Abstract
Increasing evidence implicates cohesin in the control of gene expression. Here we report the first analysis of cohesin-dependent gene regulation in fission yeast. Global expression profiling of the mis4-367 cohesin loader mutant identified a small number of upregulated and downregulated genes within subtelomeric domains (SD). These 20- to 40-kb regions between chromosome arm euchromatin and telomere-proximal heterochromatin are characterized by a combination of euchromatin (methylated lysine 4 on histone H3/methylated Tysine 9 on histone H3 [H3K4me]) and heterochromatin (H3K9me) marks. We focused our analysis on the chromosome 1 right SD, which contains several upregulated genes and is bordered on the telomere-distal side by a pair of downregulated genes. We find that the expression changes in the SD also occur in a mutant of the cohesin core component Rad21. Remarkably, mutation of Rad21 results in the depletion of Swi6 binding in the SD. In fact, the Rad21 mutation phenocopied Swi6 loss of function: both mutations led to reduced cohesin binding, reduced H3K9me, and similar gene expression changes in the SD. In particular, expression of the gene pair bordering the SD was dependent both on cohesin and on Swi6. Our data indicate that cohesin participates in the setup of a subtelomeric heterochromatin domain and controls the expression of the genes residing in that domain.
After DNA replication, sister chromatids are held together until the onset of anaphase by a large protein complex termed cohesin (20, 31, 32). Cohesion between sister chromatids is essential for their bilateral attachment to spindle microtubules and for faithful segregation into daughter cells during mitosis. Cohesin is a ring-shaped complex comprising four subunits: Smc1, Smc3, Scc3, and Mcd1/Scc1 (Psm1, Psm3, Psc3, and Rad21 in fission yeast) (for reviews, see references 37 and 41). Strong experimental evidence indicates that cohesion is ensured topologically by the cohesin ring encircling the two sister chromatids, although other modes of cohesin interaction with chromosomes may coexist (for reviews, see references 23a and 37). Cohesin is loaded onto chromosomes by the cohesin-loading complex Scc2-Scc4 (Mis4-Ssl3 in fission yeast) (4, 13). The distribution of cohesin on chromosomes is not random. In budding and fission yeasts, cohesin is enriched at telomeres, pericentromeric regions, and so-called cohesin-associated regions (CARs) on chromosome arms. In fission yeast, the recruitment of cohesin at mating-type, pericentromeric, and telomeric sites depends on the heterochromatin protein 1 (HP1) ortholog Swi6, which interacts with the cohesin component Psc3 (5, 38) and the loading factor Mis4 (15). Swi6 binds to methylated lysine 9 on histone H3 (H3K9me), the heterochromatin mark brought about by the Clr4 methyltransferase (3, 36), and is also involved in the spreading of this heterochromatin mark (22).
It is becoming increasingly clear that in addition to its central role in sister chromatid cohesion, cohesin is also involved in various other aspects of chromosome biology, in particular the regulation of gene expression (for reviews, see references 10, 41, and 54). Several metazoan developmental defects are associated with mutations in components of the cohesin network and apparently do not result from an alteration in sister chromatid cohesion. The Cornelia de Lange syndrome (CdLS) is caused by heterozygous mutations in the cohesin-loading factor SCC2 or within the cohesin subunit SMC1 or SMC3 (14, 27, 35, 50). Similarly, in fly, heterozygous mutations in the Scc2 homolog Nipped-B cause body-patterning defects during development (42, 43). In these models, hypomorphic defects in the cohesin pathway can lead to extensive modifications in gene expression (30, 44). The finding that mutations in the cohesin complex alter gene expression and differentiation in postmitotic fly neurons provided a direct demonstration of an interphase function of cohesin (40, 47). Inactivation of the cohesin complex in budding yeast also led to modifications in the expression of a small number of genes that showed significant clustering in the same chromosomal regions (48). More generally, cohesin distribution with respect to gene architecture reveals a relation between cohesin positioning and gene transcription, even if this distribution appears to differ somewhat in yeasts, flies, and mammals (19, 28, 33, 39, 45, 52). In fission yeast, both the loader complex Mis4-Ssl3 and cohesin show a preferential association with active promoters and are enriched in intergenic regions of convergent gene pairs (45). A clear picture of how cohesin modulates gene expression has yet to emerge. The mechanistic modalities of this regulation may well differ depending on the organism and the loci considered. Cohesin has been found to play a role in the nuclear localization of DNA sequences and to interact with factors mediating long-range DNA-DNA interactions and chromatin looping (17, 25, 52). In fission yeast, cohesin has also been found to play a role in preventing transcriptional read-through at convergent gene pairs (21).
In a previous study, we reported that inactivation of the cohesin-loading machinery in G1-arrested cells leads to the dissociation of cohesin from chromatin both at centromeres and at chromosome arm sites (6). We exploited this situation to ask whether such a loss of cohesin would have an impact on gene expression on a genome-wide scale in fission yeast. We found that gene expression modifications were restricted to genes residing in subtelomeric domains located between chromosome arm euchromatin and telomere-proximal heterochromatin. A detailed analysis of one such subtelomeric region revealed that cohesin is involved in setting up heterochromatin in this subtelomeric domain.
MATERIALS AND METHODS
Strains and media.
Media were prepared as described in reference 34. Cells were cultured in EMM2 synthetic medium. The strains used in this study are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| 2760 | h− ura4+-pREP2-res1 | 6 |
| 2758 | h− ura4+-pREP2-res1 mis4-367 | 4 (mis4-367) |
| 3393 | h− ura4+-pREP2-res1 mis4-GFP-LEU2 | 4 (mis4-GFP-LEU2) |
| 3212 | h− ura4+-pREP2-res1 rad21-45 | 7 (rad21-45) |
| 3452 | h− ura4+-pREP2-res1 swi6Δ::his1+ his1-102 | 1 (swi6Δ::his1+) |
| 3397 | h− ura4+-pREP2-res1 clr4Δ::LEU2 | Robin Allshire (clr4Δ::LEU2) |
| 3395 | h− ura4+-pREP2-res1 mis4-HA-LEU2 | 4 (mis4-HA-LEU2) |
| 3588 | h− ura4+-pREP2-res1 mis4-HA-LEU2 rad21-45 | |
| 3557 | h− ura4+-pREP2-res1 mis4-HA-LEU2 swi6Δ::his1+ his1-102 |
G1 arrest by C-terminal Res1 overexpression.
Cells were G1 arrested as described in reference 6. Briefly, cells bearing nmt-res1Cter integrated at the ura4 locus (ura+-pREP2res1) were grown at 25°C to late-log phase in EMM2 containing 20 μM thiamine in order to repress the expression of the C-terminal fragment of Res1 (cycling conditions). Cells were harvested, washed, and inoculated into fresh EMM2 without thiamine at a density of 3 × 105 cells/ml. Cell proliferation ceased after seven doublings. Once arrested, cells were shifted to 37°C for 2 h. Cell cycle arrest before and after the temperature shift was monitored by measuring DNA content by flow cytometry and the septation index by calcofluor staining.
Genome-wide expression analysis.
Three independent inocula of the wild type and the mis4-367 mutant were grown at 25°C, G1 arrested, and shifted to 37°C for 2 h. Cells were harvested before and after the temperature shift to proceed to total-RNA extraction. Total RNA was extracted using the standard hot-phenol method (26, 46), followed by DNase I treatment and a cleanup step on a RNeasy Mini Kit column (catalog no. 79254 and 74104; Qiagen). Poly(A)+ RNAs were labeled using the GeneChip One-Cycle Target Labeling kit (catalog no. 900493) from Affymetrix. Transcriptome data were analyzed with Stratagene ArrayAssist Expression software. Microarray data were subjected to probe summarization and were compared using four different algorithms (probe logarithmic intensity error [PLIER], RMA, GCRMA, and Li-Wong); mean fold changes were calculated; and those above a threshold of 1.7-fold were considered. The mutant and the wild type were compared at both 25 and 37°C.
Reverse transcription (RT)-QPCR.
One microgram (1 μg) of total RNA was reverse transcribed using Superscript II (Invitrogen) and oligo(dT) primers. cDNAs were analyzed using forward and reverse primers selected to amplify sequences close to the 3′ end of mRNA. The primer sequences are listed in Table 2. Real-time PCR was performed in the presence of SYBR green (Abgene) on a Stratagene Mx3000P cycler. Primer efficiency was determined using genomic DNA. The relative amount of act1 cDNA measured by quantitative PCR (QPCR) was used as an internal standard for normalization.
TABLE 2.
Primer sequences
| Locus | Sequence |
|
|---|---|---|
| Forward | Reverse | |
| imr1 | TTTTGGACAGAATGGATGGA | GCGGAGTAAGGTTAATCACG |
| dg1 | ACGGCATCGCTTGTACTTTT | TGAGGTTCATGATGGGTTCA |
| SPAC869.10c | TACCTTCACCGCCGGTAACT | AGCTTCAGCTTCCGCAACTC |
| SPAC186.01 | AACTAATGGTGCCGGTCGAG | CAAGCACTTGAAAAACCATTGC |
| SPAC186.02c | CAAGTCGGAGGTTGTGCAAT | ATCGGACGCACTCTTCAATG |
| SPAC186.04c | GCGAAGAAAACCCAACAAGC | TCATCGTTTACTCTGATCCGTGA |
| SPAC186.05c | AAATTTTCCCGGGCTTTCAT | TCCGACAATCACCGCTACC |
| SPAC186.06 | GGGAGTGGAGCTGGATCAGT | CGCCACCAACATGAATATCG |
| SPAC750.07c | TGTTACGTGGCAAGGCAGAC | CCCAAATGTGACAGCCAAAA |
| act1 | CCGATCGTATGCAAAAGGAG | AATGGATCCACCAATCCAGA |
ChIP.
Chromatin immunoprecipitations (ChIPs) were performed as described previously (4). In brief, 2 × 108 cells were fixed with 1/10 culture volume of fixative (33% formaldehyde, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 50 mM Tris-HCl [pH 8.1]) for 30 min at 20°C. Cells were lysed in ChIP lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] sodium deoxycholate). Chromatin was sheared to 0.5- to 1-kb fragments using a Bioruptor sonicator (Diagenode) (7 cycles of 30 s on and 30 s off at maximum power, performed 3 times) and was clarified by centrifugation. Immunoprecipitation was carried out overnight at 4°C in IP buffer (1% [wt/vol] Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1]). Immune complexes were collected using ChIP-Adembeads (catalog no. 04240 or 04340; Ademtech) or protein G agarose beads (Roche). Cross-links were reversed overnight at 65°C in 10 mM Tris-HCl (pH 8.1)-1 mM EDTA-0.5% (wt/vol) sodium dodecyl sulfate (SDS) containing 0.3 mg/ml proteinase K. Nucleic acids were extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and were recovered by NaCl-ethanol precipitation. ChIP quantifications were performed by real-time PCR. The primer list is available upon request. Antibodies against green fluorescent protein (GFP) (catalog no. A-11122; Invitrogen), Swi6 (catalog no. 14898; Abcam), H3K9me2 (catalog no. 1220; Abcam), and H3K4me2 (catalog no. 07-030; Millipore) were used. Anti-Psm1 antibodies were made by Proteogenics. Rabbits were immunized with a recombinant peptide corresponding to the 631 N-terminal residues expressed in Escherichia coli. Rabbit serum was then affinity purified against the recombinant protein using an N-hydroxysuccinimide (NHS)-activated HiTrap column (Amersham, GE Healthcare) according to the protocol provided by the manufacturer. In brief, 1.5 mg of purified Psm1(1-631) was bound to a 1-ml NHS-activated HiTrap column, and 15 ml of serum was applied to the column; after extensive washing, antibodies were eluted in 0.1 M glycine-HCl (pH 2.5). Eluted fractions were rapidly neutralized with 0.1 volume of 1 M Tris-HCl (pH 8.5).
Immunoprecipitations were performed in duplicate. Immunoprecipitation percentages were calculated as the ratio of immunoprecipitated DNA to total input DNA.
Microarray data accession number.
The microarray data determined in this study have been deposited in the GEO database under accession number GSE23602.
RESULTS
Inactivation of the cohesin loader Mis4 in G1-arrested cells specifically affects gene expression in subtelomeric regions.
In a previous study, it was shown that when G1-arrested cells carrying the thermosensitive mis4-367 allele are shifted to 37°C, cohesin dissociates from chromatin both at the imr centromeric region and at various cohesin-associated regions (CARs) on chromosome arms (6).
We analyzed genome-wide modifications in gene expression in that experimental situation, with the rationale that potential changes should correspond to a direct effect of cohesin dissociation rather than resulting from the loss of the sister chromatid cohesion function per se. Cells were arrested in late G1 by titrating out the Cdc10 transcription factor by overexpression of the C-terminal fragment of its binding partner Res1 (2). Total RNA was extracted from mis4-367 cells and otherwise isogenic mis4+ cells that had been kept at the permissive temperature of 25°C or had been transferred to 37°C for 2 h.
After RNA labeling and hybridization on Affymetrix GeneChip Yeast Genome 2.0 microarrays, pairwise comparisons were performed. Table 3 lists the genes showing ≥1.7-fold changes in the pairwise comparisons; 25 genes showing changes in expression were identified. Of those, 13 are located in subtelomeric regions. Another 6 genes are encoded by the mitochondrial DNA, and the remaining 6 genes are located in arm regions. RT-QPCR experiments were conducted to assess the validity of the genome-wide screen. The expression changes for all 13 subtelomeric genes were confirmed by RT-QPCR. Nine genes were upregulated, and 4 were downregulated; changes in expression were generally in the range of 2- to 3-fold. Expression modifications for the mitochondrial and chromosome arm genes analyzed were not confirmed (Table 3). The proteins encoded by the subtelomeric genes did not show any obvious functional relationship.
TABLE 3.
List of genes with modified expression in mis4-367 cellsa
| Category and systematic name | Name | Function | Fold change | Reproducibilityb | Location |
|---|---|---|---|---|---|
| Downregulated in mis4-367 at 25°C | |||||
| SPAC869.07c | α-Galactosidase (predicted) | 4.24 | Y | Subtelomeric | |
| SPMIT.01 | cox1 | Cytochrome c oxidase (subunit 1) | 2.97 | N | Mitochondrial |
| SPAC186.02c | 2-Hydroxyacid dehydrogenase (predicted) | 2.93 | Y | Subtelomeric | |
| SPAC186.01 | Glycoprotein (predicted) | 2.66 | Y | Subtelomeric | |
| SPMIT.04 | cox3 | Cytochrome c oxidase (subunit 3) | 2.21 | ND | Mitochondrial |
| SPAC27D7.09c | Predicted N-terminal signal sequence | 2.09 | N | Arm region | |
| SPMIT.05 | cob | Cytochrome b | 2.08 | ND | Mitochondrial |
| SPAC750.07c | 66294 domain | 2.06 | Y | Subtelomeric | |
| SPMIT.10 | atp9 | ATPase (subunit 9) | 1.78 | ND | Mitochondrial |
| SPCC737.04 | B8647-5 | Pfam-B_8647 domain | 1.71 | N | Arm region |
| SPMIT.11 | cox2 | Cytochrome c oxidase (subunit 2) | 1.70 | ND | Mitochondrial |
| Upregulated in mis4-367 at 25°C | |||||
| SPAC186.06 | Hypothetical protein | 2.49 | Y | Subtelomeric | |
| SPBCPT2R1.02 | Hypothetical protein | 2.48 | Y | Subtelomeric | |
| SPAC186.05c | Hypothetical protein | 2.42 | Y | Subtelomeric | |
| SPAC186.04c | Putative pseudogene, similar to N-terminal portion of transmembrane channel | 2.09 | Y | Subtelomeric | |
| SPAC212.02 | Hypothetical protein | 1.94 | Y | Subtelomeric | |
| SPAC750.05c | Telomeric duplication | 1.75 | Y | Subtelomeric | |
| SPAC977.04 | Pseudogene | 1.70 | Y | Subtelomeric | |
| Downregulated in mis4-367 at 37°C | |||||
| SPMIT.06 | Hypothetical protein | 3.47 | N | Mitochondrial | |
| SPMIT.04 | cox3 | Cytochrome c oxidase (subunit 3) | 3.34 | N | Mitochondrial |
| SPAC186.01 | Glycoprotein (predicted) | 2.43 | Y | Subtelomeric | |
| SPBC11C11.12 | Possible pseudogene | 2.02 | ND | Arm region | |
| SPAC186.02c | 2-Hydroxyacid dehydrogenase (predicted) | 1.98 | Y | Subtelomeric | |
| SPAPB8E5.05 | mfm1 | M-factor precursorc | 1.88 | ND | Arm region |
| SPBPJ4664.03 | mfm3 | M-factor precursorc | 1.78 | ND | Arm region |
| SPCC188.12 | spn6 | Septin | 1.70 | N | Arm region |
| Upregulated in mis4-367 at 37°C | |||||
| SPAC186.04c | Putative pseudogene, similar to N-terminal portion of transmembrane channel | 2.47 | Y | Subtelomeric | |
| SPAC186.05c | Hypothetical protein | 2.05 | Y | Subtelomeric | |
| SPAC186.06 | Hypothetical protein | 1.91 | Y | Subtelomeric | |
| SPAC186.08c | l-Lactate dehydrogenase (predicted) | 1.79 | Y | Subtelomeric | |
| SPBPB2B2.08 | Sequence orphan | 1.71 | Y | Subtelomeric |
Genes with at least a 1.7-fold difference between mis4-367 and wild-type cells.
Tested by RT-QPCR. Y, yes; N, no; ND, not determined.
See reference 25a.
The genes whose expression is modified in mis4-367 cells are located in a specific 20- to 40-kb subtelomeric domain forming a transition zone between bona fide telomeric heterochromatin and the euchromatic arm regions. The telomeric heterochromatin is characterized by a high level of H3K9me and almost undetectable H3K4me. In the 20- to 40-kb subtelomeric domain, the level of H3K9me is lower, and the H3K4me mark can be detected, but to a lower degree than that in bona fide euchromatin (11). This subtelomeric boundary is distinct from centromeric or mating-type euchromatin-heterochromatin boundaries, where there is a very sharp transition from high H3K9me to undetectable H3K9me (11).
The cohesin loader Mis4 and cohesin are present at the chromosome 1 right (Chr1-R) subtelomeric region.
Considering that a mutation in the cohesin loader Mis4 affects gene expression in subtelomeric regions, we wondered if the cohesin-loading complex and cohesin itself were actually present in those regions and, if so, whether cohesin binding was affected in mis4-367 cells.
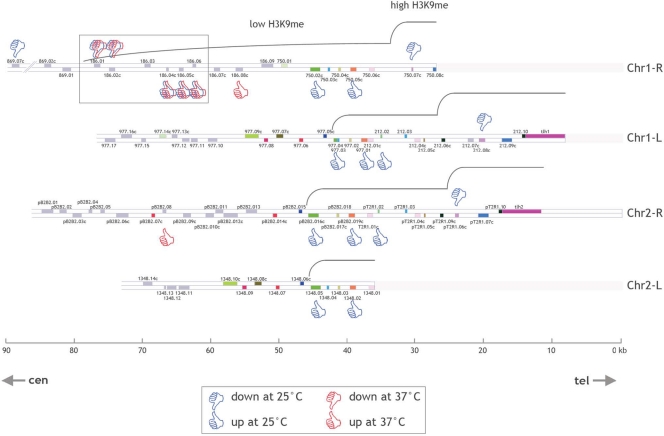
In fission yeast, the subtelomeric genes generally exist as almost identical copies on the different subtelomeres, making it impractical to assign gene expression changes to a specific subtelomeric locus. We focused on the Chr1-R subtelomeric region, where a set of 5 adjacent genes (SPAC186.01, SPAC186.02c, SPAC186.04c, SPAC186.05c, and SPAC186.06) can be specifically analyzed because of their low degree of homology with sequences from other subtelomeres (Fig. 1). Our attention was also drawn to this gene set because expression of the 5 genes was modified in the mis4-367 mutant both at 25°C and at 37°C (Table 3). SPAC186.01 and SPAC186.02c form a convergent gene pair located at the telomere-distal end of the subtelomeric domain and are downregulated in mis4-367 cells, while the 3 telomere-proximal adjacent genes are upregulated. We also analyzed the expression of these 5 genes in cycling cells, which are mainly (∼80%) in G2, and found that the pattern of gene expression was modified in the same way as in G1-arrested cells (data not shown).
FIG. 1.
Location map of open reading frames within the subtelomeric regions of chromosomes 1 and 2 of S. pombe according to GeneDB (http://old.genedb.org/genedb/pombe/). Open reading frames are represented by gray rectangles. In addition, the color coding of the open reading frames represents highly homologous paralogs residing in different subtelomeres. Genes identified in the present transcriptome analysis as up- or downregulated at 25 or 37°C in the mis4-367 mutant versus the wild type are indicated. Note that when up- or downregulation implicates genes with high levels of homology (shown by the same color in the different subtelomeres), it is not known whether the expression of one or several members of the gene family is modified. Low- and high-H3K9me regions are represented according to reference 11 and the Schizosaccharomyces pombe Epigenome Home Page, developed by Shiv Grewal's lab (http://pombe.nci.nih.gov/genome/). The present study is focused on a set of five genes (SPAC186.01, SPAC186.02c, SPAC186.04c, SPAC186.05c, and SPAC186.06 [boxed]) located at the Chr1-R subtelomeric heterochromatin boundary.
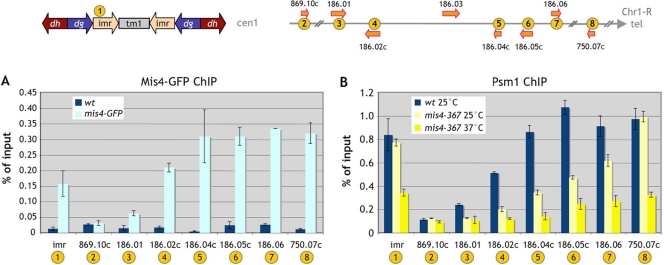
ChIP experiments were performed to assess the presence of the cohesin loader Mis4 and the cohesin subunit Psm1 at the Chr1-R subtelomeric domain. We found that Mis4-GFP was present in that region (Fig. 2A). Similarly, based on immunoprecipitation of chromatin-bound Psm1, cohesin was also found at these subtelomeric sites (Fig. 2B). Both for Mis4-GFP and for Psm1, there was a trend of increased binding toward the telomere. Mis4-GFP and cohesin were abundant in that region, and the amounts were comparable to those found at the imr centromeric domain.
FIG. 2.
Mis4 and Psm1 are present at the Chr1-R subtelomere, and Psm1 binding is affected in mis4-367 cells even at 25°C. (A) Cells were G1 arrested at 25°C. Mis4-GFP chromosomal association was assessed by chromatin immunoprecipitation. wt, wild type. (B) In G1-arrested cells at 25 and 37°C, the binding of cohesin to chromatin was monitored by Psm1 immunoprecipitation at the centromere 1 innermost region (imr); the Chr1-R arm locus SPAC869.10c, located 30 kb upstream of the subtelomeric heterochromatin region; the five Chr1-R subtelomeric loci SPAC186.01, SPAC186.02c, SPAC186.04c, SPAC186.05c, and SPAC186.06; and the telomere-proximal SPAC750.07c gene. Error bars represent the standard errors of the means from two independent experiments.
The association of cohesin with pericentric and telomere-proximal heterochromatin was unmodified in mis4-367 cells at 25°C and dropped only at 37°C. In contrast, cohesin binding on the five Chr1-R subtelomeric genes (SPAC186.01, SPAC186.02c, SPAC186.04c, SPAC186.05c, and SPAC186.06) was already affected by the mis4-367 mutation at 25°C and was further decreased at 37°C (Fig. 2B). The gene sets showing expression changes at 25 and 37°C overlap extensively (Table 3). No simple linear relationship between the reduction in cohesin binding and gene expression changes was apparent. Fold changes in gene expression did not increase upon further dissociation of cohesin at 37°C. This suggests that the cohesin depletion caused by the mis4-367 mutation at 25°C is sufficient to induce the observed modifications of gene expression.
Gene expression and Swi6 binding in the subtelomeric region are affected by a mutation of the cohesin core component Rad21.
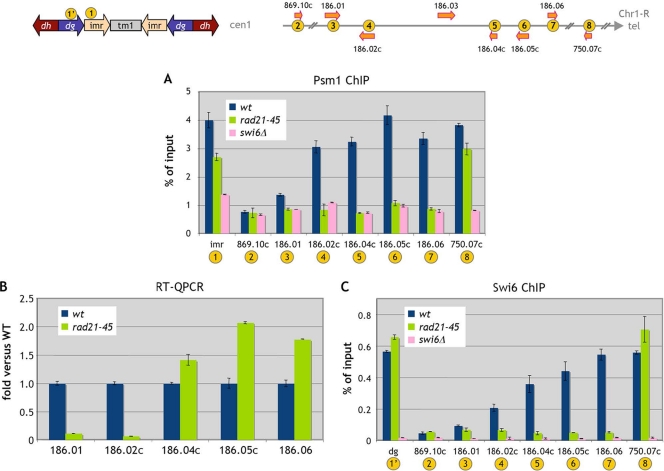
We next asked whether a mutation in a component of the cohesin core complex would have a similar effect. The rad21-45 hypomorphic mutant is deficient in DNA double-strand break repair induced either by gamma rays or by UV irradiation (7). The rad21-45 mutation consists of a T-to-C transition, which changes a nonpolar isoleucine at position 67 to a polar threonine. The Rad21-45 protein is permanently hypophosphorylated, while the wild-type protein becomes hyperphosphorylated during S phase, when cohesion between sister chromatids is established (8, 49). In the rad21-45 mutant, cohesin binding decreases dramatically in the subtelomeric domain and to a lesser extent at imr and a telomere-proximal site (Fig. 3A). Cohesin binding in the subtelomeric domain was reduced similarly in a swi6Δ background (Fig. 3A), indicating that cohesin binding is Swi6 dependent in that region, as it is at the centromere (5, 38).
FIG. 3.
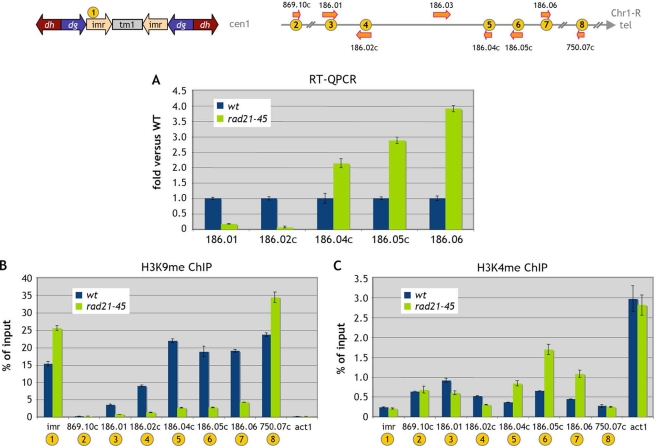
Gene expression and the presence of Psm1 and Swi6 in the Chr1-R subtelomeric region are modified in the core cohesin rad21-45 mutant. Cells were G1 arrested at 25°C. (A) The binding of Psm1 to chromatin in the rad21-45 mutant was analyzed by Psm1 immunoprecipitation. Error bars represent the standard errors of the means from duplicate samples. wt, wild type. (B) Gene expression was analyzed by RT-QPCR. Data are expressed as the fold change versus the wild type. Error bars represent the standard errors of the means from two independent biological replicates. (C) Swi6 chromosomal association in rad21-45 cells was assessed by ChIP analysis. Error bars represent the standard errors of the means from duplicate samples.
We then analyzed gene expression in the Chr1-R subtelomeric region in rad21-45 cells and found that gene expression is qualitatively affected in the same way as in the mis4-367 mutant (Fig. 3B), with downregulation of SPAC186.01 and SPAC186.02c and upregulation of the three adjacent telomere-proximal genes. Thus, both a mutation in the cohesion loader and a mutation in a cohesin core component lead to expression changes in the subtelomeric domain. This subtelomeric domain is heterochromatic (11). Thus, we wondered whether the effects of the rad21-45 mutation on gene expression might be related to modifications of heterochromatin marks in that region. We therefore analyzed Swi6 binding in that region (Fig. 3C). In wild-type cells, Swi6 was found over the entire region, with a trend of increased binding toward the telomere-proximal direction. Remarkably, Swi6 binding was drastically reduced over the entire region in the rad21-45 mutant. We also noted a slight increase in the level of Swi6 binding at the centromere and telomere.
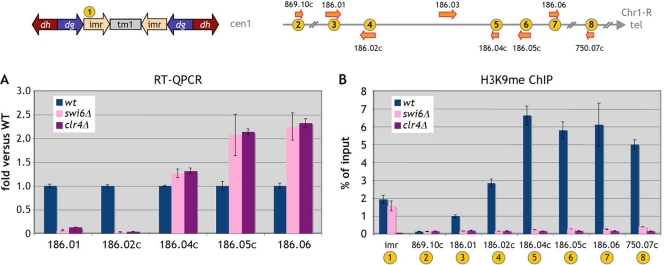
Since the rad21-45 mutation affects Swi6 binding over the subtelomeric domain, we asked whether deletion of the swi6 gene would have the same effect on gene expression. Indeed, in a swi6Δ background, the same gene expression changes occurred (Fig. 4A). The three telomere-proximal genes were upregulated, while expression from the SPAC186.01-SPAC186.02c gene pair became nearly undetectable. The same effect was observed in a clr4Δ mutant incapable of H3K9 methylation (Fig. 4A). Interestingly, the H3K9me mark was totally erased from the subtelomeric domain in a swi6Δ mutant background (Fig. 4B), indicating that H3K9me within that region is Swi6 dependent.
FIG. 4.
Swi6 or Clr4 inactivation leads to the same expression changes as the rad21-45 mutation in the Chr1-R subtelomeric region. Cells were G1 arrested at 25°C. (A) Gene expression was assessed by RT-QPCR in swi6Δ and clr4Δ mutant cells. Data are expressed as the fold change versus the wild type (wt). Error bars represent the standard errors of the means from two independent biological replicates. (B) The presence of the H3K9me mark in swi6Δ and clr4Δ mutant cells was determined by ChIP analysis. Error bars represent the standard errors of the means from duplicate samples.
A mutation in the cohesin core component Rad21 abolishes H3K9me in the subtelomeric domain.
In order to further assess the effect of a mutation in the cohesin core complex on chromatin structure in the subtelomeric region, we analyzed the effects of the rad21-45 mutation on the H3K9me and H3K4me marks in that region. We chose to conduct the experiment in cycling cells, since the effect of the rad21-45 mutation on gene expression in the subtelomeric domain was similar to those detected in G1-arrested cells (Fig. 5A). When we analyzed the H3K9me and H3K4me marks in the subtelomeric domain, we found that, in agreement with previous reports, both H3K9me and H3K4me are present within this region (Fig. 5B and C). Mutation of Rad21 led to a sharp decrease in the H3K9me mark throughout the region, while an increase in H3K9me was apparent at imr and the SPAC750.07c telomeric site, consistent with the Swi6 binding data (Fig. 3C). The H3K4me mark was also affected within the subtelomeric domain (Fig. 5C), and this modification was consistent with the gene expression changes for the 5 subtelomeric genes analyzed. Namely, the convergent gene pair whose expression was decreased showed a reduction in the H3K4me mark, while the three upregulated telomere-proximal genes displayed an increase in the H3K4me mark.
FIG. 5.
The rad21-45 mutation affects the distribution of heterochromatin and euchromatin marks, both of which are present at the Chr1-R subtelomeric region. Cells were cultured under cycling conditions at 25°C. (A) RT-QPCR analysis of gene expression in cycling rad21-45 mutant cells. Data are expressed as the fold change versus the wild type (wt). Error bars represent the standard errors of the means from two independent biological replicates. (B and C) H3K9me (B) and H3K4me (C) features of heterochromatin and euchromatin, respectively, were analyzed by ChIP at the Chr1-R subtelomeric region. Error bars represent the standard errors of the means from duplicate samples.
We conclude that mutation of the Rad21 cohesin subunit phenocopies Swi6 loss of function. Swi6 binding is strongly reduced over the entire subtelomeric domain, leading to the loss of H3K9me. The telomere-distal gene pair is downregulated, while the telomere-proximal genes are upregulated.
A particular allele of the cohesin loader Mis4 leads to Swi6 enrichment in the subtelomeric domain.
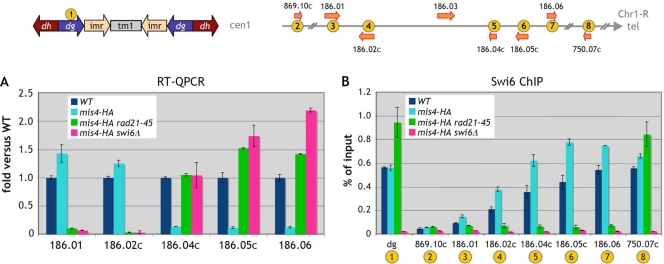
Serendipitously, we noted that a hemagglutinin (HA)-tagged mis4 allele had a profound impact on subtelomeric gene expression. The expression of the five genes of interest was affected, but in the opposite direction from that in mis4-367 cells. The SPAC186.01-SPAC186.02c gene pair was slightly upregulated, while SPAC186.04c, SPAC186.05c, and SPAC186.06 were downregulated (Fig. 6A). When we analyzed Swi6 binding in cells harboring this mis4-HA allele, we found that Swi6 binding was increased over the 5 subtelomeric genes (Fig. 6B). This effect of the mis4-HA allele on gene expression was abrogated when the mis4-HA allele was combined with the swi6Δ or rad21-45 mutation (Fig. 6A), underscoring the requirement for both cohesin and Swi6 to bring about this modification in subtelomeric gene expression. Likewise, the rad21-45 allele was epistatic over the mis4-HA allele with respect to Swi6 binding in the subtelomeric domain, suggesting that the Swi6 enrichment caused by the mis4-HA allele depends on cohesin. How the mis4-HA allele can increase Swi6 binding is not known. The interesting point is that an increased amount of Swi6 in the subtelomeric domain leads to expression changes opposite those associated with decreased Swi6 binding. This strengthens the conclusion that expression from the telomere-distal gene pair relies on a heterochromatic context, whereas telomere-proximal genes behave in the opposite way.
FIG. 6.
The expression of SPAC186.01 and SPAC186.02c is dependent on Swi6. Cells were G1 arrested at 25°C. (A) Gene expression was assessed by RT-QPCR in cells bearing a mis4-HA allele and in mis4-HA rad21-45 and mis4-HA swi6Δ double mutant cells. Data are expressed as the fold change versus the wild type (wt). Error bars represent the standard errors of the means from two independent biological replicates. (B) Swi6 chromosomal association at the Chr1-R subtelomere in mis-HA cells was determined by ChIP analysis. dg, centromere 1 outer repeat. Error bars represent the standard errors of the means from duplicate samples.
DISCUSSION
Cohesin and gene expression in Schizosaccharomyces pombe.
We find that in G1-arrested S. pombe cells, inactivation of the cohesin loader Mis4 leads to modifications in the expression of a limited set of genes located in subtelomeric regions. For Saccharomyces cerevisiae, it was recently reported that inactivation of the cohesin subunit Scc1 in late G1 leads to changes of 1.5-fold or more in the expression of 29 genes (48). Regulated genes were not enriched in genes from subtelomeric loci. Budding yeast lacks a Swi6/HP1 homolog, which may explain the difference in results between the two species. Nevertheless, as in the present study, there was significant positional clustering of the cohesin-dependent genes. Still, both in budding yeast and in fission yeast, alteration of the cohesin network appears to lead to relatively modest changes in gene expression. It remains to be established whether the role of cohesin in gene expression in S. pombe is indeed limited to the subtelomeric gene set identified. The present study focused on steady-state gene expression. It would be of interest to analyze the effect of cohesin inactivation in a situation of extensive transcriptional reprogramming, such as entry into the meiotic cycle or stress conditions. Since cohesin is required for transcription termination in G2 (21), inactivation of Mis4 or cohesin core units in G2 should lead to additional changes in gene expression. At any rate, we found that the expression changes in the subtelomeric region analyzed also occur in cycling cells, which correspond mainly (∼80%) to G2 cells.
SPAC186.01 and SPAC186.02c form a subtelomeric Swi6-dependent gene pair.
The cohesin-dependent expression changes that we have detected are ultimately all linked to a decrease in Swi6/H3K9me. Upregulation of the three telomere-proximal genes is well explained by a decrease in Swi6/H3K9me, which is usually correlated with a loss of transcriptional gene silencing in fission yeast. For the adjacent gene pair, the situation is less expected, since the expression of these two genes is positively correlated with Swi6/H3K9me levels. These genes can be qualified as Swi6 dependent, since in the absence of Swi6, their expression is reduced or abolished, and when Swi6 binding is increased, their expression increases. A role of HP1 in favoring gene expression has been documented in Drosophila, where HP1 was found to be required for the expression of genes located in heterochromatic regions (heterochromatic genes) (for a review, see reference 51). Transcriptional profiling in Su(var)2-5 mutants identified hundreds of genes that rely on HP1 for expression. Similarly, one study with S. pombe reported that 19 genes were downregulated >1.5-fold in a swi6 mutant background (53). How this induction function of Swi6 on SPAC186.01 and SPAC186.02c genes might be mediated can only be speculated at present. It has been proposed that Swi6 might mediate enhanced gene expression by recruitment of Epe1, a JmjC domain protein that counteracts heterochromatization (24).
Transcriptional read-through occurring in convergent gene pairs can result in the formation of double-stranded RNAs, leading to Swi6 binding and consequently cohesin recruitment (21). SPAC186.01 and SPAC186.02c form a convergent gene pair, and transcriptional read-through does occur in G1 in the SPAC186.01-SPAC186.02c gene pair (S. Dheur, unpublished results), so this site might represent a cohesin and Swi6 recruitment region. Interestingly, several convergent gene pairs are found at the other subtelomeres immediately adjacent to the low-H3K9me domain (Fig. 1). It is possible that these convergent gene pairs form a Swi6 recruitment site allowing for extension of the heterochromatin from the adjacent high-H3K9me telomeric heterochromatin, reminiscent of the functional cooperation between silencers and distant protosilencers in S. cerevisiae (9, 16). Deletion of a 10.7-kb region spanning SPAC186.01 and SPAC186.02c leads to the upregulation of the three adjacent telomere-proximal genes (Dheur, unpublished). This preliminary evidence suggests that this gene pair might indeed participate as a cis-acting factor in the regulation of gene expression in the subtelomeric domain.
Cohesin is involved in subtelomeric heterochromatin formation.
Our study establishes an interdependency of Swi6 and cohesin binding in the subtelomeric low-H3K9me region. In that region, as described for centromeric sites, cohesin binding is dependent on Swi6, but reciprocally and more surprisingly, mutations in the cohesin complex lead to depletion of Swi6 binding specifically in that region. One interesting feature of the subtelomeric genes we have studied is that the H3K9me and H3K4me marks in that region coincide rather than (as is most generally the case) being exclusive. In a comprehensive analysis, Cam et al. found that H3K9me and H3K4me marks coexist at the left Chr1 subtelomere but are largely exclusive in all other genomic regions analyzed (11). This situation is reminiscent of the bivalent chromatin domains described in metazoans, in which H3K4me and H3K27me marks coexist. These bivalent chromatin domains often correspond to developmentally regulated genes, particularly in embryonic stem cells (18). In this context, it is noteworthy that cohesin was found to be involved in the regulation of bivalent genes in Drosophila (44). It is possible that H3K9me subtelomeric heterochromatin protects S. pombe subtelomeres from recombination between nearly identical paralogs, while this low level of H3K9me still permits the expression of the corresponding genes when needed. The SPAC186.01c gene encodes an adhesin associated with a flocculation phenotype (29). In S. cerevisiae, adhesin genes of the FLO family also reside in subtelomeric locations, where they are subject to metastable silencing (23).
In the rad21-45 mutant, concomitant with the decrease in Swi6/H3K9me at the subtelomere, we detect a significant increase in Swi6/H3K9me at the centromere and telomere (Fig. 3C and 5B). Similarly, an S. pombe partial aneuploid bearing one extra chromosome displays a depletion of Swi6 in the subtelomeric region and a concomitant transcriptional dysregulation of subtelomeric genes (12). It was suggested that the subtelomeric fraction of Swi6 might be specifically depleted by titration of Swi6 to supernumerary stable heterochromatic sites (the telomeres and centromere of the extra chromosome). Like other silencing factors, Swi6 appears to be present in limiting amounts, and there appears to be a balance between the subtelomeric and other heterochromatic Swi6 pools.
How does cohesin participate in subtelomeric heterochromatin maintenance? The simplest hypothesis is certainly that cohesin acts locally on Swi6 binding in the subtelomeric region. Within centromeric heterochromatin, cohesin is recruited in a Swi6-dependent manner, presumably through direct contact with the cohesin subunit Psc3 (38). In addition, Swi6 was found to interact with the cohesin loader Mis4 (15). The mechanism by which cohesin helps to maintain Swi6 at the subtelomeres might similarly involve Psc3 or an interaction between Swi6 and the loader Mis4. Why, then, is the subtelomeric Swi6 pool sensitive to cohesin inactivation while the centromeric pool is not? Swi6 binds H3K9me. Importantly, we found here that the subtelomeric H3K9me is Swi6 dependent, a situation reminiscent of that described for the spreading of H3K9me at the mating-type locus (22). This interdependency establishes a negative cooperativity relationship between Swi6 binding and H3K9me maintenance, so that even a slight destabilization of Swi6 might cooperatively lead to a complete collapse of subtelomeric heterochromatin. The cohesin-Swi6 and/or Mis4-Swi6 interaction might for this reason be particularly critical in the subtelomeric region. Alternative hypotheses in which the depletion of subtelomeric Swi6/H3K9me would be a consequence of a more complex effect of cohesin inactivation on euchromatin/heterochromatin distribution cannot be excluded.
Acknowledgments
We thank Guillaume Drutel for sharing his expertise in transcriptomic data analysis. We thank Laurent Maillet for helpful discussions.
This work was supported by the Centre National de la Recherche Scientifique, l'Université Victor Segalen Bordeaux 2, la Région Aquitaine, and grants from l'Association pour la Recherche sur le Cancer and l'Agence Nationale de la Recherche (BLAN06-2_135754).
Footnotes
Published ahead of print on 28 December 2010.
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J.-P. Javerzat, and G. Granston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Ayté, J., et al. 1995. The Schizosaccharomyces pombe MBF complex requires heterodimerization for entry into S phase. Mol. Cell. Biol. 15:2589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, P., et al. 2006. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr. Biol. 16:875-881. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, P., et al. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, P., et al. 2008. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 27:111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birkenbihl, R. P., and S. Subramani. 1992. Cloning and characterization of rad21, an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20:6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenbihl, R. P., and S. Subramani. 1995. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J. Biol. Chem. 270:7703-7711. [DOI] [PubMed] [Google Scholar]
- 9.Boscheron, C., et al. 1996. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 15:2184-2195. [PMC free article] [PubMed] [Google Scholar]
- 10.Bose, T., and J. L. Gerton. 2010. Cohesinopathies, gene expression, and chromatin organization. J. Cell Biol. 189:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cam, H. P., et al. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37:809-819. [DOI] [PubMed] [Google Scholar]
- 12.Chikashige, Y., et al. 2007. Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct. Funct. 32:149-161. [DOI] [PubMed] [Google Scholar]
- 13.Ciosk, R., et al. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243-254. [DOI] [PubMed] [Google Scholar]
- 14.Deardorff, M. A., et al. 2007. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 80:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, T., et al. 2009. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 106:8998-9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fourel, G., E. Lebrun, and E. Gilson. 2002. Protosilencers as building blocks for heterochromatin. Bioessays 24:828-835. [DOI] [PubMed] [Google Scholar]
- 17.Gard, S., et al. 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 187:455-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaspar-Maia, A., et al. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460:863-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glynn, E. F., et al. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2:E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gullerova, M., and N. J. Proudfoot. 2008. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132:983-995. [DOI] [PubMed] [Google Scholar]
- 22.Hall, I. M., et al. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 23.Halme, A., S. Bumgarner, C. Styles, and G. R. Fink. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405-415. [DOI] [PubMed] [Google Scholar]
- 23a.Huang, C. E., M. Milutinovich, and D. Koshland. 2005. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaac, S., et al. 2007. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 175:1549-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagey, M. H., et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Kjaerulff, S., J. Davey, and O. Nielsen. 1994. Analysis of the structural genes encoding M-factor in the fission yeast Schizosaccharomyces pombe: identification of a third gene, mfm3. Mol. Cell. Biol. 14:3895-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köhrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 27.Krantz, I. D., et al. 2004. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 36:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lengronne, A., et al. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linder, T., et al. 2008. Two conserved modules of Schizosaccharomyces pombe Mediator regulate distinct cellular pathways. Nucleic Acids Res. 36:2489-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, J., et al. 2009. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7:e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Losada, A., M. Hirano, and T. Hirano. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 33.Misulovin, Z., et al. 2008. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117:89-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 35.Musio, A., et al. 2006. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 38:528-530. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 37.Nasmyth, K., and C. H. Haering. 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43:525-558. [DOI] [PubMed] [Google Scholar]
- 38.Nonaka, N., et al. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89-93. [DOI] [PubMed] [Google Scholar]
- 39.Parelho, V., et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422-433. [DOI] [PubMed] [Google Scholar]
- 40.Pauli, A., et al. 2008. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell 14:239-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters, J. M., A. Tedeschi, and J. Schmitz. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev. 22:3089-3114. [DOI] [PubMed] [Google Scholar]
- 42.Rollins, R. A., M. Korom, N. Aulner, A. Martens, and D. Dorsett. 2004. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 24:3100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rollins, R. A., P. Morcillo, and D. Dorsett. 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152:577-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaaf, C. A., et al. 2009. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One 4:e6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, C. K., N. Brookes, and F. Uhlmann. 2009. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 10:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuldiner, O., et al. 2008. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell 14:227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skibbens, R. V., J. Marzillier, and L. Eastman. 25 April 2010. Cohesins coordinate gene transcriptions of related function within Saccharomyces cerevisiae. Cell Cycle 9:1601-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomonaga, T., et al. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14:2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tonkin, E. T., T. J. Wang, S. Lisgo, M. J. Bamshad, and T. Strachan. 2004. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 36:636-641. [DOI] [PubMed] [Google Scholar]
- 51.Vermaak, D., and H. S. Malik. 2009. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 43:467-492. [DOI] [PubMed] [Google Scholar]
- 52.Wendt, K. S., et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796-801. [DOI] [PubMed] [Google Scholar]
- 53.Wirén, M., et al. 2005. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 24:2906-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, A. J., A. F. Severson, and B. J. Meyer. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11:391-404. [DOI] [PMC free article] [PubMed] [Google Scholar]