Abstract
Early thymocytes possess multilineage potential, which is progressively restricted as cells transit through the double-negative stages of T-cell development. DN1 cells retain the ability to become natural killer cells, dendritic cells, B cells, and myeloid cells as well as T cells, but these options are lost by the DN3 stage. The Notch1 signaling pathway is indispensable for initiation of the T-cell lineage and inhibitory for the B-cell lineage, but the regulatory mechanisms by which the T-cell fate is locked in are largely undefined. Previously, we discovered that the E-protein transcription factor HEBAlt promoted T-cell specification. Here, we report that HEB−/− T-cell precursors have compromised Notch1 function and lose T-cell potential. Moreover, reconstituting HEB−/− precursors with Notch1 activity enforced fidelity to the T-cell fate. However, instead of becoming B cells, HEB−/− DN3 cells adopted a DN1-like phenotype and could be induced to differentiate into thymic NK cells. HEB−/− DN1-like cells retained GATA3 and Id2 expression but had lower levels of the Bcl11b gene, a Notch target gene. Therefore, our studies have revealed a new set of interactions between HEB, Notch1, and GATA3 that regulate the T-cell fate choice in developing thymocytes.
T cells develop in the thymus from bone marrow-derived hematopoietic stem cells. The earliest T-cell progenitors are identified within the CD4− CD8− double-negative (DN) subset, which can be subdivided into four subsets based on the expression of CD44 and CD25 (22). The most immature subset of cells is referred to as DN1 (CD44+ CD25−). This subset is heterogeneous and possesses a differential capacity to generate T cells (3, 35). DN1 cells become specified to the T lineage upon Notch1-Delta-like (Notch1-DL) interactions in the thymus, resulting in transition to the DN2 (CD44+ CD25+) stage and culminating in the DN3 (CD44− CD25+) stage, when T-lineage commitment occurs. At the DN2 to DN3 stages, lymphocyte and T-cell-specific factors, such as Rag-1, Rag-2, pTα, CD3ɛ, and interleukin-7 (IL-7) receptor alpha (IL7Rα; CD127), are upregulated (45), and the TCRβ genes are rearranged and assembled into pre-T-cell receptor (pre-TCR) complexes. Signaling through the pre-TCR is required for survival, proliferation, and further differentiation of DN3 cells, and this progression is known as β selection (26).
At the DN2 stage, T-cell precursors express IL7Rα and c-kit (CD117) and are thus responsive to both IL-7 and stem cell factor (SCF). However, by the late DN3 stage, both of these receptors are downregulated, rendering DN3 cells dependent solely on pre-TCR and Notch1-DL signaling for survival (14). As a consequence, the cells that do not undergo β selection die, unless they have committed to the TCRγδ lineage. Following β selection, the cells transit to the DN4 (CD44− CD25−) stage, upregulate CD4 and CD8, and become double-positive (DP; CD4+ CD8+) cells. At the DP stage, TCRα gene rearrangements take place, which lead to pairing of TCRα with TCRβ chains into a TCR complex and further selection processes that generate CD4+ or CD8+ single-positive (SP) cells.
Early thymocytes possess multilineage potential, which is progressively restricted as cells transit through the DN stages. In addition to T cells, DN1 cells have limited potential to become B cells, natural killer (NK) cells, dendritic cells (DC), or myeloid cells (2, 6, 11, 13). The generation of these cells depends heavily on the availability and dosages of specific transcription factors and a timely interplay between them. DN1 cells express GATA3, PU.1, and Id2, and upon Notch-DL signaling upregulate Bcl11b (33) and the E protein HEBAlt (34, 48). Increasing the expression level of PU.1 or GATA3, however, blocks T-cell development (4), resulting in the generation of myeloid cells (5) or mast cells (44), respectively. Compromised Notch1-DL signaling, on the other hand, leads to intrathymic B-cell development at the expense of T cells (36, 37). Furthermore, deficiency in Bcl11b or overexpression of Id factors is known to promote NK cell development (25, 27, 32, 33). It is clear that E-protein transcription factors are important determinants of hematopoietic choice (16). However, the role of each specific E protein in lineage determination is not well defined.
The E proteins constitute a basic helix-loop-helix (bHLH) family of transcription factors that function as heterodimers or homodimers and bind to E-box elements on DNA. The family consists of three genes, each encoding two transcription factors: the E2A gene (which encodes E47 and E12), the E2-2 gene (E2-2Can and E2-2Alt), and the HEB gene (HEBCan and HEBAlt) (30). Gene knockout studies have demonstrated the importance of E2A in B-cell development (8) and HEB in early T-cell development (9, 10), while E2-2 factors have been linked to plasmacytoid DC development (15). Recently, HEB factors were shown to be important in the generation of invariant NKT cells as well (17). We have shown that HEBAlt uniquely promotes T-cell specification (48) and that the loss of HEB has detrimental effects on β selection (12). Here, we report that a subset of HEB−/− T-cell precursors with compromised Notch1 and pre-TCR signaling survives inappropriately in an IL-7-dependent manner. However, unlike Notch1-deficient cells, HEB−/− cells were unable to become B cells. Instead, they inappropriately modulated expression of GATA3, Bcl11b, and Id2 and could be redirected to the thymic NK cell lineage. Our studies thus reveal new sets of interactions between HEB, Notch1, and GATA3 during early T-cell development that impact differentiation, fate choice, and survival.
MATERIALS AND METHODS
Mice, cells, and cell lines.
WT, HEB+/−, Rag-1−/−, and HEB+/− Rag-1−/− mice, all on the C57BL/6 background, were maintained in the comparative research facility at Sunnybrook Research Institute (SRI). All animal protocols were approved by the animal care committee at SRI. Rag-2EGFP (FVB) reporter mice were purchased from the Jackson Laboratory and bred with HEB+/− mice to generate HEB+/− Rag-2EGFP (C57BL/6/FVB) mice. OP9-DL1 and OP9-green fluorescent protein (GFP) cells (obtained from J. C. Zúñiga-Pflücker, SRI and University of Toronto), fetal liver cocultures, and fetal thymic organ cultures (FTOC) were maintained in OP9 medium (40).
Isolation of fetal liver hematopoietic stem cells.
Since HEB−/− mice are embryonic lethal on the C57BL/6 background, timed matings were set up between HEB+/− mice (or HEB+/− Rag-2EGFP or HEB+/− Rag−/− mice) to generate embryonic day 14.5 (E14.5) fetal livers and thymic lobes. Fetal liver cells of like genotypes (wild type [WT] or null) were pooled and lineage depleted with antibodies against F4/80, Gr1, Ter119, and CD19 using magnetically activated cell sorting (Miltenyi Biotec). Lineage-negative (Lin−) cells were cultured overnight in OP9 medium supplemented with 10 ng/ml of IL-7, Flt3L, and SCF (R&D Systems).
Fetal liver cell sorting and flow cytometry.
Lin− (CD11b, Gr1, F4/80, Ter119) Sca-1+ c-kit+ (LSK) populations were sorted using a FACSDiVa or FACSAria cell sorter (Becton Dickinson). For some experiments, LSK cells were cryopreserved in 90% fetal bovine serum (FBS) and 10% dimethyl sulfoxide (DMSO) before coculture, which had no detectable impact on phenotype. Flow cytometry was performed using a FACSCalibur or LSRII system (Becton Dickinson). Data were analyzed with FlowJo (Tree Star) software.
Differentiation of cells in vitro.
LSK and DN3 cells were cocultured with OP9-DL1 cells supplemented with 5 ng/ml of IL-7 and Flt3L, unless otherwise indicated (see Fig. 6 and Table 1). For gene expression studies, OP9-DL1 cell-derived DN3 (CD4− CD8− CD27+ CD44− CD25+) subsets were sorted after day 7 or 8 of coculture. The DN3 cells were replated on OP9-DL1 cells, and after 7 additional days in coculture, the DN1-like (CD4− CD8− Thy-1+ CD44+ CD25−) cells were sorted. In some experiments, CD45 was used in place of CD27 or Thy-1 as a marker of hematopoietic lineage cells. In the retroviral transduction experiments, T-cell precursors at day 7 of OP9-DL1 cell coculture were transduced by spin infection as previously described (48). Cells were cultured overnight on OP9-DL1 cells and sorted for the GFP+ CD45+ cells, which were incubated on OP9-DL1 cells for up to 19 days. For thymic NK (tNK) cell potential assays, coculture-generated DN1-like cells were seeded onto OP9-GFP cells and cultured with 5 ng/ml IL-7 and 20 ng/ml IL-15 (tNK cytokines). In IL-7-dependence experiments, coculture-derived HEB−/− Rag−/− DN3 cells were seeded at a density of 2 × 104 cells/well of a 6-well plate in suspension cultures supplemented with 5 ng/ml of IL-7 or without IL-7. For the cytokine profiling assay, DN1-like cells were cultured with the tNK cell cytokines on OP9-GFP cells for 7 days. Medium was collected and tested using a Proteome Profiler mouse cytokine array (R&D Systems). Pictures of the cocultures were taken using an Axiovert 25 microscope (Zeiss) and AxioVisionLE software (Zeiss).
TABLE 1.
Percentages of cells on day 4 of culture with or without IL-7
| Cell type | % cells at indicated stage on day 4a |
|||
|---|---|---|---|---|
| DN4-like |
DN1-like |
|||
| −IL-7 | +IL-7 | −IL-7 | +IL-7 | |
| HEB+/+ Rag−/− | — | — | — | — |
| HEB−/− Rag−/− | — | 64.3 ± 11.2 | — | 35.7 ± 11.2 |
Shown are results ± standard deviations for cultures with or without IL-7. DN4-like cells, CD27+ CD4− CD8− CD44− CD25−; DN1-like cells, CD27+ CD4− CD8− CD44+ CD25−; −IL-7, without IL-7; +IL-7, with IL-7; —, none surviving.
Genomic DNA extraction and PCRs.
Genomic DNA was isolated using a DNeasy kit (Qiagen). For analysis of Dβ2-Jβ2 rearrangements, DN3 cells were sorted from coculture-derived T-cell precursors from WT and HEB−/− progenitors, and C57BL/6 and Rag-1−/− thymocytes. Thymi were disaggregated mechanically. Following the isolation of genomic DNA, 50 ng of template was amplified with Taq polymerase (MBI/Fermentas) and a PTC-255 Peltier thermal cycler (MJ Research). Primers and conditions were as previously described (38, 51).
Reverse transcriptase and real-time PCRs.
Total RNA was isolated from sorted cell populations with Trizol (Invitrogen) and converted into cDNA using Superscript III (Invitrogen). Quantitative real-time PCR reactions were performed with SYBR green (Bio-Rad) and 2.5 μM gene-specific primers. For a list of the primers, see Table S1 in the supplemental material. Reactions were run and analyzed using an Applied Biosystems 7000 sequence detection system.
Statistics.
Cellularity, cell percentages, and gene expression values were analyzed by Student's t test with one- or two-tailed analysis (as specified in the figure legends). Data are means, and error bars show standard errors of the means. When only two experiments were done, error bars show standard deviations.
RESULTS
HEB deficiency allows conversion of DN3 precursors to a DN1-like state.
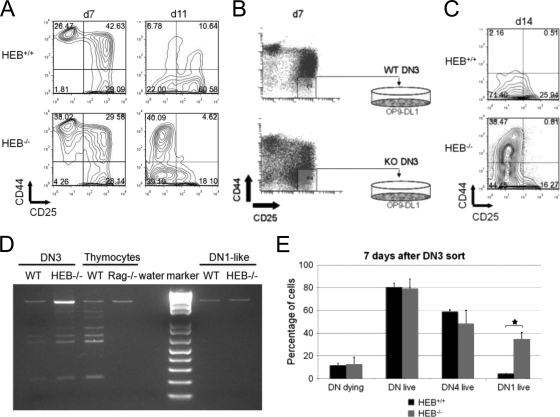
We (12) and others (9) have shown that HEB deficiency inhibits T-cell development at the β-selection checkpoint. However, the fate of the HEB−/− precursors that fail β selection has not been previously determined. To address this question, we monitored the kinetics of T-cell differentiation in WT versus HEB−/− precursors using the OP9-DL1 coculture system (41). Since the HEB-null allele is embryonic lethal, we were restricted to using fetal progenitors. Timed matings were set up between HEB+/− mice to obtain HEB−/− E14.5 fetal livers, which contain the early hematopoietic progenitor cells included within the LSK cell population. LSK cells were sorted and cocultured with OP9-DL1 cells, which express the Notch ligand Delta-like 1 and support T-cell development in vitro (41). After 7 days of coculture, both WT and HEB−/− precursors were passing through the DN2 and DN3 cell stages of T-cell development (Fig. 1A). By day 11 of coculture, DN3 and DN4 cells were present in WT cultures, and these cells ultimately differentiated into DP cells (data not shown). HEB−/− precursors, however, did not become DP cells (12). Moreover, HEB−/− precursors did not generate DP cells in fetal thymic organ culture or from fetal thymocytes cultured on OP9-DL1 cells (see Fig. S1A in the supplemental material). Instead, HEB−/− precursors generated cells that phenotypically resembled DN1 and DN4 cells (Fig. 1A) (see Fig. S1B in the supplemental material).
FIG. 1.
HEB deficiency allows conversion of DN3 precursors to a DN1-like state. E14.5 fetal liver cells were lineage (Lin [Ter119, Gr1, F4/80, CD19, CD11b]) depleted and sorted for the Sca-1+ c-kit+ (LSK) cell population. LSK cells were cultured on OP9-DL1 cells in the presence of 5 ng/ml IL-7 and Flt3L. (A) Developmental progression to the DN stages was assessed at day 7 and day 11 of coculture. The cells were gated on the Thy-1+ populations. (B) Coculture-derived DN3 cells (CD27+ CD4− CD8− CD44− CD25+) were sorted at day 7 and replated onto fresh OP9-DL1 cells supplemented with 5 ng/ml IL-7 and Flt3L. (C) Developmental progression to the DN4 stage was assessed 7 days later. (D) Genomic DNA was isolated from the OP9-DL1 coculture-derived DN3 cells (day 7 coculture; see Fig. S2A in the supplemental material) and DN1-like cells (day 14 coculture; see Fig. S2B in the supplemental material) and used in analysis of TCRβ D-J rearrangements by PCR. WT (C57BL/6) thymocytes were used as a positive control, while Rag-1−/− thymocytes were used as a negative control. (E) Coculture-derived DN3 cells were sorted at day 7 and replated onto fresh OP9-DL1 cells supplemented with 5 ng/ml IL-7 and Flt3L. Seven days later, the cells were scored for dying cells (DAPI− [4′,6′-diamidino-2-phenylindole-negative] annexin V+) and live cells (DAPI− annexin V−). Total DN (CD45+ CD4− CD8−), DN4 (CD45+ CD4− CD8− CD44− CD25−) or DN1-like (CD45+ CD4− CD8− CD44+ CD25−) cell percentages are shown as means ± standard errors (n = 3 experiments). The star indicates statistical significance. P value, 0.045 for DN1-like live cells (two-tailed Student's t test).
To determine whether the DN1-like cells arose from an expansion of the DN1 population present at day 7 or from DN3 cells converting to a DN1-like phenotype, we sorted DN3 cells from day 7 cocultures and plated them on fresh OP9-DL1 cells (Fig. 1B). Seven days later, HEB−/− DN3 cells had given rise to DN1-like and DN4-like cells, whereas the WT cultures had mostly DN3 and DN4 cells (Fig. 1C). Therefore, a portion of the DN1-like population in the HEB−/− cultures represented diverted DN3 precursors, indicating a potential defect in T-lineage commitment. The intriguing emergence of the DN1-like population from HEB−/− DN3 cells prompted us to assess the cell surface expression of CD19, NK1.1, CD11b, and TCRγδ. However, none of these lineage markers were observed (data not shown).
DN1-like cells arise from HEB−/− DN3 cells with the TCRβ gene in a germ line configuration.
To assess whether DN1-like cells arose directly from DN3 cells that had undergone TCRβ gene rearrangements, we sorted DN3 cells from day 7 OP9-DL1 cocultures, which were replated onto fresh OP9-DL1 cells and cultured for 7 additional days (day 14). The DN3 cells from day 7 cocultures and the DN1-like cells from day 14 cocultures (see Fig. S2A and S2B, respectively, in the supplemental material) were sorted, and a genomic PCR was performed to examine the rearrangement status of the TCRβ genes. While HEB−/− DN3 cells displayed Dβ-Jβ rearrangements, the TCRβ genes of the DN1-like cells were in germ line configuration (Fig. 1D). It is worth noting that a small population of DN1-like cells with germ line TCRβ genes was also generated in the WT cocultures. Therefore, the DN1-like population arose from DN3 cells that had not undergone TCRβ gene rearrangements.
Cells that fail β selection normally die by apoptosis. Since HEB−/− cells cannot efficiently pass through β selection, we assessed the percentages of apoptotic cells in the WT versus HEB−/− cultures arising from sorted DN3 cells 7 days later. However, we did not observe higher rates of apoptosis in HEB−/− cultures, even between the DN4 populations (Fig. 1E), which presumably contained cells failing β selection. We also observed no decrease in overall cell numbers (data not shown). However, some of the DN4-like cells were likely in transit toward becoming DN1-like, and the DN1-like cells in HEB−/− cultures showed significant increases in survival (Fig. 1E). Therefore, it was possible that preferential survival of HEB−/− cells without rearranged TCRβ genes accounted for the similar percentages of total DN live cells in HEB−/− and WT cultures.
Complete diversion of HEB−/− Rag-1−/− precursors to the DN1-like state.
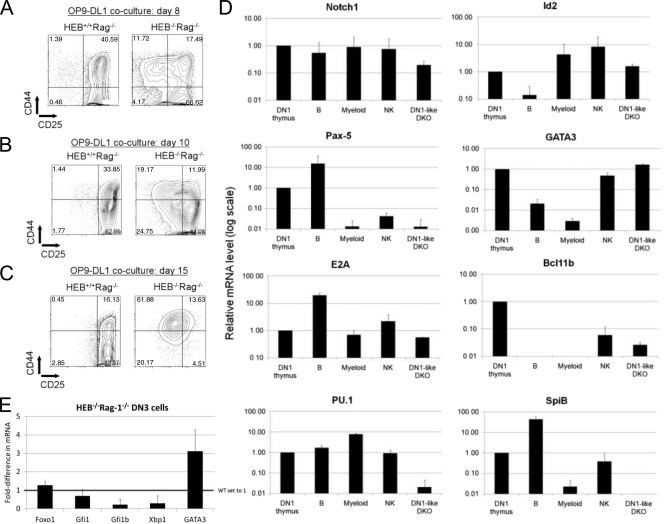
Our results suggested that the majority of HEB−/− precursors with TCRβ gene rearrangements died, whereas those without rearrangements generated DN1-like cells. To separate these two populations, we first used Rag-1−/− precursors. Rag-1−/− cells lack recombination machinery and therefore do not initiate gene rearrangements. Moreover, they are fully blocked in development at the DN3 stage. Hence, we bred HEB+/− Rag-1−/− mice to generate HEB−/− Rag-1−/− embryos, and we cultured E14.5 fetal liver LSK cells on OP9-DL1 cells for up to 15 days. By day 8 of the coculture, most of the HEB+/+ Rag-1−/− cells had become DN2- to DN3-stage cells, while approximately 10% of the HEB−/− Rag-1−/− cells remained at the DN1 stage, consistent with a partial block in this transition (Fig. 1A and 2A) (48). At day 10, however, it appeared that the HEB−/− Rag-1−/− DN3 cells were downregulating CD25 and upregulating CD44 (Fig. 2B), and by day 15, nearly all cells had adopted the DN1-like phenotype (Fig. 2C). Sorting and replating of HEB−/− Rag-1−/− DN3 cells also produced the same population of DN1-like cells, indicating that it was not merely an expansion of the enriched DN1 population present early in the cultures (data not shown). In contrast, the HEB+/+ Rag-1−/− cells remained primarily at the DN2 and DN3 stages of T-cell development.
FIG. 2.
Diversion of HEB−/− Rag-1−/− precursors to the DN1-like state reveals maintenance of GATA3 and Id2 expression. E14.5 fetal liver-derived HEB−/− Rag-1−/− LSK cells were lineage depleted and cultured on OP9-DL1 cells as described in the legend to Fig. 1. (A to C) Developmental progression was assessed at the indicated time points by flow cytometry. Lymphocytes were gated on the CD45+ CD4− CD8− fraction. (D) DN3 cells were sorted at day 7 of OP9-DL1 coculture and replated on OP9-DL1 cells to generate DN1-like cells. Seven days later, DN1-like cells were sorted and used for RNA isolation. cDNA from DN1-like HEB−/− Rag-1−/− (DKO) cells was subjected to real-time quantitative PCR analysis. DKO cells were compared with the DN1 cells isolated from a WT thymus (CD45+ CD4− CD8− CD44+ CD25−), B cells (CD19+), myeloid cells (CD11b+), and NK cells (CD49b+). Samples were normalized to β-actin levels. All samples are shown on a log scale relative to the DN1 thymus value, which was set to 1. Data are means ± standard deviations (n = 2). (E) DN3 cells were sorted at day 7 of OP9-DL1 coculture and used for RNA isolation. cDNA from HEB+/+ Rag-1−/− and HEB−/− Rag-1−/− cells was analyzed by real-time quantitative PCR. Samples were normalized to β-actin levels. Sample values are shown relative to that for HEB+/+ Rag-1−/−, which was set to 1. Data are means ± standard deviations (n = 2).
Phenotypically, DN1-like cells resembled DN1 and DN2 intermediate stages of T-cell development, but they could not give rise to T cells. To assess the molecular nature of these cells, we sorted HEB−/− Rag-1−/− DN1-like cells and analyzed the expression of several key lineage regulatory genes (46). The mRNA levels of these genes in HEB−/− Rag-1−/− (DN1-like DKO) were compared with the levels found in B cells, myeloid cells, NK cells, and thymic DN1 cells (Fig. 2D). Levels of Pax5, PU.1 and SpiB, which can bias cells toward alternative lineages, were decreased in HEB−/− Rag1−/− DN1-like cells compared to levels in the WT DN1 cells. E2A, Id2, and GATA3 mRNA levels were similar in DN1 thymocytes and DN1-like cells. Bcl11b and Notch1 expression, however, was lower in DN1-like cells than in DN1 thymocytes. Therefore, the molecular signature indicated that DN1-like cells were not the same as WT DN1 thymocytes and that they did not appear to be turning into myeloid or B cells.
Since the DN1-like cells arose from the DN3 cells, we set out to determine whether the expression of key T-lineage genes was perturbed in sorted HEB−/− Rag-1−/− DN3 cells from day 7 cocultures (Fig. 2E). Expression of Notch1 mRNA was slightly lower (data not shown). However, expression of GATA3 mRNA was elevated in HEB−/− Rag-1−/− DN3 cells compared to that in the control HEB+/+ Rag-1−/− DN3 cells. Interestingly, it has been previously shown that expression of GATA3 is increased in the absence of E2A (28) and that E2A decreases expression of GATA3 by upregulating Gfi1b (52). We therefore assessed the expression of Gfi1b and found that it was lower in HEB−/− Rag-1−/− DN3 cells than in the control cells. Since this profile was similar to that for the E2A−/− cells, we also examined the expression of additional E2A target genes (28, 43). The level of expression of Xbp-1 was also slightly lower in the HEB−/− Rag-1−/− cells than in the control, similar to the case for the E2A−/− cells. However, expression of Foxo1, which is induced by E2A, and that of Gfi1, which is downregulated by E2A, were not perturbed in HEB−/− Rag-1−/− cells. Therefore, while E2A and HEB share a subset of target genes, E2A appears to regulate other genes independently of HEB.
Loss of HEB does not perturb Rag-2 expression at the DN2 and DN3 stages.
The molecular phenotype of DN1-like cells arising from DN2- and DN3-stage cells in HEB−/− Rag-1−/− cultures indicated that HEB might be involved in the maintenance of T-lineage genes and in limiting survival of cells without TCRβ gene rearrangements. The germ line status of the TCRβ gene in DN1-like cells (Fig. 1D) suggested that Rag activity may have been compromised in HEB−/− cells. To further track the fates of HEB−/− DN3 cells, we used Rag-2EGFP transgenic reporter mice (54). In these mice, the expression of the enhanced GFP (EGFP) transgene is driven by the Rag-2 promoter, thus marking both DN and DP stages of TCR rearrangement. HEB+/− mice were bred with Rag-2EGFP transgenic mice, and HEB+/− Rag-2EGFP mice were mated to obtain E14.5 HEB−/− Rag-2EGFP and HEB+/+ Rag-2EGFP fetal liver cells. LSK cells derived from these mice were cultured on OP9-DL1 cells for 7 days, and DN subsets were assessed for the expression of Rag-2-driven EGFP by using a fluorescence-activated cell sorter (FACS).
The majority of the DN2 and DN3 cells in both WT and HEB−/− cocultures expressed EGFP, confirming that Rag-2 was expressed appropriately in the majority of the HEB−/− cells (Fig. 3A and C). Furthermore, sorted HEB−/− EGFP+ DN3 cells displayed Dβ-Jβ gene rearrangements, showing that the recombination machinery in these cells was functional (Fig. 3B). The percentages of EGFP+ cells in the DN1 and DN4 subsets varied somewhat between experiments. However, there tended to be a lower percentage of EGFP+ DN1 cells in the HEB−/− background (Fig. 3C). The levels of cellularity of DN3 EGFP+ and EGFP− cells were similar between WT and HEB−/− cells at day 7 of coculture (Fig. 3D).
FIG. 3.
Loss of HEB does not perturb Rag-2 expression at the DN2 and DN3 stages. (A) Day 7 of LSK culture on OP9-DL1 cells. Lymphocytes were gated on the CD45+ CD4− CD8− cells. Transgenic EGFP expression is driven by the Rag-2 promoter. (B) Genomic DNA was isolated from the OP9-DL1 coculture-derived WT and HEB−/− EGFP+ DN3 cells and used in analysis of TCRβ D-J rearrangements by PCR. C57BL/6 thymocytes were used as a positive control, while Rag-1−/− thymocytes were used as a negative control. (C) Percentages of EGFP+ DN subsets obtained from WT and HEB−/− cells at day 7 of OP9-DL1 cocultures. Data are means ± standard errors of the means (n = 4). (D) Cell numbers of EGFP+ and EGFP− DN3 cells at day 7 of OP9-DL1 cocultures. Data are means ± standard errors of the means (n = 4). WT, HEB+/+; KO, HEB−/−.
Selective diversion of DN3 cells lacking Rag-2 expression to the DN1-like state.
To track the fate of cells that had or had not expressed Rag-2 at the DN3 stage, we sorted EGFP+ DN3 cells (Fig. 4A, green) and EGFP− DN3 cells (Fig. 4A, gray) from day 7 WT and HEB−/− cocultures. These separated populations were then placed in OP9-DL1 cocultures for 7 additional days to test their likelihood of becoming DN1-like. After 7 days in culture, the EGFP+ DN3 cells (green) had given rise to two populations, (i) those that maintained EGFP expression (Fig. 4A, orange) and (ii) those that lost EGFP expression (Fig. 4A, teal). The EGFP− DN3 cells had also given rise to two populations, (i) those that gained EGFP expression (Fig. 4A, pink) and (ii) those that remained EGFP− (Fig. 4A, black).
FIG. 4.
Selective diversion of DN3 cells lacking Rag-2 expression to the DN1-like state. (A) Schematic representation of the Rag-2-driven EGFP expression in Rag-2EGFP T-cell precursors (as shown in Fig. 3). E14.5 fetal liver LSK cells were cultured on OP9-DL1 cells for 7 days to generate DN3 cells. Sorted EGFP+ (green circle) and EGFP− (gray circle) DN3 cells were cultured separately on OP9-DL1 cells supplemented with 5 ng/ml IL-7 and Flt3L for an additional 7 days. The progenies of these cells were detected by flow-cytometric analysis. Orange circle, EGFP+ cells arising from the EGFP+ cells. Teal circle, EGFP− cells arising from the EGFP+ cells. Pink circle, EGFP+ cells arising from the EGFP− cells. Black circle, EGFP− cells arising from the EGFP− cells. (B) Percentages of total EGFP+ and EGFP− cells arising from the day 7 sort. (C) Percentages of DN1-like cells within the populations defined in panel A. (D) Percentages of DP cells within the populations defined in panel A. For data shown in panels B to D, error bars show standard deviations (n = 2). WT, HEB+/+; KO, HEB−/−.
We first examined the modulation of EGFP expression in cultures arising from sorted DN3 cells. About half of the EGFP+ sorted WT cells maintained EGFP, whereas the majority of the equivalent HEB−/− population lost EGFP (and thus Rag-2) expression (Fig. 4B, Progeny of EGFP+ parent). Next, we examined EGFP expression in cultures derived from EGFP− DN3 cells (Fig. 4B, Progeny of EGFP− parent). Approximately 20% of the WT cells upregulated EGFP, consistent with the progression to the DP stage (Fig. 4D). However, the majority of the cells derived from EGFP− DN3 cells remained EGFP− (Fig. 4B). None of the HEB−/− DN3 cells developed to the DP stage (Fig. 4D), as expected.
We next determined the percentages of DN1-like cells arising in these cultures. Only small percentages of EGFP+ DN3 cells that maintained EGFP developed into DN1-like cells, with or without HEB (Fig. 4C, orange). However, approximately 40% of the HEB−/− cells that had downregulated EGFP became DN1-like (Fig. 4C, teal), indicating an increased likelihood of EGFP+ DN3 cells losing T-lineage identity in the absence of HEB. Cells that gained EGFP expression (Fig. 4C, pink) were also more likely to become DN1-like when HEB was absent. Nearly all the cells in the HEB−/− cultures derived from EGFP− cells that remained EGFP− became DN1-like (Fig. 4C, black). Interestingly, WT DN3 cells lacking EGFP expression also survived and developed into DN1-like cells (Fig. 4C, black), suggesting that they are normally inhibited from proliferating by T-cell precursors (Fig. 1 and 2).
Notch1 signaling is defective in HEB-deficient cells.
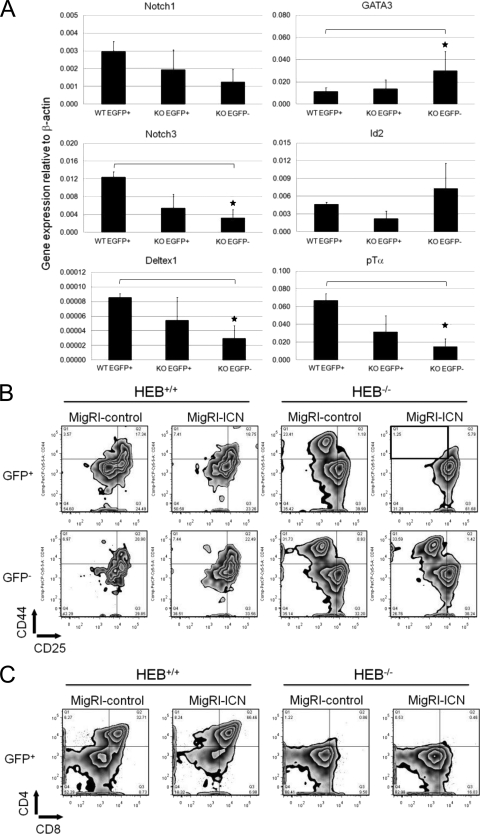
HEB and E2A are known regulators of the Notch1 receptor (53), and our results suggested that the expression and/or function of Notch1 might be compromised in HEB−/− DN3 cells. We therefore sorted EGFP+ (Fig. 4A, green) and EGFP− (Fig. 4A, gray) DN3 cells from HEB−/− cultures, and EGFP+ DN3 cells from WT cultures, at day 7 of coculture and compared their gene expression profiles (Fig. 5A). Notch1 expression was slightly lower in HEB−/− EGFP+ and HEB−/− EGFP− DN3 cells than in WT cells. However, the Notch1 target genes Notch3 and Deltex1 were expressed at significantly lower levels in the HEB−/− EGFP− cells, suggesting that Notch1 signaling was defective. pTα, a known target of HEB and Notch1 signaling, was also significantly lower in HEB−/− EGFP− DN3 cells. GATA3 and lck (data not shown) were expressed in these DN3 cells, consistent with a T-lineage program. Furthermore, the transcript levels of GATA3 were significantly higher in HEB−/− EGFP− DN3 cells than in WT DN3 cells (Fig. 5A). The expression of Id2 was not perturbed, while the expression of PU.1, C/EBPα, and Pax5 remained low (data not shown). WT EGFP− cell numbers were not consistently high enough to perform gene expression analysis; however, a preliminary characterization revealed patterns of expression similar to those of the HEB−/− EGFP− DN3 cells (data not shown).
FIG. 5.
Constitutively active Notch signaling restores T-lineage fidelity in HEB−/− cells. (A) LSK cells from HEB+/+ Rag-2EGFP and HEB−/− Rag-2EGFP fetal livers were cultured on OP9-DL1 cells for 7 days. Lymphocytes were gated on the CD45+ CD4− CD8− fraction and sorted for the EGFP+ and EGFP− DN3 cells, which were used in real-time quantitative reverse transcription-PCR (RT-PCR) analysis. Data are means ± standard errors of the means (n = 3). Stars indicate statistical significance. The P values were 0.0351 for Notch3, 0.0315 for pTα, and 0.0001 for GATA3 (one-tailed Student's t test). WT, HEB+/+; KO, HEB−/−. (B) Day 7 cocultures were transduced with either MigRI-control or MigRI-ICN constructs and incubated overnight. The next day, CD45+ GFP+ cells were sorted and cultured on OP9-DL1 cells with 5 ng/ml IL-7 and Flt3L for an additional 7 days. GFP+ and GFP− lymphocytes were gated on the CD45+ CD4− CD8− fraction. (C) Progression to the DP and SP stages was assessed at day 19 of coculture. Lymphocytes were gated on the GFP+ fraction. Data are representative of results of two independent experiments.
Constitutively active Notch signaling restores T-lineage fidelity.
These results revealed that Notch1 expression and signaling were affected by the loss of HEB. Therefore, we assessed whether commitment to the T-cell lineage could be rescued by the enforced expression of constitutively active intracellular Notch1 (ICN). Day 7 cocultures were transduced with either MigRI-control or MigRI-ICN retroviral constructs, and GFP+ transduced cells were sorted and replated on OP9-DL1 cells. Eight days later, the cocultures were assessed by flow cytometry for their developmental progression to the DN and DP stages (Fig. 5B and C). As expected, WT GFP+ cells from either control- or ICN-transduced cultures had progressed to the DN4 and later stages of T-cell development, whereas HEB−/− cells did not differentiate to the DP stage, even in the presence of ICN (Fig. 5C). HEB−/− cells transduced with control constructs generated DN1-like cells as expected; however, HEB−/− cells expressing ICN did not (Fig. 5B). Furthermore, HEB−/− cells that had downregulated GFP expression, and hence downregulated ICN expression, did acquire a DN1-like phenotype, indicating that sustained Notch1 signaling was required to maintain T-lineage fidelity. Overall, our results indicate that Notch1 signaling is perturbed in HEB−/− T-cell precursors, and this failure contributes to the loss of T-lineage identity and commitment.
HEB-deficient DN3 cells depend on IL-7 for survival.
Our results showed that both WT and HEB−/− T-cell precursors are capable of giving rise to DN1-like cells but that only HEB−/− DN1-like cells persist and accumulate in the fetal thymus (see Fig. S1B in the supplemental material). Enhanced proliferation and IL-7 dependence have been reported for HEB−/− E2A−/− thymocytes (50). Therefore, we assessed whether the emergence and survival of the DN1-like population was dependent on IL-7. HEB+/+ Rag-1−/− and HEB−/− Rag-1−/− LSK cells were placed in OP9-DL1 cocultures for 7 days, and DN3 cells were sorted and cultured in media with or without IL-7 for 4 days. Consistent with a previous report (14), HEB+/+ Rag-1−/− DN3 cells did not survive upon withdrawal of Notch-DL signals whether IL-7 was present or absent (Table 1). In contrast, HEB−/− Rag-1−/− DN3 cells survived, but only in the presence of IL-7. Importantly, DN3 cells were still diverted to the DN1-like state.
The persistence of DN1-like cells in vitro is in contrast to a previous finding (9) that HEB−/− DN cells remained predominantly at the DN3 stage. To assess whether this difference was due to the presence of a well-defined thymic structure, we injected coculture-derived DN3 cells into the thymus of sublethally irradiated recipients. Seven days later, the thymi were assessed for the presence of DN1 and DN4 cells (see Fig. S3 in the supplemental material). In agreement with results reported by Barndt et al. (9), DN1 cells did not accumulate in the postnatal thymus. This is consistent with the restricted availability of IL-7 in the cortex of the adult thymus (55). In contrast, the abundance of IL-7 in the fetal thymus (55) and OP9-DL1 cocultures would have supported the emergence and persistence of DN1-like cells in vitro.
HEB−/− DN1-like cells generate thymic NK cells.
In addition to playing an essential role in T-cell development, GATA3 has been implicated in thymic NK (tNK) cell development (47). The generation of tNK cells is strictly dependent on GATA3 and IL7Rα. We therefore assessed the ability of DN1-like cells to become tNK cells by culturing them on OP9-GFP cells with tNK cell cytokines. Since HEB+/+ Rag-1−/− cultures failed to produce a robust DN1-like population (as shown in Fig. 2A to C), we were unable include them in these experiments. By day 4 of coculture, DN1-like cells began to upregulate CD122, an immature NK cell marker (data not shown), while by day 7, approximately 50% of the cells expressed CD122 (Fig. 6 B). Moreover, by day 10 of coculture, 70% of the cells had upregulated DX5, a mature NK cell marker (Fig. 6A). The DX5+ cells also expressed high levels of IL7Rα, qualifying them as tNK cells. We also assessed tNK cell development in FTOC without the addition of exogenous cytokines (Fig. 6C). HEB+/+ Rag-1−/− FTOC contained approximately 18% NK cells, which is appropriate for the Rag-deficient background (31). In contrast, HEB−/− Rag-1−/− FTOC had approximately 75% NK cells. IL7Rα levels were intermediate in both cultures. These results show that although there is some residual tNK potential in HEB+/+ Rag-1−/− cells, tNK potential is greatly enhanced in the absence of HEB.
FIG. 6.
HEB−/− Rag-1−/− DN1-like cells have the potential to generate tNK cells. (A and B) DN1-like cells were generated from sorted DN3 cells plated onto OP9-GFP cells and incubated with 5 ng/ml IL-7 and 20 ng/ml IL-15. Upregulation of DX5, IL7Rα, and CD122 was monitored 7 and 10 days later by flow cytometry. Lymphocytes were gated on the CD45+ fraction. (C) Intact E14.5 fetal thymic lobes were cultured in fetal thymic organ culture for 7 days. Development of NK cells was assessed by flow cytometry. The plots were gated on the CD45+ fraction. (D) Pictures of HEB−/− Rag-1−/− DN1-like cells cocultured with OP9-GFP cells were taken at ×10 magnification, using an Axiovert 25 microscope and AxioVisionLE software at day 7. The top picture shows a healthy stromal monolayer at day 7 of coculture, while the bottom picture displays NK cells and disintegrated stromal cells. (E) Culture medium from day 7 of HEB−/− Rag-1−/− DN1-like cell coculture with OP9-GFP cells (as shown in panel D) was subjected to cytokine profiling. Culture medium collected at day 7 of OP9-GFP culture (as shown in panel D) provided background signals. Data are means ± standard deviations of duplicate readings.
To further verify the NK cell phenotype, DN1-like HEB−/− Rag-1−/− cells were cultured on OP9-GFP cells with tNK cytokines for up to 10 days. At day 7, nearly all of the stromal cells were killed in cocultures containing DN1-like cells (Fig. 6D), suggesting that NK cells were already in an activated state. Furthermore, we assessed the culture medium for the presence of inflammatory cytokines. NK cells in our cocultures produced gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Fig. 6E), consistent with the tNK cell phenotype as defined in a previous report (47).
DISCUSSION
Previously, E-box factors have been shown to reinforce multiple T-cell checkpoints. E2A deficiency led to inappropriate traversal through β selection (20), whereas dual E2A and HEB deficiencies resulted in altered CD4+ and CD8+ SP cell delineation (20, 29). HEB deficiency, in contrast, inhibited T-cell development at the β-selection checkpoint (12). Herein, we show that HEB is critical for early T-cell development. HEB deficiency in DN3 cells resulted in two separate phenomena. First, the lack of HEB permitted the escape from death in cells without DNA rearrangements. Second, cells lacking HEB retained lineage plasticity and were able to develop into tNK cells. Therefore, our studies have revealed specific roles for HEB factors in the maintenance of T-cell identity and the commitment to the T-lineage fate.
Prior to passing the pre-TCR signaling checkpoint, wild-type DN cells survive via a cytokine-dependent pathway. At the pre-TCR checkpoint, however, wild-type DN3 cells not rescued by pre-TCR signaling die by p53-induced cell death (24). We have previously shown that HEB−/− cells are defective at pre-TCR signaling (12). Since DN3 cells lacking HEB were capable of rearranging TCRβ genes, hence introducing DNA damage, this is likely a pathway by which a majority of the HEB−/− DN3 cells died. However, very little is known about the pathways that induce cell death in DN3 cells without gene rearrangements. One pathway by which such cells may be eliminated in the thymus involves the activity of FoxO3a, which upregulates Bim to induce apoptosis (34). This pathway would provide an additional quality-control step to ensure that only appropriate DN3 cells survive and compete for the limited thymic niches. However, HEB−/− DN3 cells still managed to escape death, even in the absence of DNA breaks. Therefore, it is plausible that HEB−/− DN3 cells might have circumvented the FoxO3a pathway by signaling through IL7R, which enhances survival through upregulation of Bcl-2 (1). Since careful balance of the proapoptotic and antiapoptotic factors is known to ensure proper selection of developing cells, it is possible that Bcl-2 levels were sufficiently high to allow survival of HEB−/− DN3 cells. Whether HEB factors directly regulate the expression of FoxO3a and/or Bcl-2 family members warrants further investigation.
HEB−/− cells were dependent on IL-7 for survival. While PU.1 is required for IL7R expression in B-cell precursors (18), the transcription factors modulating IL7Rα expression in T cells are still largely unknown. IL7Rα is upregulated at the transition from DN1 to DN2 and then is downregulated at the transition from DN3 to DN4. Recently, IL7Rα expression was found to be upregulated by Notch1 signaling in human T-cell development (23). This may be the case for mouse T lymphopoiesis as well, since the CSL/RBP-Jκ site in the IL7Rα regulatory region is well conserved (23). E2A and HEB may also be involved in the induction of IL7Rα expression. E2A−/− mice displayed a partial block at the transition from DN1 to DN2 (7), and fetal thymocytes from E2A−/− mice exhibited a dose-dependent decrease in expression of IL7Rα (28). On the other hand, the hyperproliferation observed for E2A−/− HEB−/− DN3 cells in response to IL-7 suggests that one or both of these factors may exert negative regulatory control on IL7R or its signaling components (50). Our results suggest that HEB alone antagonizes IL7R-dependent survival across β selection. Therefore, Notch1 and E proteins may provide positive signals for IL7Rα expression during the DN2 stage, whereas the DN3 stage may require negative regulators to be activated downstream of E proteins to turn off IL7Rα expression and render DN3 cells dependent on pre-TCR signaling.
While our studies showed that HEB factors may be involved in the regulation of survival, they have also revealed independent roles for HEB in the proper execution of a T-cell program. Many T-cell genes, such as the lck, GATA3, and TCRβ genes, were expressed in HEB−/− DN3 cells (12). This suggests that every effort was made to turn on genes associated with an early T-cell developmental program. However, Notch1 signaling was impaired in HEB−/− DN3 cells. Consistent with a role for HEB in regulating Notch1, Id overexpression has been shown to inhibit Notch1 signaling (49) and influence the T/NK cell fate decision (42). Furthermore, Notch1 signaling is critical for expression of Bcl11b, which reduces NK cell potential (27, 32, 33). In a different study, E-box elements in the regulatory region of Notch1 were responsive to both E2A and HEB factors and Id3-mediated repression of E-protein activity was shown to be responsible for downregulation of Notch1 receptor expression at the β-selection checkpoint (53). Moreover, Notch1 transcript levels were reduced in a dose-dependent manner in E2A−/− lymphoid cell-primed multipotent progenitors (19). Therefore, these studies combined indicate a critical role for HEB and E2A in Notch1 function during early T-cell development.
Our results suggest that one role of HEB factors could be to maintain appropriate levels of GATA3 during early T-cell development. GATA3 overexpression has been previously shown to abort T-cell development (4) and to be linked to Notch1 downregulation, which redirected cells into the mast cell lineage when an appropriate inductive environment was provided (44). In addition, GATA3 is also expressed in thymic NK cells (47). Our results support a model in which HEB restricts thymic NK cell potential by limiting the expression of both GATA3 and IL7Rα. Interestingly, the expression patterns of GATA3 and IL7Rα overlap during DN2 and DN3 stages of early T-cell development. While GATA3 expression persists throughout T-cell development, IL7Rα is downregulated by the DN4 stage (46). Hence, their timely intersection would suggest that thymic NK cells and T cells can be generated in equivalent proportions from a bipotent T/NK cell precursor. However, HEB expression, as well as limited IL-7 availability within a thymus, may shift the balance in favor of T-cell over tNK cell generation. Indeed, injection of DN3 cells lacking HEB directly into the thymus suppressed the outgrowth of NK and DN1-like cells. Further work will be necessary to determine whether any of the genes that are perturbed in the absence of HEB, such as the GATA3 or IL7Rα genes, are direct targets of HEB.
It is intriguing that no B cells, DC, or mast cells were generated from HEB−/− cells, particularly since perturbations in Notch1 signaling have led to the generation of B cells and DC in the thymus (21, 37). One possible explanation is that even a low level of Notch1 signaling coupled with a high level of GATA3 expression was sufficient to interfere with the expression of the key regulators of alternative lineages. Another possibility is that the regulatory regions of the Pax5, SpiB, and PU.1 genes were permanently silenced or that lineage-specific cytokine receptors were downregulated even in the absence of HEB. Our results thus indicate that a decrease in Notch1 signaling is not the sole consequence of HEB deficiency.
We therefore propose a working model of interactions between E2A, HEB, Notch1, and GATA3 at the DN3 stage of T-cell development as shown in Fig. 7. Both HEB and E2A positively regulate Notch1 expression, whereas Delta-Notch1 signaling, in turn, induces the expression of HEBAlt (48). Previous studies have shown that E2A negatively regulates GATA3 by inducing Gfi1b (52). Here, we provide the first evidence that HEB also provides a negative input for GATA3 expression. HEB-deficient cells also have decreased levels of Gfi1b, suggesting that HEB may downregulate GATA3 indirectly through Gfi1b. HEB may also regulate GATA3 directly, but more work is needed to assess this possibility. Interestingly, neither E2A nor HEB can compensate for the loss of the other, suggesting that the proper regulation of GATA3 expression is dependent on the total dosage of E proteins or on the presence of E2A/HEB heterodimers. One possibility is that HEBAlt is uniquely required to prevent diversion to the tNK cell lineage. However, our preliminary analysis of the HEB−/− Rag-1−/− precursors carrying an HEBAlt transgene under the control of the lck promoter indicated that the HEBAlt transgene alone was unable to fully suppress tNK cell potential (unpublished observation). This is in contrast to its ability to rescue T-cell development to the DP stage (12) and suggests unique roles for HEBCan in early T-cell development. Overall, our study reveals that HEB factors ensure appropriate survival and limited hematopoietic lineage choice in the thymus. Furthermore, it provides strong evidence for an intimate link among GATA3, Notch1, and HEB factors in regulating the developmental program leading to the generation of T cells.
FIG. 7.
Model of proposed HEB, GATA3, and Notch1 interactions. Both HEB and E2A positively regulate Notch1 expression, whereas Notch1-DL signaling, in turn, induces the expression of HEBAlt. These interactions establish a positive-feedback loop between HEB and Notch1-DL signaling. E2A negatively regulates GATA3 indirectly through Gfi1b. HEB also provides a negative input for GATA3 expression, possibly through Gfi1b. Neither E2A nor HEB can compensate for the loss of the other for the proper regulation of GATA3. In the absence of HEB, Gfi1b expression is decreased and GATA3 expression is increased, while the expression of Notch1 target genes (arrow pointing to ICN) is decreased. Green arrow, positive regulation; red blunt arrow, negative regulation; dashed blunt arrow, partial inhibition; gray arrows, disrupted connections in the absence of HEB.
Supplementary Material
Acknowledgments
We thank J. Carlyle for critical reviews of the manuscript. We are also grateful to P. Rajkumar and A. Moore for assistance in genotyping of mice and G. Knowles and A. Khandani for sorting expertise.
This work was supported by research grants from the Canadian Institute for Health Research (MOP82861) and the Leukemia Research Fund to M.K.A. and by an Ontario Graduate Scholarship to M.B.
Footnotes
Published ahead of print on 28 December 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akashi, K., M. Kondo, U. von Freeden-Jeffry, R. Murray, and I. L. Weissman. 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell 89:1033-1041. [DOI] [PubMed] [Google Scholar]
- 2.Akashi, K., L. I. Richie, T. Miyamoto, W. H. Carr, and I. L. Weissman. 2000. B lymphopoiesis in the thymus. J. Immunol. 164:5221-5226. [DOI] [PubMed] [Google Scholar]
- 3.Allman, D., et al. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168-174. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, M. K., et al. 2002. Definition of regulatory network elements for T cell development by perturbation analysis with PU.1 and GATA-3. Dev. Biol. 246:103-121. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, M. K., A. H. Weiss, G. Hernandez-Hoyos, C. J. Dionne, and E. V. Rothenberg. 2002. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16:285-296. [DOI] [PubMed] [Google Scholar]
- 6.Ardavin, C., L. Wu, C. L. Li, and K. Shortman. 1993. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature 362:761-763. [DOI] [PubMed] [Google Scholar]
- 7.Bain, G., et al. 1997. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 17:4782-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bain, G., et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 9.Barndt, R., M. F. Dai, and Y. Zhuang. 1999. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J. Immunol. 163:3331-3343. [PubMed] [Google Scholar]
- 10.Barndt, R. J., M. Dai, and Y. Zhuang. 2000. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 20:6677-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhandoola, A., H. von Boehmer, H. T. Petrie, and J. C. Zúñiga-Pflücker. 2007. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity 26:678-689. [DOI] [PubMed] [Google Scholar]
- 12.Braunstein, M., and M. K. Anderson. 2010. Developmental progression of fetal HEB−/− precursors to the pre-T-cell stage is restored by HEBAlt. Eur. J. Immunol. 40:3173-3182. [DOI] [PubMed] [Google Scholar]
- 13.Carlyle, J. R., and J. C. Zúñiga-Pflücker. 1998. Lineage commitment and differentiation of T and natural killer lymphocytes in the fetal mouse. Immunol. Rev. 165:63-74. [DOI] [PubMed] [Google Scholar]
- 14.Ciofani, M., and J. C. Zúñiga-Pflücker. 2005. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6:881-888. [DOI] [PubMed] [Google Scholar]
- 15.Cisse, B., et al. 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochrane, S. W., Y. Zhao, R. S. Welner, and X. H. Sun. 2009. Balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions. Blood 113:1016-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Cruz, L. M., J. Knell, J. K. Fujimoto, and A. W. Goldrath. 2010. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 11:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeKoter, R. P., H. J. Lee, and H. Singh. 2002. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity 16:297-309. [DOI] [PubMed] [Google Scholar]
- 19.Dias, S., R. Mansson, S. Gurbuxani, M. Sigvardsson, and B. L. Kee. 2008. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity 29:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel, I., C. Johns, G. Bain, R. R. Rivera, and C. Murre. 2001. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194:733-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feyerabend, T. B., et al. 2009. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity 30:67-79. [DOI] [PubMed] [Google Scholar]
- 22.Godfrey, D. I., J. Kennedy, T. Suda, and A. Zlotnik. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150:4244-4252. [PubMed] [Google Scholar]
- 23.Gonzalez-Garcia, S., et al. 2009. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7Rα gene expression in early human thymopoiesis and leukemia. J. Exp. Med. 206:779-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guidos, C. J., et al. 1996. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 10:2038-2054. [DOI] [PubMed] [Google Scholar]
- 25.Heemskerk, M. H., et al. 1997. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med. 186:1597-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman, E. S., et al. 1996. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 10:948-962. [DOI] [PubMed] [Google Scholar]
- 27.Ikawa, T., et al. 2010. An essential developmental checkpoint for production of the T cell lineage. Science 329:93-96. [DOI] [PubMed] [Google Scholar]
- 28.Ikawa, T., H. Kawamoto, A. W. Goldrath, and C. Murre. 2006. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 203:1329-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, M. E., and Y. Zhuang. 2007. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 27:860-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kee, B. L. 2009. E and ID proteins branch out. Nat. Rev. Immunol. 9:175-184. [DOI] [PubMed] [Google Scholar]
- 31.Laurent, J., N. Bosco, P. N. Marche, and R. Ceredig. 2004. New insights into the proliferation and differentiation of early mouse thymocytes. Int. Immunol. 16:1069-1080. [DOI] [PubMed] [Google Scholar]
- 32.Li, L., M. Leid, and E. V. Rothenberg. 2010. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329:89-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, P., et al. 2010. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal, M., et al. 2008. Regulation of lymphocyte progenitor survival by the proapoptotic activities of Bim and Bid. Proc. Natl. Acad. Sci. U. S. A. 105:20840-20845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porritt, H. E., et al. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20:735-745. [DOI] [PubMed] [Google Scholar]
- 36.Pui, J. C., et al. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299-308. [DOI] [PubMed] [Google Scholar]
- 37.Radtke, F., et al. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10:547-558. [DOI] [PubMed] [Google Scholar]
- 38.Rodewald, H. R., K. Kretzschmar, S. Takeda, C. Hohl, and M. Dessing. 1994. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 13:4229-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothenberg, E. V., J. E. Moore, and M. A. Yui. 2008. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8:9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt, T. M., et al. 2004. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat. Immunol. 5:410-417. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt, T. M., and J. C. Zúñiga-Pflücker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17:749-756. [DOI] [PubMed] [Google Scholar]
- 42.Schotte, R., et al. 2010. Synergy between IL-15 and Id2 promotes the expansion of human NK progenitor cells, which can be counteracted by the E protein HEB required to drive T cell development. J. Immunol. 184:6670-6679. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz, R., I. Engel, M. Fallahi-Sichani, H. T. Petrie, and C. Murre. 2006. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl. Acad. Sci. U. S. A. 103:9976-9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taghon, T., M. A. Yui, and E. V. Rothenberg. 2007. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 8:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taghon, T. N., E. S. David, J. C. Zúñiga-Pflücker, and E. V. Rothenberg. 2005. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 19:965-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tydell, C. C., et al. 2007. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J. Immunol. 179:421-438. [DOI] [PubMed] [Google Scholar]
- 47.Vosshenrich, C. A., et al. 2006. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 7:1217-1224. [DOI] [PubMed] [Google Scholar]
- 48.Wang, D., et al. 2006. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J. Immunol. 177:109-119. [DOI] [PubMed] [Google Scholar]
- 49.Wang, H. C., S. S. Perry, and X. H. Sun. 2009. Id1 attenuates Notch signaling and impairs T-cell commitment by elevating Deltex1 expression. Mol. Cell. Biol. 29:4640-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wojciechowski, J., A. Lai, M. Kondo, and Y. Zhuang. 2007. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J. Immunol. 178:5717-5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfer, A., A. Wilson, M. Nemir, H. R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta lineage thymocytes. Immunity 16:869-879. [DOI] [PubMed] [Google Scholar]
- 52.Xu, W., and B. L. Kee. 2007. Growth factor independent 1B (Gfi1b) is an E2A target gene that modulates Gata3 in T-cell lymphomas. Blood 109:4406-4414. [DOI] [PubMed] [Google Scholar]
- 53.Yashiro-Ohtani, Y., et al. 2009. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 23:1665-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, W., et al. 1999. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature 400:682-687. [DOI] [PubMed] [Google Scholar]
- 55.Zamisch, M., et al. 2005. Ontogeny and regulation of IL-7-expressing thymic epithelial cells. J. Immunol. 174:60-67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.