Abstract
Alternative splicing of the precursor mRNA (pre-mRNA) of human parvovirus B19 (B19V) plays a key role in posttranscriptional regulation of B19V gene expression. We report that the central exon of the B19V pre-mRNA is defined by three GAA motif-containing exonic splicing enhancers and a G/GU-rich intronic splicing enhancer that lies adjacent to the second donor site. Moreover, targeting of morpholino antisense oligonucleotides to the two splicing enhancers surrounding the second donor site led to a significant reduction in splicing at this donor site during B19V infection of permissive CD36+ erythroid progenitor cells.
Human parvovirus B19 (B19V) is the only human parvovirus that has been confirmed to cause diseases in humans (23). All of the B19V mRNA transcripts are generated from the processing of a single precursor mRNA (pre-mRNA), which is transcribed from a single promoter at map unit 6 (P6), through alternative splicing and polyadenylation (12). All the spliced B19V mRNA transcripts contain the central exon (exon 2), which spans the A1-1/A1-2 to D2 splice site (Fig. 1). Therefore, splicing at the D2 donor site is a central step in control of B19V pre-mRNA processing. While analyzing the sequence of exon 2 with the program ESEfinder, version 3.0 (1, 19) (Fig. 2 A), we found a number of SR protein-binding GAA motifs in exon 2 and a G/GU-rich region that lies directly 3′ of the D2 donor site, which suggested that exonic splicing enhancers/intronic splicing enhancers (ESEs/ISEs) facilitate the definition of exon 2. SR proteins are serine/arginine-rich proteins that bind to ESEs/ISEs and function to promote exon inclusion during pre-mRNA processing (5, 8). Identification of the cis-acting sequences that facilitate recognition of the D2 donor site will eventually reveal mechanisms that ensure appropriate expression levels of the capsid proteins VP1 and VP2, as well as the small nonstructural 11-kDa protein (11kDa), during B19V infection.
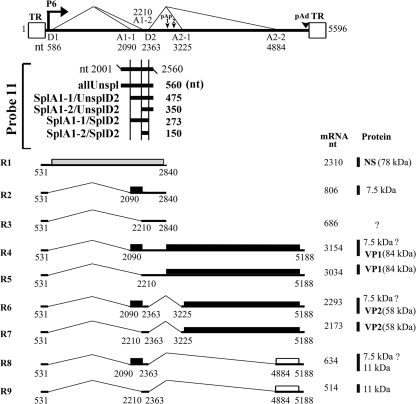
FIG. 1.
B19V transcripts and the probe used for RNase protection assays. Top, the B19V genome is depicted, with the locations of the terminal repeats (TR), the P6 promoter, the first intron donor (D1) and acceptor (A1-1 and A1-2) sites, the second intron donor (D2) and acceptor (A2-1 and A2-2) sites, the internal polyadenylation site (pAp), and the distal polyadenylation site (pAd) indicated. For each splice site, the nucleotide position is indicated. Middle, the antisense probe used in this study, probe 11, is shown with starting and ending nucleotides indicated (nt 2001 to 2560). Bands protected by this probe are diagramed, with their respective designations shown to the left and their length (in nucleotides) indicated to the right. More specifically, “allUnspl” represents all B19V mRNAs that are not spliced at any of the splice sites that lie between nt 2001 and 2560; “SplA1-1/UnsplD2” and “SplA1-2/UnsplD2” represent B19V mRNAs that are spliced at the A1-1 and A1-2 sites, respectively, but not at the D2 site; “SplA1-1/SplD2” represents B19V mRNAs that are spliced from the A1-1 to D2 sites, and “SplA1-2/SplD2” represents B19V mRNAs that are spliced from the A1-2 to the D2 sites. Bottom, the major transcripts (R1 to R9) and the proteins they encode are shown, along with their lengths and molecular masses (in kilodaltons), respectively. Question marks indicate that proteins corresponding to these transcripts have not been identified or confirmed. All of the nucleotide numbers for the B19V genome that are used in this study refer to the B19V J35 isolate DNA with GenBank accession no. AY386330 (24).
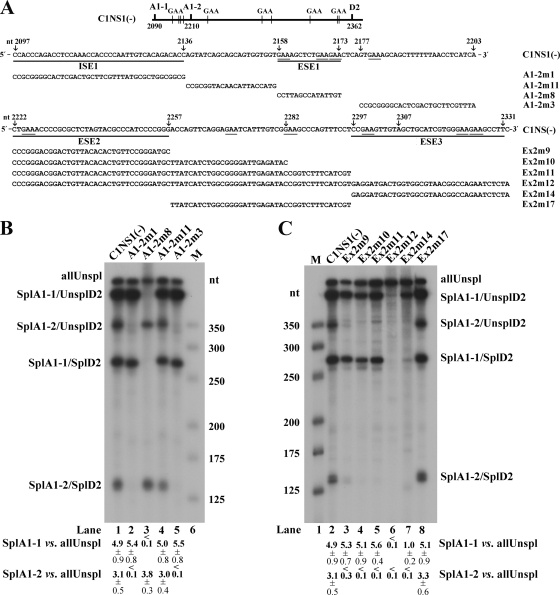
FIG. 2.
Exonic splicing enhancers (ESEs) in the central exon of the B19V genome. (A) Top, the B19V central exon (exon 2) is depicted, with the locations of splice sites and GAA motifs indicated. Middle, exon 2 sequence between the A1-1 and A1-2 sites (nt 2097 to 2203), with mutations in this region indicated (arrow plus nucleotide number) above the sequence, the identified ISE1 and ESE1 underlined, and mutant constructs tested in RNase protection assays identified and also shown below, in panel B. Bottom, exon 2 sequence between the A1-2 and D2 sites (nt 2222 to 2331), with mutations in this region indicated (arrow plus nucleotide number) above the sequence, identified ESE2 and ESE3 underlined, and mutant constructs tested in RNase protection assays identified and also shown below, in panel C. (B and C) RNase protection assays assessing the enhancer activities of the presence of ESE1 and ISE1 (B) and ESE2 and ESE3 (C). For these RNase protection assays, the C1NS1(−) plasmid and its mutation-containing derivatives (shown in panel A) were transfected into COS-7 cells. At 48 h posttransfection, total RNA was isolated and protected using probe 11. Transfections, RNA isolation, and RNase protection assays were performed as described previously (6, 11, 18). The protected bands are identified and labeled as the products described in the legend to Fig. 1. The size maker was made as previously described (16). A representative protection assay from at least three independent experiments is shown. Ratios of SplA1-1 (SplA1-1/UnsplD2 + SplA1-1/SplD2) to allUnspl and of SplA1-2 (SplA1-2/UnsplD2 + SplA1-2/SplD2) to allUnspl are shown, respectively, for each protection assay; some are given as averages and standard deviations.
A GAA-rich sequence in the region between the A1-1 and A1-2 sites is essential for splicing at the A1-1 3′ splice site.
We first generated a mutant form of the B19V DNA that disrupts the GAA-rich sequence (Fig. 2A, A1-2m8). RNAs generated by transfecting this mutant into COS-7 cells were analyzed by an RNase protection assay using probe 11 (Fig. 1). Transfection of cells with this mutant form resulted in failed production of B19V mRNAs spliced at the A1-1 site but did not change the level of the B19V mRNAs spliced at the A1-2 site (Fig. 2B, lane 3). This result supports the idea that the GAA-rich sequence (5′GAAAGCTCTGAAGAA3′; underlining indicates GAA motif) functions as an ESE (ESE1) and that it facilitates splicing at the proximal splice site (the upstream A1-1 acceptor site). This and all other mutant DNAs transfected into COS-7 cells in this study were generated from the parent plasmid, the C1NS1(−) plasmid (22), which contains a simian virus 40 (SV40) replication origin and therefore replicates in COS-7 cells, leading to a B19V transcription profile resembling that in permissive cells during B19V infection (6, 20, 22).
To confirm that ESE1 plays a unique role in facilitating splicing at the A1-1 site, we made three additional mutants affecting the region between the A1-1 and A1-2 sites, the A1-2m1, A1-2m11, and A1-2m3 mutants. In the A1-2m1 and A1-2m3 mutants, which affected sequences just following the A1-1 site and the polypyrimidine tract of the A1-2 site, respectively, splicing at the A1-2 site was abolished, whereas splicing at the A1-1 site was unaffected (Fig. 2B, lanes 2 and 5). However, in the A1-2m11 mutant that affected sequences spanning nucleotides (nt) 2137 to 2157, just 5′ of ESE1, splicing at either the A1-1 or the A1-2 site remained unchanged compared to that in the C1NS1(−) parent (Fig. 2B, lane 4). Thus, the region between the A1-1 site and ESE1 (nt 2097 to 2136) is critical for splicing of the B19V pre-mRNA at the A1-2 site and thus behaves like an ISE (ISE1).
A GAA-rich sequence in exon 2 is required to define this exon.
We next carried out random mutagenesis of the appreciably longer exon 2 sequence between the A1-2 and D2 sites. Probe 11-based protection of B19V mRNAs generated from transfection of the Ex2m12 mutant revealed that only an unspliced RNA was generated (Fig. 2C, lane 6), supporting our hypothesis that cis-acting elements are present in this region of exon 2. Further mutations from the 5′ end basically knocked out or significantly reduced splicing from the A1-2 site but did not significantly affect splicing from the A1-1 site (Fig. 2C, lanes 3, 4, and 5). In contrast, mutations lacking the 3′ end of the exon abolished all splicing from both the A1-1 and A1-2 sites to the D2 site, as well as most of the splicing of any kind involving the A1-2 site (Fig. 2C, lane 7, SplA1-2/UnsplD2), but not general splicing involving the A1-1 site (Fig. 2C, lane 7, SplA1-1/UnsplD2). Taken together, these results suggest that the 5′ end of exon 2, specifically the nt 2222 to 2257 fragment, serves as an ESE (ESE2) that facilitates excision of the B19V pre-mRNA at the A1-2 site and that the 3′ end of exon 2, specifically the fragment encompassing nt 2297 to 2331, serves as an ESE (ESE3) that facilitates recognition of the D2 donor site. As control, the mutant Ex2m17, which affected sequence spanning nt 2258 to 2296 in the middle of exon 2, produced all B19V mRNAs at levels similar to those from the C1NS1(−) parent (Fig. 2C, lane 8).
Identification of a G/GU-rich intronic splicing enhancer adjacent to the D2 donor site.
The D2 donor site is a nonconsensus U1 snRNP-binding site (25). We hypothesized that the G/GU-rich region, which lies directly 3′ of the D2 site (Fig. 3 A), acts as an ISE to facilitate splicing of the B19V pre-mRNA.
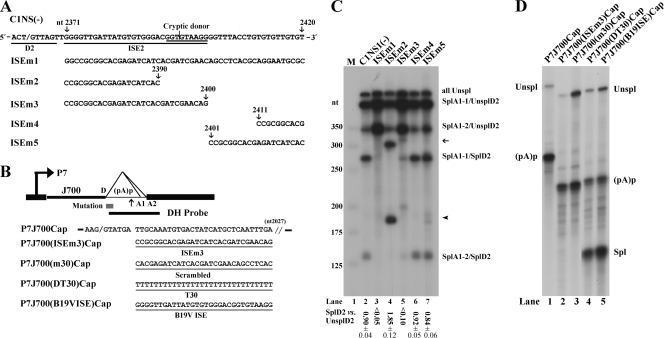
FIG. 3.
Intronic splicing enhancer (ISE) immediately 3′ of the D2 donor site. (A) The D2 site and its neighboring G/GU-rich sequence in the B19V genome are depicted, with the locations of five mutations in this region indicated (arrows plus nucleotide number). The D2 site, identified cryptic donor site, and identified ISE2 sequence are underlined, and the mutant constructs used for RNase protection assays in panel B are shown below. (B) The test construct P7J700Cap (previously described in reference 15) is diagramed with the location of the DH probe indicated (16). The AAV5 donor (D) site and the sequence to nt 2027 of the AAV5 genome (3) are shown. P7J700Cap and its derivatives, P7J700(ISEm3)Cap, P7J700(m30)Cap, P7J700(DT30)Cap, and P7J700(B19VISE)Cap, were constructed by mutating the sequence between nt 2097 and 2027 as shown. (C) RNase protection assays showing the functionality of the ISE2 and the cryptic donor. RNase protection assays of RNAs generated in COS-7 cells following transfection of the C1NS1(−) parent and of its mutant forms are shown with identities of the bands indicated to the right. A representative protection assay from at least three independent experiments is shown. The ratio of SplD2 to UnsplD2 is shown for each protection assay; some are given as averages and standard deviations. SplD2 includes signals from bands of SplA1-1/SplD2 and SplA1-2/SplD2, and UnsplD2 includes signals from bands of SplA1-1/UnsplD2, SplA1-2/UnsplD2, and allUnspl. The arrow and arrowhead show RNAs spliced from the A1-1 and A1-2 sites to the cryptic donor, respectively. (D) RNase protection assays showing that the B19V ISE2 promotes AAV5 RNA when lengths of the 5′ exon increase. For these experiments, total RNA isolated from plasmid-transfected COS-7 cells was subjected to RNase protection assays using probe DH (shown in panel B), as previously described (15). Unspl, RNA unspliced at the AAV5 donor site; Spl, RNA spliced at the AAV5 donor site; and (pA)p, RNA polyadenylated at the AAV5 (pA)p site.
We first mutated the entire G/GU-rich sequence. RNase protection of B19V mRNAs generated from transfection of mutant ISEm1 showed that splicing at the D2 site was completely abolished (Fig. 3C, lane 3). Further mutations at the 3′ end of the G/GU-rich sequence (ISEm4 and ISEm5) did not drastically affect splicing at the D2 site (Fig. 3C, lanes 6 and 7). However, the ISEm3 mutant, in which sequences at the 5′ end of the G/GU-rich sequence are changed, abolished splicing at the D2 site drastically (Fig. 3C, lane 5), much as did ISEm1, which spans the entire region (Fig. 3C, lane 3). These results strongly suggest that the G/GU-rich sequence (nt 2371 to 2400) adjacent to the D2 site is a strong ISE (ISE2). Notably, mutation of the G/GU-rich region encompassed by nt 2371 to 2390 blocked splicing at the D2 site, yet activated a high level of splicing at a cryptic donor site located at nt 2395 (Fig. 3C, lane 4). This result suggests that the ISE2 also plays a critical role in controlling splicing of the B19V pre-mRNA at a precise position.
We next examined the function of the B19V ISE2 in a heterologous system, specifically, in the context of the adeno-associated virus type 5 (AAV5) intron. Construct P7J700Cap was used as the parent plasmid (15, 17). Transfection of this construct failed to produce a spliced RNA (Fig. 3D, lane 1) due to the fact that the 5′exon is relatively long (15, 17). Transfection of such a construct with the B19V ISE2 inserted 3′ of the AAV5 donor site led to abundant expression of the spliced RNA (Fig. 3D, lane 5). As a positive control, we tested a U stretch, which binds the TIA protein to facilitate U1 snRNP binding to the donor site at the same location (4), and the U stretch supported efficient splicing at this donor site (Fig. 3D, lane 4), as we previously reported (15). Conversely, the insertion of either a scrambled sequence or the ISEm3 mutant sequence into this site did not facilitate splicing of the P7-generated pre-mRNA at the AAV5 donor site (Fig. 3D, lanes 2 and 3). We hypothesize that the B19V ISE2 may function in a way to increase the affinity of the U1 snRNP to the D2 donor site.
The B19V ESE3 and ISE2 are functional during B19V infection of CD36+ EPCs.
To confirm that the above-identified ESE and ISE elements play a role in processing of the B19V pre-mRNA during infection of CD36+ erythroid progenitor cells (EPCs) (6, 21), we designed morpholino antisense oligonucleotides (MOs) that target ESE3 and ISE2, as well as the respective negative-control oligonucleotides (Gene Tools, LLC). An MO targeting the D2 site was used as a positive control (Fig. 4 A). MOs have been used to knock down gene expression by targeting ISEs/ESEs to block splicing of the pre-mRNA (9, 10, 13).
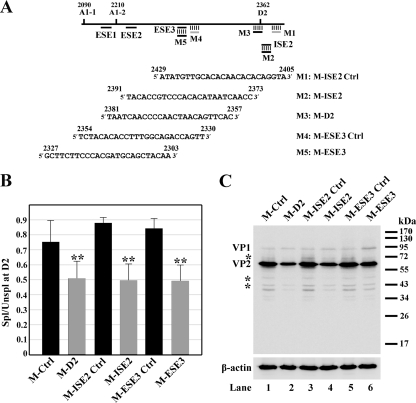
FIG. 4.
ESE3- and ISE2-mediated enhancement of splicing during B19V infection as assessed by a morpholino approach. (A) Top, B19V exon 2 is depicted with the positions of splice sites and targets of the morpholino oligonucleotides indicated. Bottom, sequences of the morpholino antisense oligonucleotides used in this experiment. (B and C) Morpholino oligonucleotides inhibited splicing of the B19V pre-mRNA at the D2 site. For these experiments, the CD36+ erythroid progenitor cells were expanded as previously described (6, 21). The cells were treated with morpholino oligonucleotides 16 h prior to B19V infection, as previously described (2). (B) At 48 h postinfection, mRNA was isolated from B19V-infected cells and quantified using qRT-PCR. The qRT-PCR method, primers, and probes for β-actin, VP2-encoding mRNA, and 11kDa-encoding mRNA have been described previously (7). For the VP1-encoding mRNA, the following primers and probe were used: forward primer, 5′-TGGGTTTCAAGCACAAGTAGTAAAA-3′; reverse primer, 5′-CCGGCAAACTTCCTTGAAAA-3′; and TaqMan probe, 5′-FAM-TGCAGCTGCCCCTGTGGCC-BHQ1-3′, where FAM is 6-carboxyfluorescein, and BHQ1 is black hole quencher 1. The absolute copy numbers of the B19V VP1 (R4+R5)-, VP2 (R6+R7)-, and 11kDa-encoding (R8+R9) mRNAs (Fig. 1) were obtained using the copy number of β-actin as an internal control. Results shown are averages and standard deviations from three independent experiments. ** denotes statistical significance (P value, <0.05) for the groups compared with their respective controls as well as M-Ctrl, a standard control oligonucleotide from GeneTools, LLC, as determined by the Student t test. The ratio of spliced to unspliced B19V mRNAs at the D2 site, as indicated by “Spl/Unspl,” was obtained by quantifying the numbers of copies of the unspliced VP1-encoding mRNA and the numbers of copies of the spliced B19V VP2- and 11kDa-encoding mRNAs. (C) At 48 h postinfection, cells were collected and analyzed by Western blotting using a monoclonal antibody against VP2 (clone R92F6; Millipore Co., MA) as described previously (14). Signals were developed using SuperSignal West Dura substrate (Thermo) and quantified with a Fujifilm LAS 4000 imager. Asterisks indicate degraded bands of VP1 and VP2.
MOs were delivered into CD36+ EPCs using the Endo-Porter peptide, as previously described (2), 16 h prior to B19V infection (6). At 48 h postinfection, we used quantitative reverse transcription-PCR (qRT-PCR) to quantify the VP1-encoding mRNA (represents mRNA unspliced at the D2 site), and the VP2- and 11kDa-encoding mRNAs (both are spliced at the D2 donor site) (Fig. 1). Application of the MOs M-ISE2 and M-ESE3, which specifically target the ISE2 and ESE3, respectively, led to a decrease in splicing at the D2 site. The ratio of spliced RNA to unspliced RNA at the D2 site (Fig. 4B, Spl/Unspl), assessed as the ratio of the collective number of copies of the VP2- and 11kDa-encoding mRNAs to VP1 encoding mRNA (Fig. 1), was reduced from an average of 0.88 to 0.50 and from 0.84 to 0.49 by the application of M-ISE2 and M-ESE3, respectively (Fig. 4B). As a result, the ratio of VP1 to VP2 as determined by Western blotting (Fig. 4C) was increased from approximately 1:30 for the control MO-treated groups to approximately 1:15 for the donor/enhancer-specific MO-treated groups (Fig. 4C, compare lanes 1, 3, and 5 to lanes 2, 4, and 6, respectively). These results confirmed that the B19V ESE3 and ISE2 that we identified by transfection of B19V nonpermissive cells are in fact required for the inclusion of exon 2 during B19V pre-mRNA processing in the context of virus infection.
In conclusion, we show that the definition of B19V exon 2 is the consequence of a balanced interplay among a weak 5′ splice donor (D2), the ISE2 just 3′ of the D2 site, the ISE1 that is adjacent to the A1-2 site, and ESE1, ESE2, and ESE3 in exon 2. We hypothesize that the SR proteins that bind to ESE3 and ISE2 facilitate recognition of the D2 donor site and further promote the binding of U1 snRNP to the D2 donor site and that the SR proteins that bind to ESE1 and ESE2 help splicing at the A1-1 and A1-2 acceptor sites, respectively, defining the central exon 2. A weak 5′ splice donor in the second intron may provide an opportunity for regulatory splicing during B19V permissive infection. Thus, our study provides the first evidence of the molecular basis of regulatory splicing during B19V pre-mRNA processing.
Acknowledgments
This work was supported by PHS grant R01 AI070723 from the National Institute of Allergy and Infectious Diseases and grant P20 RR016443 from the Centers of Biomedical Research Excellence program, National Center for Research Resources.
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1.Cartegni, L., J. Wang, Z. Zhu, M. Q. Zhang, and A. R. Krainer. 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31:3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, A. Y., et al. 2010. The small 11kDa nonstructural protein of human parvovirus B19 plays a key role in inducing apoptosis during B19 virus infection of primary erythroid progenitor cells. Blood 115:1070-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiorini, J. A., F. Kim, L. Yang, and R. M. Kotin. 1999. Cloning and characterization of adeno-associated virus type 5. J. Virol. 73:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forch, P., O. Puig, C. Martinez, B. Seraphin, and J. Valcarcel. 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21:6882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan, W., et al. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J. Virol. 82:9951-9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan, W., S. Wong, N. Zhi, and J. Qiu. 2009. The genome of human parvovirus B19 virus can replicate in non-permissive cells with the help of adenovirus genes and produces infectious virus. J. Virol. 83:9541-9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manley, J. L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 9.Morcos, P. A. 2007. Achieving targeted and quantifiable alteration of mRNA splicing with morpholino oligos. Biochem. Biophys. Res. Commun. 358:521-527. [DOI] [PubMed] [Google Scholar]
- 10.Moulton, J. D., and Y. L. Yan. 2008. Using morpholinos to control gene expression. Curr. Protoc. Mol. Biol. 2008:Unit 26.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 6:1107-1119. [DOI] [PubMed] [Google Scholar]
- 12.Ozawa, K., et al. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 61:2395-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popplewell, L. J., C. Trollet, G. Dickson, and I. R. Graham. 2009. Design of phosphorodiamidate morpholino oligomers (PMOs) for the induction of exon skipping of the human DMD gene. Mol. Ther. 17:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu, J., F. Cheng, L. R. Burger, and D. Pintel. 2006. The transcription profile of Aleutian mink disease virus in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J. Virol. 80:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu, J., F. Cheng, and D. Pintel. 2007. Distance-dependent processing of adeno-associated virus type 5 RNA is controlled by 5′ exon definition. J. Virol. 81:7974-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu, J., R. Nayak, G. E. Tullis, and D. J. Pintel. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 76:12435-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu, J., and D. J. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by the distance between the promoter and the intron and is inhibited by U1 small nuclear RNP binding to the intervening donor. J. Biol. Chem. 279:14889-14898. [DOI] [PubMed] [Google Scholar]
- 18.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 181:22-34. [DOI] [PubMed] [Google Scholar]
- 19.Smith, P. J., et al. 2006. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 15:2490-2508. [DOI] [PubMed] [Google Scholar]
- 20.St. Amand, J., C. Beard, K. Humphries, and C. R. Astell. 1991. Analysis of splice junctions and in vitro and in vivo translation potential of the small, abundant B19 parvovirus RNAs. Virology 183:133-142. [DOI] [PubMed] [Google Scholar]
- 21.Wong, S., et al. 2008. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J. Virol. 82:2470-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoto, Y., J. Qiu, and D. J. Pintel. 2006. Identification and characterization of two internal cleavage and polyadenylation sites of parvovirus B19 RNA. J. Virol. 80:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young, N. S., and K. E. Brown. 2004. Parvovirus B19. N. Engl. J. Med. 350:586-597. [DOI] [PubMed] [Google Scholar]
- 24.Zhi, N., Z. Zadori, K. E. Brown, and P. Tijssen. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318:142-152. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang, Y., and A. M. Weiner. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46:827-835. [DOI] [PubMed] [Google Scholar]