 and 61, and spots 285 and 97.
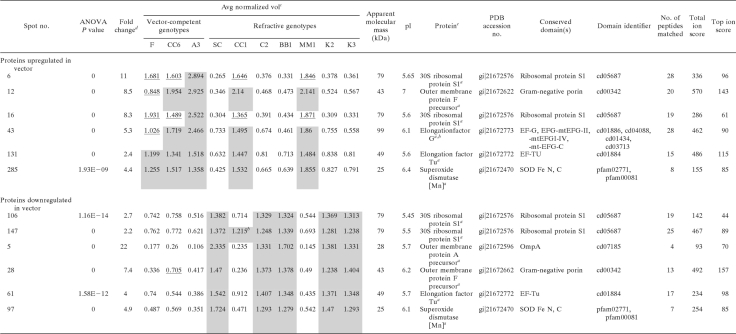
The identities of the proteins in these spots were obtained using mass spectrometry coupled to homology-based searching of the predicted gene models from the A. pisum (38) and Buchnera (66) genomes deposited in the NCBI nr database. The 18 spots listed in Table 2 contained aphid proteins involved in sugar digestion, cell cycle, energy metabolism, membrane and protein trafficking, and the cytoskeleton. For some of these, e.g., spots 58 and 122, MALDI MS-MS/MS analysis by direct spotting of the trypsin digests did not yield enough MS/MS spectra, and thus we performed LC-MS/MS to increase the resolution and sensitivity by including an online first-dimension LC separation coupled to electrospray ionization on either a 4000QTrap, Synapt HDMS, or LTQ-Orbitrap Velos mass spectrometer. The identities of the proteins in spots 3 and 74 could not be determined using these methods, and these are candidates for S. graminum-specific proteins, since no match could be found in the available aphid and endosymbiont databases. The 12 spots listed in Table 3 contained only six different Buchnera proteins. Thus, the same proteins were found in multiple spots (for example, the 30S ribosomal subunit was identified in spots 6, 16, 106, and 147), a common occurrence in 2-D electrophoresis.
Multiple different proteins can coresolve in the same 2-D spot (78), and therefore a change in spot volume may be due to the addition or subtraction of a protein or to a change in expression level of one or more proteins in the spot. It is imperative that the protein composition of spots be determined for multiple genotypes to confirm that the protein has an expression profile consistent with the relative quantification of DIGE data across the genotypes. We determined the proteins in the spots of interest for two to seven different genotypes and reported the protein identification(s) found upregulated or downregulated in the competent genotypes. For instance, spot 30 (Table 2) was upregulated in all competent genotypes and two refractive genotypes. For two abundant proteins, homer and 6-phosphogluconate dehydrogenase, only homer was detected in transmission-competent genotypes by DDA mass spectrometry, and therefore homer is a good candidate for involvement in virus transmission. Similarly, spot 121 (Table 2) contained a melanization pathway regulator, SERPIN-4. The spot was upregulated in all but two genotypes. Multiple different SERPIN-4 proteins were detected in this spot for the refractive genotypes, but only one form of SERPIN-4 (Protein Data Bank [PDB] accession no. gi|193617837) was detected in the competent genotypes. Therefore, the presence of multiple proteins per spot is an alternative explanation to gene additivity for the increase in spot volume observed for the refractive genotypes.
Homology-based database interrogation reaches its limits if there are no close matches in the database, if multiple matches exist in the database with no unique tryptic peptides, or if peptides from the sample match multiple different proteins. For example, for spots 46 and 122 from the competent genotypes (Table 2), peptides were identified in both spots that matched two A. pisum AAA ATPases: the homologue (PDB accession no. gi|193713874) of nuclear valosin-containing protein and the homologue (PDB accession no. gi|193617621) of the yeast protein CDC48, an ATPase in the endoplasmic reticulum that controls the fate of ubiquitinated substrates. Also identified in these spots were a homologue of sorting nexin 9 (PDB accession no. gi|193697739) and peroxinectin (PDB accession no. gi|193681143) (not shown). Spots 46 and 122 from the refractive genotype BB1 contained only peroxinectin (not shown), and an increase in peroxinectin expression for the two refractive genotypes may explain the increase in spot volume. The matches to the A. pisum homologues were above the MASCOT threshold cutoff scores for identity or extensive homology for peptides matching all of the proteins described (Table 2). We hypothesize that the actual protein resolved in spots 46 and 122 may be expressed from an S. graminum-specific gene or that it is not found in its entirety in our protein databases, possibly because the A. pisum genome is not fully annotated.
Coupling transmission barrier phenotype to DIGE analysis identifies distinct proteins correlated with CYDV-RPV movement through the gut or ASG.
Previously, we demonstrated that the gut and ASG are potential barriers to CYDV-RPV transmission in S. graminum and that these barriers segregate independently in refractive F2 genotypes (8). We categorized the refractive genotypes into three groups: the refractive type 1 (RT-1) genotypes have primarily a gut barrier, refractive type 2 (RT-2) genotypes have primarily an ASG barrier, and refractive type 3 (RT-3) genotypes have both a gut and an ASG barrier (Table 1). Thus, we predicted that it would be possible to identify proteins whose expression pattern could be correlated with the presence of a specific barrier in refractive genotypes by using the RT-1 and RT-2 genotypes. Spots in these categories may or may not be expressed in the RT-3 genotypes, which possess both barriers to transmission, to various degrees.
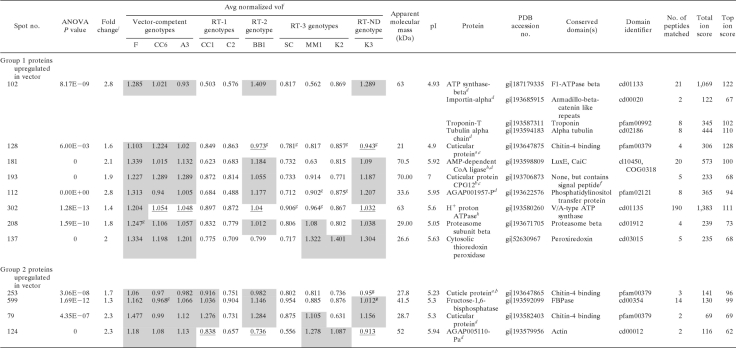
To identify proteins that may facilitate virus transmission across the gut, we used correlation analysis to identify proteins that were upregulated in genotypes competent for virus movement in the gut (the competent genotypes and the RT-2 genotype) and downregulated in the RT-1 genotypes, which possess a gut barrier. Two groups of spots were identified as covarying in expression in the F2 genotypes (Table 4 ). Group 1 included eight spots (102, 128, 181, 193, 112, 302, 208, and 137) that were upregulated in gut-competent genotypes but not in the RT-1 genotypes. This strong correlation indicates that these are likely protein candidates for acting at the gut to facilitate virus transmission. Among the proteins in these spots were two cuticle proteins (spots 128 and 193), one of which has a predicted signal peptide (spot 193), phosphatidylinositol transfer protein (PITP; spot 112), coenzyme A (CoA) ligase (spot 181), the H+ proton ATPase (spot 302), the proteasome beta subunit (spot 208), and a cytosolic thioredoxin peroxidase (spot 137). At least four proteins were identified in abundance in spot 102 across multiple competent genotypes. The homologues in the A. pisum genome for two of these (alpha tubulin [PDB accession no. gi|193594183] and ATP synthase-beta [PDB accession no. gi|187179335]) have nearly identical pIs (5.01 and 5.06, respectively), although they differ slightly in mass (50 kDa and 56 kDa, respectively). Since these are both highly abundant proteins and had nearly identical electrophoretic migratory coordinates on the 2-D gel, we were unable to either resolve them into separate 2-D gel spots or determine which of the two proteins is correlated with virus transmission. None of these four proteins was detected in gel spots analyzed by mass spectrometry from the identical 2-D coordinate for the refractive genotype C2 (data not shown).
TABLE 4.
Proteins predicted to be involved in virus transmission in the guts of Schizaphis graminum aphids based on the coupling of genetic and DIGE data
a Protein found differentially expressed as a molecular weight isoform in virus transmission-competent and refractive aphid genotypes.
b Protein found differentially expressed as a pI isoform in virus transmission-competent and refractive aphid genotypes.
c Protein ID differs in competent and refractive samples, indicating multiple proteins with similar molecular sizes and pIs per spot. The protein identified in competent genotypes is likely responsible for the observed fold change.
d ESI-MS/MS on a Synapt HDMS instrument was used for protein identification.
e MALDI-MS/MS was used for protein identification.
f Interpro Scan SignalP hidden Markov model.
g Averaged normalized value was elevated but not statistically significant.
h ESI-MS/MS on an LTQ-Orbitrap Velos instrument was used for protein identification.
i Mean separations of log-normalized volumes are shown via shading and underlining, with shading indicating higher expression levels on a spot-by-spot basis (ANOVA P value, <0.05). Sample spot volumes were normalized using a background-adjusted abundance ratio to the Cy2 internal standard and were adjusted by a gain factor calculated for each sample.
j The fold change value reported is the maximum change between the mean normalized volumes between genotypes.
Group 2 included four spots (253, 599, 79, and 124) (Table 4) that were all upregulated in the gut-competent genotypes but were also upregulated in CC1. Proteins in these spots included two cuticle proteins, fructose 1,6-biphosphatase, and actin. The weaker correlation of these spots in the gut-refractive genotypes indicates that these proteins may be necessary for virus translocation at the gut but are not sufficient to confer a complete transmission phenotype or that they are not involved in translocation across the gut.
Correlation analysis was used to identify protein spots that may block virus translocation in the gut tissues. We predicted that such spots would be upregulated in RT-1 genotypes possessing a gut barrier and turned off in competent and RT-2 genotypes. No protein spots with this expression profile were identified.
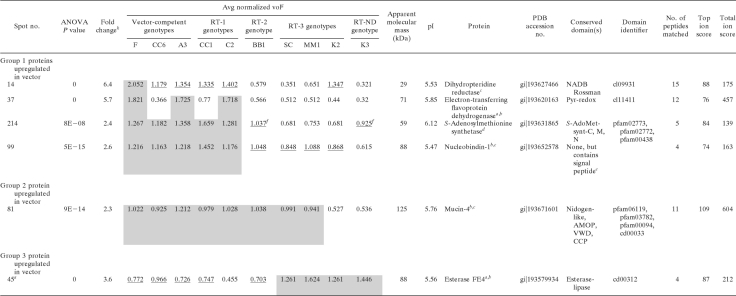
Correlation analysis was used to identify protein candidates involved in virus transmission in aphid tissues distal to the gut in the circulative transmission pathway (Table 5). These tissues include the hemolymph and the two known transmission barriers in the ASG, the basal lamina and the plasmalemma (29). Such proteins would be expressed in transmission-competent genotypes, the RT-1 genotypes (CC1 and C2), and possibly the RT-3 genotypes but not in the RT-2 genotype BB1. The RT-1 genotypes possess a gut barrier to transmission but are transmission competent when virus is injected into the hemolymph (8). Remarkably, C2 transmits virus that is injected into the hemolymph even more efficiently than transmission-competent genotypes do. Since there are three potential barriers distal to the gut, i.e., hemolymph, ASG basal lamina, and ASG plasmalemma, all RT-2 and RT-3 genotypes (and K3, with an unknown barrier [RT-ND]) were treated as independent groups in the correlation analysis to maximize protein discovery. Accordingly, three groups of protein spots were identified (Table 5). Group 1 contained four spots that were downregulated in both RT-2 and K3 genotypes relative to the competent and RT-1 genotypes. Group 2 contained one spot that was downregulated in K3 relative to the competent and RT-1 genotypes. Among these five spots, spot 14, containing the melanization cascade regulator dihydropteridine reductase (DHR), stood out as the most promising candidate protein linked to virus transmission competency in the ASG because it was upregulated in the competent genotypes and the RT-1 genotypes, which are competent for transmission at the ASG. Group 3 contained an esterase-like protein, spot 45, which was downregulated in competent, RT-1, and RT-2 genotypes and upregulated in RT-3 and RT-ND genotypes.
TABLE 5.
Proteins predicted to be involved in virus transmission in the accessory salivary glands of Schizaphis graminum aphids based on the coupling of genetic and DIGE data
a ESI-MS/MS on a 4000QTrap instrument was used for protein identification.
b Multiple proteins were identified in the spot.
c ESI-MS/MS on a Synapt HDMS instrument was used for protein identification.
d MALDI-MS/MS was used for protein identification.
e Interpro Scan SignalP hidden Markov model.
f Average normalized volume appears upregulated, but difference is not statistically significant.
g Mean separations of log-normalized volumes are shown via shading and underlining, with shading indicating higher expression levels on a spot-by-spot basis (ANOVA P value, <0.05). Sample spot volumes were normalized using a background-adjusted abundance ratio to the Cy2 internal standard and were adjusted by a gain factor calculated for each sample.
h The fold change value reported is the maximum change between the mean normalized volumes between genotypes.
MS/MS analysis of the gel-extracted peptides from spot 81 (Table 5) identified numerous tryptic fragments within only the first 750 amino acids of the protein. These peptides spanned two highly conserved domains, a nidogen domain found in basement membrane proteins with binding affinity for laminin and collagen (3, 4, 24, 69) and an extracellular adhesion-associated domain contained in mucin 4 and other proteins (AMOP) domain found in mucin 4 (12) and other proteins that function in cell adhesion (61). No peptides were found to match the C-terminal domain of the A. pisum homologue of the protein, from amino acid 751 through the predicted C terminus, amino acid 1285, by MS/MS analysis of multiple trypsin digestions of technical and biological gel spot replicates (data not shown). Quite possibly, this domain in S. graminum has diverged to the extent that no matches were obtained using our stringent search criteria. The A. pisum homologue has two additional domains in the C terminus, a von Willebrand factor type D domain and a complement control protein module, which is the domain that shares homology with the mammalian CR2 receptor. At this time, we cannot confirm if these domains also exist in the S. graminum homologue.
Aphid and Buchnera protein isoforms linked to virus transmission.
One of the hallmarks of 2-D electrophoresis is its ability to resolve protein isoforms. There were more than 100 differentially expressed spots in the parental genotypes Sg-F and Sg-SC that contributed more than 86% of their total proteome variation (13). Sixteen of these spots were discovered to be expressed as protein isoform pairs that vary in pI (Fig. 3; Table 6). The two spots of each pair were in close proximity to each other on the DIGE gels and were expressed differentially between competent and refractive parent genotypes (Fig. 3). Seven of the eight pairs consisted of one spot that was upregulated in the refractive parent, Sg-SC, and one spot that was upregulated in the competent parent, Sg-F, i.e., the parents were homozygous for each of these isoform variants (Table 6). The two spots of the eighth pair (spots 102 and 27) were both present in the Sg-SC proteome, but only spot 102 (troponin-T) was present in the Sg-F proteome (Table 6).
FIG. 3.
Numerous proteins associated with the CYDV-RPV transmission-competent phenotype in the competent parent and competent F2 genotypes had an isoform inherited from the maternal lineage that differed in pI. (A) pH 3 to 10, nonlinear, 24-cm DIGE gel of protein extracts from competent and refractive genotypes. Boxes highlight areas of the gel containing pI isoforms segregating in the Schizaphis graminum populations that differ in CYDV-RPV vectoring capacity. Proteins from transmission-competent genotypes are labeled with Cy3 (green), and proteins from the refractive genotype are labeled with Cy5 (red). (B) From the area in box 1, pI isoforms of troponin-T. (C) From the area in box 2, pI isoforms of replication protein A 70-kDa subunit. (D) From the area in box 2, pI isoforms of CoA ligase. (E) From the area in box 2, pI isoforms of electron-transferring flavoprotein dehydrogenase. (F) From the area in box 3, pI isoforms of GAPDH. In some instances (E), one isoform had a pI and molecular weight similar to those of another protein and could not be resolved from that comigrating protein, which was not differentially expressed (yellow signal resulting from signals in both Cy3 and Cy5 channels). The gel was visualized using a Typhoon variable-mode imager (GE Healthcare) according to the manufacturer's instructions on imaging cyanine dyes.
TABLE 6.
Expression of protein isoforms in Schizaphis graminum F2 and parental genotypes that are linked to virus transmission
a The virus transmission-competent isoform (C) is spot 181, the transmission-refractive isoform (R) is spot 22, and the candidate protein is CoA ligase.
b C isoform is spot 193, R isoform is spot 52, and candidate protein is a cuticle protein (CP).
c C isoform is spot 64, R isoform is spot 493, and candidate protein is RepA.
d C isoform is spot 102, R isoform is spot 27, and candidate protein is troponin-T.
e C isoform is spot 86, R isoform is spot 42, and candidate protein is GAPDH.
f C isoform is spot 88, R isoform is spot 345, and candidate protein is the delta subunit of ATP synthase (ATPD).
g C isoform is spot 37, R isoform is spot 269, and candidate protein is ETF-dehydrogenase (ETF-DH).
h C isoform is spot 14, R isoform is spot 13, and candidate protein is DHR.
i Expression of protein isoform pairs was translated into + (for upregulated) and − (for downregulated). Shading indicates that the genotype expresses the competent isoform. DIGE quantification data for each genotype are provided in Tables 2, 3, and 4 for C isoforms and in Table S1 in the supplemental material for R isoforms, except for spot 52, for which data are shown in Table 2.
Data supporting the segregating pattern for one of the isoform pairs (spots 14 and 13; DHR) can be found in Fig. 4. In the F2 genotypes where only one isoform of dihydropteridine reductase was expressed (Fig. 4, RT-2 and RT-ND columns), the expression levels were similar to expression in the parent Sg-SC. In F2 genotypes that expressed both isoforms, each isoform was expressed at a lower level than that in either of the parents (Fig. 4, competent, RT-1, and RT-3 columns; also see Fig. S1D and E in the supplemental material). This type of pattern prompted us to hypothesize that the charge-distinct isoform pairs were allelic variants differing only in pI and originating from the same locus, as has previously been observed in other organisms (27, 45, 54, 71). To test whether these isoform pairs could be allelic variants, we used mass spectrometry to identify the proteins in the isoform pairs. For most pairs, the same protein was found in both spots associated with the competent or refractive parent (Table 6), indicating that the expression of each isoform pair was derived from loci that each produced two charge-distinct isoforms (Fig. 3). Spot 102 from Sg-F and Sg-SC contained a number of different proteins (Table 4), including troponin-T, whereas the paired spot, spot 27, present only in Sg-SC, contained only troponin-T (see Table S1 in the supplemental material).
FIG. 4.
Three patterns of protein isoform expression were observed in the parent and F2 S. graminum genotypes. (A) Expression (y axis) of one isoform pair, dihydropteridine reductase, in the parents, a competent F2 genotype, and each type of refractive F2 genotype. The Sg-F isoform (blue) and the Sg-SC isoform (red) segregate in the F2 population as allelic variants. The percent intensity of each isoform was calculated by dividing the intensity of each isoform by the summed total intensity for the total protein produced from the locus. F2 genotypes expressing both isoforms expressed each isoform at a lower level than that in genotypes expressing only one isoform. (B) 3-D gel spot image of each isoform in the pair.
Each of the eight protein isoform pairs was useful in predicting the transmission competence and the principal barrier of the F2 genotypes. F2 genotypes contained either the isoform found in the competent parent, Sg-F, or the isoform found in the refractive parent, Sg-SC, or both isoforms were expressed codominantly (Table 6). Based on the expression of the transmission-competent isoform in the F2 genotypes, the transmission-competent isoforms of CoA ligase, the cuticle protein, RepA, and troponin-T were best correlated with virus translocation across the gut. The competent isoform was not expressed in the RT-1 genotypes with a gut barrier and was expressed in the RT-2 genotype with the ASG barrier. Conversely, the expression pattern of DHR was best correlated with virus translocation across the ASG, since it was expressed similarly in the competent genotypes and in the RT-2 genotype but not in the RT-1 genotypes (Table 6). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ETF-DH, and ATP-D expression was not correlated with a specific barrier but did correlate with transmission ability. Homozygous expression of the competent isoforms of GAPDH and ETF-DH correlated with transmission ability, whereas homozygous expression of the refractive isoform or heterozygous expression of both isoforms correlated with poor transmission efficiency (Table 6).
Surprisingly, the expression of several other isoform pairs whose segregation pattern correlated with transmission originated from the proteome of the bacterial endosymbiont Buchnera aphidicola (Table 3; Fig. 5). The S. graminum F2 genotypes possessed Buchnera isoforms matching those in either Sg-F or Sg-SC (Table 3), i.e., two proteome types were defined by pI differences in numerous proteins (Table 3). For simplicity, we call the collective group of Buchnera protein isoforms in Sg-F the competent Buchnera proteome type and the collective group of Buchnera protein isoforms in Sg-SC the refractive Buchnera proteome type. The Buchnera proteins in the F2 transmission-competent genotypes and the refractive genotypes CC1 and MM1 had the same pI and expression levels of the Buchnera proteins expressed by the competent parent (Table 3), Sg-F. The remaining refractive F2 genotypes, BB1, K2, K3, and C2, expressed Buchnera proteins that shared a pI and expression profile similar to that of the refractive parent Sg-SC (Table 3), except for spot 147 in genotype CC1 (Table 3). Thus, the refractive genotypes expressed either the competent or refractive Buchnera proteome type, but all competent genotypes expressed only the competent Buchnera proteome type (Table 3).
FIG. 5.
The pIs of numerous Buchnera proteins in transmission-competent F2 genotypes matched those in the competent male parent Sg-F, not the refractive maternal parent, even though Buchnera species are inherited transovarially through the maternal line. For OMP-F, there are a total of five isoforms found in the aphid genotypes. (A) Sg-SC is labeled with Cy3 (green), and Sg-F is labeled with Cy5 (red). Two of the isoforms (1 and 2) are visible in the refractive parent Sg-SC; isoform 1 is specific to Sg-SC, and isoform 2 is also found in the competent parent Sg-F, although the expression level is lower than that in the Sg-SC sample. The yellow staining for isoform 1 is due to the comigration of a different protein, not OMP-F. Four isoforms (isoforms 2 to 5) are visible in the competent paternal Sg-F genotype, including three specific, more-basic isoforms (3 to 5) and the one isoform also found in Sg-SC (2) (see Fig. S2 in the supplemental material for images of each channel independently). (B) Sg-SC is labeled with Cy3, and the transmission-competent F2 genotype CC6 is labeled with Cy5. Three isoforms are found in the competent F2 progeny, namely, two of the basic isoforms (3 and 4) and the shared isoform, 2, which is expressed more highly in CC6 than in Sg-F. Similar patterns of isoform inheritance were observed for the Buchnera proteins EF-Tu (C and D) and SOD (E and F). (C) EF-Tu expression in the parental genotypes. Sg-SC is labeled with Cy3, and Sg-F is labeled with Cy5. (D) EF-Tu expression in the competent F2 progeny CC6 (Cy5) unexpectedly matches the paternal Sg-F isoform (Cy5) (C). The yellow signal observed over the Sg-SC isoform indicates a low level of expression of the more acidic isoform in Sg-SC, and expression of this isoform of EF-Tu in Sg-SC was confirmed by mass spectrometry (not shown). (E) SOD expression in the parental genotypes. Sg-SC is labeled with Cy3, and Sg-F is labeled with Cy5. (F) SOD expression in the competent F2 progeny CC6 (Cy5) matches the paternal Sg-F isoform (Cy5) (E). The gel was visualized using a Typhoon variable-mode imager (GE Healthcare) according to the manufacturer's instructions on imaging cyanine dyes.
Buchnera is inherited through the maternal line, so we expected the Buchnera proteome type in the F2 genotypes to be the same as that expressed in the maternal parent, Sg-SC, but this was not observed. Buchnera proteins identified as differentially expressed between competent and refractive genotypes that were subsequently identified to be pI isoforms included elongation factor Tu (EF-Tu; spots 61 and 131) (Table 2), 30S ribosomal protein S1 (spots 6, 106, and 147) (Table 2), superoxide dismutase (SOD; spots 97 and 285) (Table 2), and outer membrane protein F (OMP-F; spots 12 and 28) (Table 2). OMP-F appeared as five different pI isoforms (Fig. 5A and B). The maternal genotype, Sg-SC, expressed one specific isoform, isoform 1, and shared another (isoform 2) with the paternal genotype, Sg-F (Fig. 5A, Cy3 spot). Sg-F also expressed three additional isoforms (isoforms 3, 4, and 5) (Fig. 5A, Cy5 spots). Surprisingly, the transmission-competent F2 genotypes expressed isoforms 2 (yellow signal in Fig. 5B), 3, and 4 (Fig. 5B, Cy5 signals). Similarly, the expression of EF-Tu (Fig. 5C and D), two isoforms of 30S ribosomal protein S1, and one isoform of SOD (Fig. 5E and F) matched the paternal, not maternal, expression pattern. Faint expression of the isoform found in competent genotypes was observed for the maternal parent (Fig. 5, note the yellow signal over isoform 1 in panels C and D). The images of these isoforms in separate channels can be found in Fig. S2 in the supplemental material. Thus, in this F2 population, although both Buchnera proteome types were derived from the female refractive lineage Sg-SC, the transmission-competent F2 genotypes expressed only the competent Buchnera proteome pattern, similar to the competent parent, Sg-F.
DISCUSSION
In this F2 S. graminum population, the regulation of aphid protein expression which was correlated with the virus transmission phenotype was likely inherited from the male, competent parent, whereas the regulation of endosymbiont protein expression which correlated with the virus transmission phenotype was likely inherited through the female, refractive line. Whether the contributions from the male or female lineage of these proteins reside in stable DNA polymorphisms that alter the charge of the protein (i.e., alleles), heritable regulation of posttranslational modifications, promoter activity, or protein stability and turnover remains to be determined. Quantitative proteomic analysis (2-D DIGE) of the F2 population facilitated three other major advances in understanding the mechanisms regulating circulative virus transmission by aphids: (i) association of proteins was linked to transmission to specific points along the circulative transmission pathway, (ii) charge-distinct isoforms were linked to vector competency, and (iii) heterogeneity in the Buchnera population was associated with vector competency.
The barrier to virus transmission segregated in the F2 genotypes, allowing differential protein expression to be linked directly to a potential site of action. Correlation analysis readily identified multiple proteins predicted to function at the gut in multiple aphid genotypes; however, fewer candidate proteins with a clear correlation with the transmission phenotype were identified in tissues distal to the gut. These data support the hypothesis that the biochemical regulation of the gut barrier is similar in these F2 genotypes. They also suggest that the biochemical control of transmission in tissues distal to the gut is complex and different among the F2 genotypes.
Correlation analysis identified two proteins predicted to function in the ASG (Table 5) that have been localized to salivary tissues in other insects. NucB1, a Golgi apparatus resident protein, has been localized specifically to salivary tissues in Drosophila melanogaster by in situ hybridization (52), and the enzyme activity of FE4-esterase was found in the saliva of Aedes aegypti (2). A number of different cuticle proteins were predicted to function at the gut (Table 4). Cuticle proteins have been shown to interact with luteoviruses (62), and therefore a direct mechanism of cuticle protein involvement in virus movement across the gut barrier is plausible. Many insect cuticular proteins that contain the chitin-4 domain have a highly conserved R&R consensus amino acid sequence that binds chitin (16, 17, 42, 57, 67). The R&R motif shares no sequence similarity to the cysteine-containing chitin-binding domain found in lectins, chitinases, and peritrophic membrane proteins of other invertebrates and plants (57). It is unlikely that a cuticle protein-chitin interaction is involved directly in transmission in the hindgut, as the hindgut of S. graminum lacks a chitin lining and an obvious peritrophic membrane (31). Thus, since the cuticle proteins differentially expressed between competent and refractive genotypes of S. graminum are primarily those with the chitin-4 domain, these proteins may have additional, uncharacterized functions in the hindgut or may serve to generate a physical barrier to transmission in the chitin-lined foregut, or even in the stylet (72). Alternatively, the differences in cuticle protein composition observed between competent and refractive genotypes may reflect underlying differences in sec-23-mediated deposition of the cuticle in these genotypes. sec-23 is a component of coat protein complex II-coated vesicles involved in endoplasmic reticulum-to-Golgi apparatus protein transport (5, 60). In the nematode Caenorhabditis elegans, sec-23 is required for cuticle secretion and embryogenesis (60). Differences in sec-23-mediated secretion between competent and refractive genotypes, as reflected by differences in the cuticle protein compositions, may also be responsible for differences in the secretion of other proteins, including virions. Differences in secretory pathway proteins were indeed observed between competent and refractory genotypes: we identified PITP (spot 112) (Table 4), a sec-14p homologue which was upregulated in transmission-competent genotypes. PITP induces local changes in lipid membrane bilayers required for vesiculation from the trans-Golgi network (64). The differential expression of PITP in aphids that are transmission competent through the gut indicates that virions may hijack existing lipid metabolism and phosphoinositide signaling pathways during their journey through gut tissues in aphids.
Analyzing different genotypes on the same gel by using 2-D DIGE eliminated differences in the spot patterns that might be due to isoelectric focusing and/or gel-to-gel variation and highlighted differences arising from protein expression changes between competent and refractive genotypes that might contribute to the virus transmission phenotype. Many of the differences can be ascribed to small changes in the pI of the proteins involved. 2-D gel analysis comparing Sitobion avenae F1 genotypes differing in virus transmission abilities of Barley yellow dwarf virus PAV (BYDV-PAV) identified a few differentially expressed proteins with certain isoforms linked to transmission ability (54). Earlier, we reported four proteins specifically expressed in the competent parent and the competent F2 progeny (77). Two of these proteins, a luciferase homologue and cyclophilin, bound to CYDV-RPV in vitro (77). Cyclophilin was identified as two isoforms, one of which was linked to the virus transmission phenotype (C. Tamborindeguy, unpublished data). In this study, CoA ligase, a luciferase homologue (spot 181), was specifically expressed in two isoforms, with one form deriving from the competent parent and one form deriving from the refractive parent. Only the competent genotypes, the RT-2 genotype, which permits virus movement across the gut barrier, and the K3 genotype, with an unknown barrier, expressed the same isoform as the competent parent. Seven other aphid proteins and four Buchnera proteins were similarly observed as isoforms on the gels, and the isoform expressed by the transmission-competent parent was the same isoform expressed by the transmission-competent F2 genotypes.
Intriguingly, both of the previously published studies investigating proteomic differences in F1 (54) and F2 (77) aphid populations and this study report that the differences in expression between transmission-competent and refractive aphids are attributed not to gross changes in the expression of numerous proteins but rather to a shift in pI of only a small number of proteins. From a reductionist, molecular biological perspective, it is tempting to ascribe the virus transmission phenotype simply to the presence or absence of one of these proteins. However, the story becomes much richer when considered from a systems biology perspective, including the additive and possibly synergistic action of the genes involved. We propose that the isoforms observed here are just a few of the isoforms involved due to the inherent nature of 2-D DIGE enabling the detection of only the most abundant proteins. Two competent genotypes contained eight of eight transmission competence-specific isoforms, and one competent genotype contained seven of eight competent isoforms (Table 6). The refractive genotypes contained only two to five of these isoforms, which does not appear to be sufficient to confer the vectoring phenotype. Collectively, these data strongly suggest that the control of luteovirus transmission in aphids is via the expression of protein isoforms, which may be allelic variants, acting by additive (8, 9) and potentially nonadditive (synergistic) mechanisms. An alternative hypothesis is that these alleles are neutral with respect to the virus transmission phenotype and that their association with the virus transmission phenotype is because they are linked to another gene (47) that has a functional role in virus transmission. Whether or not the genes coding for these proteins are epistatic or genetically linked to each other or to other genes involved in circulative transmission in S. graminum remains to be determined.
The third major insight gained from this study was the surprising observation that the pIs of the bacterial Buchnera proteins did not align with the maternal genotype for the competent F2 genotypes, which may reveal a novel role for Buchnera in virus transmission by aphids. Buchnera species are obligate intercellular endosymbionts with a reduced genome that deliver essential amino acids to the aphid that are otherwise lacking in the diet (21). They are inherited transovarially through the maternal lineage during embryogenesis (21). Analysis of the parental genotypes revealed two different Buchnera proteomes distinguished by the pIs of a number of highly abundant Buchnera proteins. Although we expected all Buchnera proteins in the F2 genotypes to be identical to those of the maternal, refractive genotype (as no splice variants would be expected from a bacterial proteome), we observed the pIs of the Buchnera proteins in the competent F2 genotypes to be identical to those in the competent male parent. Given that male aphids cannot transmit the endosymbiont to their offspring (21), these data indicate that genetic heterogeneity of Buchnera exists in the maternal genotype. Upon closer inspection and in furtherance of the genetic heterogeneity hypothesis, the maternal parent genotype (Sg-SC) did possess faint spots on the 2-D gels (Fig. 5C and D) which corresponded to the pIs observed for the competent genotypes. We subsequently confirmed one of these to contain the Buchnera protein EF-Tu by using mass spectrometry (data not shown). However, there was only one predominant isoform in the competent parent and the F2 offspring, suggesting a strong bottlenecking effect during sexual reproduction for a single genotype of Buchnera in the maternal lineage. In other words, the females used in the initial cross had a genetically diverse Buchnera population that was predominated by one strain, but the dominant strain was not inherited by every F2 individual. We can eliminate the possibility that transmission-competent Sg-F genotype females contaminated the crosses, such as an Sg-F female being crossed to an Sg-F male, because protein isoforms observed only in Sg-SC (for example, the Sg-SC isoform of CoA ligase or RepA) were expressed codominantly in transmission-competent F2 genotypes. Bacterial endosymbiont genetic heterogeneity in a single host was recently discovered in host vesicomyid clams by DNA sequencing and is proposed to facilitate gene transfer between divergent endosymbiont lineages (65). To our knowledge, it has not been reported for aphids.
We do not propose that any of the Buchnera proteins we identified as differentially expressed are involved directly in virus movement within the aphid, since these distinct proteome phenotypes are likely the result of the high rate of nonsynonymous mutations observed for Buchnera (75). Many of the proteins identified are likely integral membrane proteins in the mycetocyte, for example OMPs A and F, and would not be expected to come into direct contact with virions during the circulative route in the aphid unless the proteins are released into the aphid hemocoel as the mycetocytes degrade with age (22). We propose two alternative hypotheses to explain the strong link between one of the Buchnera populations and the vectoring phenotype. The first hypothesis is that the distinct pIs identified in transmission-competent genotypes serve as biomarkers for a specific genotype of Buchnera that the aphid must possess to efficiently transmit luteoviruses from plant to plant. The idea is that the Buchnera proteins facilitate the ability of the aphid to support virus transmission. In line with this hypothesis would be a new model for the aphid transmission of luteoviruses that requires the aphids to have a transmission-competent genotype of Buchnera in addition to the full suite of aphid proteins required for virus transmission. The proteins from Buchnera are good candidates for those derived from transgressive segregation of genes facilitating virus transmission observed in the transmission-competent F2 genotypes. Recent work demonstrated a similar supportive role of endosymbiotic bacteria in whiteflies. Circulative plant virus transmission efficiency is correlated with whiteflies harboring only a particular species of bacterial endosymbiont (30). In aphids, the presence of the Buchnera isoforms was insufficient to confer the vectoring phenotype in the absence of the aphid protein isoforms linked to vector competency (for example, in the refractive genotypes CC1 and MM1), suggesting that synergistic interactions between the aphid and bacterial endosymbiont proteomes may be required for circulative virus transmission by aphids. A second hypothesis is that the genes that control virus transmission in aphids overlap with those that regulate the partitioning of these two strains of Buchnera during reproduction. In line with the latter hypothesis, the Buchnera proteins become mere statistical biomarkers for vector competence without causally promoting that competence. For example, if the genes that regulate the membrane trafficking events for the movement of the virus in competent aphids also influence the interactions between Buchnera and the aphid, this may explain the linkage between one genotype of Buchnera and the virus transmission phenotype. In support of this hypothesis, we identified a vasa-like protein (spot 76) that was upregulated in competent genotypes and the refractive genotype MM1, and these all contain the same Buchnera type. In A. pisum, the expression of vasa mRNA is upregulated at the stage of embryogenesis when the bacterial endosymbionts are partitioned into the germ line (11).
Until now, the proposed role of Buchnera in virus transmission has been limited to the involvement of the chaperone protein GroEL (21, 31) in virion stability. Our data neither support nor refute GroEL involvement, and we did not observe GroEL to be expressed differentially between competent and refractive aphids. A major evolutionary implication of our findings is that aphid hosts and maternally inherited Buchnera may be in evolutionary conflict, which would further predict that genetically distinct strains of Buchnera will have other profound, observable phenotypic effects on S. graminum physiology beyond transmission of luteoviruses as they compete with each other for transmission into progeny. In other words, rival Buchnera strains competing for transmission into offspring might engage in a tug-of-war (58) that is costly to the aphid host. Indeed, endosymbiotic bacteria in other insects are well known to manipulate host physiology and behavior. In the ant Camponotus fellah, the primary endosymbiotic bacterium Blochmannia modulates the ant immune response and colony growth rate (19). In the most extreme example of endosymbiotic manipulation of insect biology, the maternally transmitted bacterium Wolbachia manipulates the sex ratio of the butterfly Acraea encedon for its own selfish interests by killing males (40). The exact role of Buchnera in virus transmission and other aspects of aphid physiology can be tested directly in our S. graminum genetic system because molecular and proteomic characterization of different Buchnera variants can be compared for aphid genotypes that are otherwise genetically similar.
The discovery power of our approach to identify proteins involved in virus transmission is derived from the analysis of multiple related F2 genotypes by DIGE. One potential limitation of the study is that we used DDA mass spectrometry, which undoubtedly resulted in undersampling of the peptides extracted from each gel spot (76). However, the presence or absence of the proteins and protein isoforms was correlated across multiple genotypes, greatly increasing the likelihood that the proteins involved in virus transmission were detected using DDA. The proteins correlated with transmission in competent genotypes and with barriers in refractive genotypes were heritable and stable in multiple F2 genotypes, confirming our earlier observations that this particular aphid phenotype is to a large extent under genetic control. Multiple proteins were identified as being expressed differentially between competent and refractive genotypes, also supporting our earlier observations that the control of luteovirus transmission is regulated by multiple different genes that are additive in effect. In refractive genotypes with a single barrier to transmission, we identified unique sets of proteins predicted to act at the gut and ASG across multiple genotypes and also found proteins that are likely required at both cellular barriers for virus translocation. Future work should focus on using targeted proteomic methods such as selective reaction monitoring (56, 70, 76) for absolute quantification of these proteins in competent and refractory S. graminum aphid genotypes and in other aphid and insect species that transmit plant viruses via the circulative pathway.
Furthermore, for the first time, we established a correlation with virus transmission competency for a distinct population of Buchnera classified on the basis of unique pI isoforms of several highly abundant Buchnera proteins. These different isoforms may derive from DNA polymorphisms in Buchnera genes or possibly from protein phosphorylation (20, 39) or other modifications by coming into contact with aphid proteins. One Buchnera strain is found only in the transmission-competent parent Sg-F, whereas this and a second Buchnera strain coexist in the refractive parent Sg-SC. Virus transmission-competent F2 genotypes always harbored the same Buchnera population, and this was the population observed in the transmission-competent parent (Sg-F). The discovery of a distinct Buchnera population that segregates in the F2 population on the basis of its proteomic phenotype and that is tightly linked with virus transmission competency indicates that tritrophic interactions between the aphid, its bacterial endosymbiont, and plant viruses are required for efficient virus transmission along the circulative pathway.
Supplementary Material
Acknowledgments
This work was supported by USDA-NRI grant 2007-04567, NSF grant DBI-0606596, and USDA-ARS CRIS projects 1907-101-16 and 1907-21000-024/25-00D.
We thank Angela Douglas and Kern Reeve for insightful discussions on the interpretation of the Buchnera proteomic data. We thank James VanEe and John Flaherty in the Cornell University Life Sciences Core Laboratories Center IT group for database and software support. We thank Sheng Zhang and Sabine Baumgart for their expert assistance with the 4000 QTrap instrument and Sheng Zhang and Celeste Ptak for their expert assistance with the Orbitrap Velos instrument. We especially thank Dawn Smith, Tom Hammond, and Mike McLaughlin for assistance in rearing aphids, maintaining the greenhouses, and data collation, respectively. We thank Kent Loeffler for assistance with the figures and cover image.
Footnotes
Published ahead of print on 15 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argentine, J. A., and A. A. James. 1995. Characterization of a salivary gland-specific esterase in the vector mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 25:621-630. [DOI] [PubMed] [Google Scholar]
- 3.Aumailley, M., et al. 1993. Nidogen mediates the formation of ternary complexes of basement membrane components. Kidney Int. 43:7-12. [DOI] [PubMed] [Google Scholar]
- 4.Aumailley, M., H. Wiedemann, K. Mann, and R. Timpl. 1989. Binding of nidogen and the laminin-nidogen complex to basement membrane collagen type IV. Eur. J. Biochem. 184:241-248. [DOI] [PubMed] [Google Scholar]
- 5.Baines, A. C., and B. Zhang. 2007. Receptor-mediated protein transport in the early secretory pathway. Trends Biochem. Sci. 32:381-388. [DOI] [PubMed] [Google Scholar]
- 6.Bencharki, B., et al. 2010. Phloem protein partners of cucurbit aphid borne yellows virus: possible involvement of phloem proteins in virus transmission by aphids. Mol. Plant Microbe Interact. 23:799-810. [DOI] [PubMed] [Google Scholar]
- 7.Brault, V., et al. 2010. Transcriptomic analysis of intestinal genes following acquisition of pea enation mosaic virus by the pea aphid Acyrthosiphon pisum. J. Gen. Virol. 91:802-808. [DOI] [PubMed] [Google Scholar]
- 8.Burrows, M. E., et al. 2006. Genetic regulation of polerovirus and luteovirus transmission in the aphid Schizaphis graminum. Phytopathology 96:828-837. [DOI] [PubMed] [Google Scholar]
- 9.Burrows, M. E., M. C. Caillaud, D. M. Smith, and S. M. Gray. 2007. Biometrical genetic analysis of luteovirus transmission in the aphid Schizaphis graminum. Heredity 98:106-113. [DOI] [PubMed] [Google Scholar]
- 10.Carolan, J. C., C. I. Fitzroy, P. D. Ashton, A. E. Douglas, and T. L. Wilkinson. 2009. The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9:2457-2467. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C. C., et al. 2007. Apvasa marks germ-cell migration in the parthenogenetic pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). Dev. Genes Evol. 217:275-287. [DOI] [PubMed] [Google Scholar]
- 12.Ciccarelli, F. D., T. Doerks, and P. Bork. 2002. AMOP, a protein module alternatively spliced in cancer cells. Trends Biochem. Sci. 27:113-115. [DOI] [PubMed] [Google Scholar]
- 13.Cilia, M., et al. 2009. A comparison of protein extraction methods suitable for gel-based proteomic studies of aphid proteins. J. Biomol. Techn. 20:201-215. [PMC free article] [PubMed] [Google Scholar]
- 14.Cilia, M., et al. 2011. Tangible benefits of the aphid Acyrthosiphon pisum genome sequencing for aphid proteomics: enhancements in protein identification and data validation for homology-based proteomics. J. Insect Physiol. 57:179-190. [DOI] [PubMed] [Google Scholar]
- 15.Cohen, J. 1988. Statistical power analysis for the behavioral sciences, 2nd ed. L. Erlbaum Associates, Hillsdale, NJ.
- 16.Cornman, R. S., et al. 2008. Annotation and analysis of a large cuticular protein family with the R&R consensus in Anopheles gambiae. BMC Genomics 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornman, R. S., and J. H. Willis. 2009. Annotation and analysis of low-complexity protein families of Anopheles gambiae that are associated with cuticle. Insect Mol. Biol. 18:607-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Arcy, C. J., and P. A. Burnett (ed.). 1995. Barley yellow dwarf, 40 years of progress. APS Press, St. Paul, MN.
- 19.de Souza, D. J., A. Bezier, D. Depoix, J. M. Drezen, and A. Lenoir. 2009. Blochmannia endosymbionts improve colony growth and immune defence in the ant Camponotus fellah. BMC Microbiol. 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutscher, J., and M. H. Saier, Jr. 2005. Ser/Thr/Tyr protein phosphorylation in bacteria—for long time neglected, now well established. J. Mol. Microbiol. Biotechnol. 9:125-131. [DOI] [PubMed] [Google Scholar]
- 21.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 22.Douglas, A. E., and A. F. G. Dixon. 1987. The mycetocyte symbiosis of aphids: variation with age and morph in virginoparae of Megoura viciae and Acyrthosiphon pisum. J. Insect Physiol. 33:109-113. [Google Scholar]
- 23.Filichkin, S. A., S. Brumfield, T. P. Filichkin, and M. J. Young. 1997. In vitro interactions of the aphid endosymbiotic SymL chaperonin with barley yellow dwarf virus. J. Virol. 71:569-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox, J. W., et al. 1991. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO J. 10:3137-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francis, F., et al. 2006. Proteomics in Myzus persicae: effect of aphid host plant switch. Insect Biochem. Mol. Biol. 36:219-227. [DOI] [PubMed] [Google Scholar]
- 26.Francis, F., et al. 2010. Tritrophic interactions among Macrosiphum euphorbiae aphids, their host plants and endosymbionts: investigation by a proteomic approach. J. Insect Physiol. 56:575-585. [DOI] [PubMed] [Google Scholar]
- 27.Garge, N., et al. 2010. Identification of quantitative trait loci underlying proteome variation in human lymphoblastoid cells. Mol. Cell. Proteomics 9:1383-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garret, A., C. Kerlan, and D. Thomas. 1996. Ultrastructural study of acquisition and retention of potato leafroll luteovirus in the alimentary canal of its aphid vector, Myzus persicae Sulz. Arch. Virol. 141:1279-1292. [DOI] [PubMed] [Google Scholar]
- 29.Gildow, F. E., and W. F. Rochow. 1980. Role of accessory salivary glands in aphid transmission of barley yellow dwarf virus. Virology 104:97-108. [DOI] [PubMed] [Google Scholar]
- 30.Gottlieb, Y., et al. 2010. The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 84:9310-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray, S., and F. E. Gildow. 2003. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41:539-566. [DOI] [PubMed] [Google Scholar]
- 32.Gray, S. M. 2008. Aphid transmission of plant viruses. Curr. Protoc. Microbiol. 2008:16B.1.1-16B.1.10. [DOI] [PubMed] [Google Scholar]
- 33.Gray, S. M., and N. Banerjee. 1999. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray, S. M., M. C. Caillaud, M. Burrows, and D. M. Smith. 2007. Transmission of two viruses that cause barley yellow dwarf is controlled by different loci in the aphid, Schizaphis graminum. J. Insect Sci. 7:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray, S. M., D. M. Smith, L. Barbierri, and J. Burd. 2002. Virus transmission phenotype is correlated with host adaptation among genetically diverse populations of the aphid Schizaphis graminum. Phytopathology 92:970-975. [DOI] [PubMed] [Google Scholar]
- 36.Hesler, L. S., W. E. Riedell, M. A. Langham, and S. L. Osborne. 2005. Insect infestations, incidence of viral plant diseases, and yield of winter wheat in relation to planting date in the northern Great Plains. J. Econ. Entomol. 98:2020-2027. [DOI] [PubMed] [Google Scholar]
- 37.Hogenhout, S. A., D. Ammar el, A. E. Whitfield, and M. G. Redinbaugh. 2008. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46:327-359. [DOI] [PubMed] [Google Scholar]
- 38.IAGC. 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jers, C., B. Soufi, C. Grangeasse, J. Deutscher, and I. Mijakovic. 2008. Phosphoproteomics in bacteria: towards a systemic understanding of bacterial phosphorylation networks. Expert Rev. Proteomics 5:619-627. [DOI] [PubMed] [Google Scholar]
- 40.Jiggins, F. M., J. P. Randerson, G. D. Hurst, and M. E. Majerus. 2002. How can sex ratio distorters reach extreme prevalences? Male-killing Wolbachia are not suppressed and have near-perfect vertical transmission efficiency in Acraea encedon. Evolution 56:2290-2295. [DOI] [PubMed] [Google Scholar]
- 41.Jolliffe, I. T. 2002. Principal components analysis, 2nd ed. Springer-Verlag, New York, NY.
- 42.Karouzou, M. V., et al. 2007. Drosophila cuticular proteins with the R&R consensus: annotation and classification with a new tool for discriminating RR-1 and RR-2 sequences. Insect Biochem. Mol. Biol. 37:754-760. [DOI] [PubMed] [Google Scholar]
- 43.Karp, N. A., and K. S. Lilley. 2007. Design and analysis issues in quantitative proteomics studies. Proteomics 7(Suppl. 1):42-50. [DOI] [PubMed] [Google Scholar]
- 44.Karp, N. A., P. S. McCormick, M. R. Russell, and K. S. Lilley. 2007. Experimental and statistical considerations to avoid false conclusions in proteomics studies using differential in-gel electrophoresis. Mol. Cell. Proteomics 6:1354-1364. [DOI] [PubMed] [Google Scholar]
- 45.Klose, J., et al. 2002. Genetic analysis of the mouse brain proteome. Nat. Genet. 30:385-393. [DOI] [PubMed] [Google Scholar]
- 46.Marchler-Bauer, A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClellan, J., and M. C. King. 2010. Genetic heterogeneity in human disease. Cell 141:210-217. [DOI] [PubMed] [Google Scholar]
- 48.Miller, W. A., and L. Rasochova. 1997. Barley yellow dwarf viruses. Annu. Rev. Phytopathol. 35:167-190. [DOI] [PubMed] [Google Scholar]
- 49.Morin, S., et al. 1999. A GroEL homologue from endosymbiotic bacteria of the whitefly Bemisia tabaci is implicated in the circulative transmission of tomato yellow leaf curl virus. Virology 256:75-84. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen, T. T., S. Boudreault, D. Michaud, and C. Cloutier. 2008. Proteomes of the aphid Macrosiphum euphorbiae in its resistance and susceptibility responses to differently compatible parasitoids. Insect Biochem. Mol. Biol. 38:730-739. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen, T. T., D. Michaud, and C. Cloutier. 2009. A proteomic analysis of the aphid Macrosiphum euphorbiae under heat and radiation stress. Insect Biochem. Mol. Biol. 39:20-30. [DOI] [PubMed] [Google Scholar]
- 52.Otte, S., S. Barnikol-Watanabe, G. Vorbruggen, and N. Hilschmann. 1999. NUCB1, the Drosophila melanogaster homolog of the mammalian EF-hand proteins NEFA and nucleobindin. Mech. Dev. 86:155-158. [DOI] [PubMed] [Google Scholar]
- 53.Pappin, D. J., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 54.Papura, D., et al. 2002. Two-dimensional electrophoresis of proteins discriminates aphid clones of Sitobion avenae differing in BYDV-PAV transmission. Arch. Virol. 147:1881-1898. [DOI] [PubMed] [Google Scholar]
- 55.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 56.Picotti, P., et al. 2010. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7:43-46. [DOI] [PubMed] [Google Scholar]
- 57.Rebers, J. E., and J. H. Willis. 2001. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem. Mol. Biol. 31:1083-1093. [DOI] [PubMed] [Google Scholar]
- 58.Reeve, H. K., S. T. Emlen, and L. Keller. 1998. Reproductive sharing in animal societies: reproductive incentives or incomplete control by dominant breeders. Behav. Ecol. 9:267-278. [Google Scholar]
- 59.Reinbold, C., E. Herrbach, and V. Brault. 2003. Posterior midgut and hindgut are both sites of acquisition of cucurbit aphid-borne yellows virus in Myzus persicae and Aphis gossypii. J. Gen. Virol. 84:3473-3484. [DOI] [PubMed] [Google Scholar]
- 60.Roberts, B., C. Clucas, and I. L. Johnstone. 2003. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol. Biol. Cell 14:4414-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossi, V., G. Beffagna, A. Rampazzo, B. Bauce, and G. A. Danieli. 2004. TAIL1: an isthmin-like gene, containing type 1 thrombospondin-repeat and AMOP domain, mapped to ARVD1 critical region. Gene 335:101-108. [DOI] [PubMed] [Google Scholar]
- 62.Seddas, P., et al. 2004. Rack-1, GAPDH3, and actin: proteins of Myzus persicae potentially involved in the transcytosis of beet western yellows virus particles in the aphid. Virology 325:399-412. [DOI] [PubMed] [Google Scholar]
- 63.Silva, E., et al. 2010. In the eye of the beholder: does the master see the SameSpots as the novice? J. Proteome Res. 9:1522-1532. [DOI] [PubMed] [Google Scholar]
- 64.Simon, J. P., et al. 1998. An essential role for the phosphatidylinositol transfer protein in the scission of coatomer-coated vesicles from the trans-Golgi network. Proc. Natl. Acad. Sci. U. S. A. 95:11181-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stewart, F. J., and C. M. Cavanaugh. 2009. Pyrosequencing analysis of endosymbiont population structure: co-occurrence of divergent symbiont lineages in a single vesicomyid host clam. Environ. Microbiol. 11:2136-2147. [DOI] [PubMed] [Google Scholar]
- 66.Tamas, I., et al. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 67.Tang, L., J. Liang, Z. Zhan, Z. Xiang, and N. He. 2010. Identification of the chitin-binding proteins from the larval proteins of silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 40:228-234. [DOI] [PubMed] [Google Scholar]
- 68.Timms, J. F., and R. Cramer. 2008. Difference gel electrophoresis. Proteomics 8:4886-4897. [DOI] [PubMed] [Google Scholar]
- 69.Timpl, R., et al. 1984. Laminin, proteoglycan, nidogen and collagen IV: structural models and molecular interactions. Ciba Found. Symp. 108:25-43. [DOI] [PubMed] [Google Scholar]
- 70.Tomazela, D. M., et al. 2010. Measurement of human surfactant protein-B turnover in vivo from tracheal aspirates using targeted proteomics. Anal. Chem. 82:2561-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Touzet, P., et al. 1995. Characterizing allelic proteins for genome mapping in maize. Electrophoresis 16:1289-1294. [DOI] [PubMed] [Google Scholar]
- 72.Uzest, M., et al. 2010. The “acrostyle”: a newly described anatomical structure in aphid stylets. Arthropod Struct. Dev. 39:221-229. [DOI] [PubMed] [Google Scholar]
- 73.van den Heuvel, J. F., M. Verbeek, and F. van der Wilk. 1994. Endosymbiotic bacteria associated with circulative transmission of potato leafroll virus by Myzus persicae. J. Gen. Virol. 75:2559-2565. [DOI] [PubMed] [Google Scholar]
- 74.Wang, Y., et al. 2010. Integrated metabonomic-proteomic analysis of an insect-bacterial symbiotic system. J. Proteome Res. 9:1257-1267. [DOI] [PubMed] [Google Scholar]
- 75.Wernegreen, J. J., and N. A. Moran. 1999. Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol. Biol. Evol. 16:83-97. [DOI] [PubMed] [Google Scholar]
- 76.Wolf-Yadlin, A., S. Hautaniemi, D. A. Lauffenburger, and F. M. White. 2007. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U. S. A. 104:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, X., et al. 2008. Coupling genetics and proteomics to identify aphid proteins associated with vector-specific transmission of polerovirus (Luteoviridae). J. Virol. 82:291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang, Y., T. W. Thannhauser, L. Li, and S. Zhang. 2007. Development of an integrated approach for evaluation of 2-D gel image analysis: impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28:2080-2094. [DOI] [PubMed] [Google Scholar]
- 79.Zwiener, C. M., S. P. Conley, W. C. Bailey, and L. E. Sweets. 2005. Influence of aphid species and barley yellow dwarf virus on soft red winter wheat yield. J. Econ. Entomol. 98:2013-2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.