Abstract
Ras GTPases were long thought to function exclusively from the plasma membrane (PM). However, a current model suggests that Ras proteins can compartmentalize to regulate different functions, and an oncogenic H-Ras mutant that is restricted to the endomembrane can still transform cells. In this study, we demonstrated that cells transformed by endomembrane-restricted oncogenic H-Ras formed tumors in nude mice. To define downstream targets of endomembrane Ras pathways, we analyzed Cdc42, which concentrates in the endomembrane and has been shown to act downstream of Ras in Schizosaccharomyces pombe. Our data show that cell transformation induced by endomembrane-restricted oncogenic H-Ras was blocked when Cdc42 activity was inhibited. Moreover, H-Ras formed a complex with Cdc42 on the endomembrane, and this interaction was enhanced when H-Ras was GTP bound or when cells were stimulated by growth factors. H-Ras binding evidently induced Cdc42 activation by recruiting and/or activating Cdc42 exchange factors. In contrast, when constitutively active H-Ras was restricted to the PM by fusing to a PM localization signal from the Rit GTPase, the resulting protein did not detectably activate Cdc42 although it activated Raf-1 and efficiently induced hallmarks of Ras-induced senescence in human BJ foreskin fibroblasts. Surprisingly, PM-restricted oncogenic Ras when expressed alone could only weakly transform NIH 3T3 cells; however, when constitutively active Cdc42 was coexpressed, together they transformed cells much more efficiently than either one alone. These data suggest that efficient cell transformation requires Ras proteins to interact with Cdc42 on the endomembrane and that in order for a given Ras protein to fully transform cells, multiple compartment-specific Ras pathways need to work cooperatively.
Ras GTPases cycle between GDP- and GTP-bound states, and when they are GTP bound, they stimulate downstream effectors to regulate a wide range of signal transduction pathways (16). Ras activation is dependent on guanine nucleotide exchange factors (GEFs), and Ras inactivation is accelerated by GTPase activating proteins (GAPs). While Ras proteins are perhaps best known for their ability to mediate growth factor signaling to promote proliferation, deregulation of which causes cell transformation and tumorigenesis, Ras proteins also regulate many antiproliferative activities, such as apoptosis, differentiation, and senescence. How Ras proteins can regulate different functions with selectivity is of great importance but poorly understood.
In mammals, there are three RAS genes, H-RAS, N-Ras, and K-RAS. Their gene products are highly similar at the N termini, which contain the major binding sites for effectors. Not surprisingly, in vitro, these Ras proteins can all efficiently interact with many effectors. Despite this, there is evidence that in vivo Ras proteins may control different functions. For example, in mice, when the WNT1 oncogene is overexpressed in the mammary gland, the resulting tumors frequently contain H-RAS mutations (31); in contrast, K-RAS mutations are frequently detected when breast tumors are induced by MYC (7). In humans, most RAS mutations in tumors are in K-RAS, whereas H-RAS and N-RAS mutations are restricted to tumors of certain tissues (16). Furthermore, different Ras isoforms are selectively overexpressed in different subtypes of breast tumors. For example, H-RAS is selectively upregulated in luminal B and HER2+/ER− subtypes of human breast cancer, while N-RAS is upregulated in the basal-like subtype (10).
To decipher how Ras proteins can control different functions, one approach focuses on their C termini, the most divergent regions in these proteins (called the hypervariable region [HVR]). The C termini of Ras proteins undergo various lipidations, which play key roles in mediating protein subcellular localization (30, 55). All Ras proteins contain a CAAX motif, the cysteine residue of which can be farnesylated to allow association with the endomembrane (EM). However, association with the plasma membrane (PM) differs among Ras proteins: K-Ras4B associates with the PM via its polylysine residues, while H- and N-Ras do so via palmitoylation at their cysteine residues immediately upstream of the CAAX box. These different modes of PM association appear to affect where these proteins are found in the cell; at steady state, while N- and H-Ras can be readily detected on the PM as well as the endomembrane (e.g., Golgi compartment), K-Ras4B is nearly exclusively found on the PM. These observations support a model whereby Ras proteins can control different functions by localizing to different compartments.
One of the earliest tests of this model came when Hancock et al. (8) demonstrated that expression of an oncogenic H-Ras that is restricted to the endomembrane (e.g., by mutating its two cysteine residues for palmitoylation to serine) can transform NIH 3T3 cells (8), which was also later confirmed in a more detailed study (5). These observations suggest that the endomembrane can be a productive compartment from which Ras proteins can signal to induce cell transformation. However, it was unclear with which endogenous effectors Ras proteins interact on the endomembrane and whether cells transformed by endomembrane-restricted Ras are tumorigenic. We have recently further investigated this model in the fission yeast Schizosaccharomyces pombe, which unlike most mammalian cells has just one Ras protein, called Ras1. Our data show that Ras1 clearly compartmentalizes to activate two separate downstream pathways: PM-restricted Ras1 selectively activates a mitogen-activated protein (MAP) kinase module to regulate mating, while endomembrane-restricted Ras1 activates Cdc42 to regulate morphogenesis, transport, mitosis, and proteasome functions (3, 30). Cdc42 in mammalian cells is well documented to play a role in trans-Golgi network transport by regulating vesicle trafficking (1), and one of its effectors is COP-γ (56), a vesicle coat protein. Furthermore, overexpressing a dominant negative Cdc42 [Cdc42(17N)] can block Ras-induced cell transformation (33, 39). It thus seems likely that Ras can also act via Cdc42 on the endomembrane in mammalian cells to influence cell transformation.
In this study, we show that cells transformed by endomembrane-restricted oncogenic H-Ras efficiently form tumors in immunodeficient mice. We further present evidence that upon growth factor stimulation, activated H-Ras can turn on a Cdc42 pathway on the endomembrane, which is critical for transformation. H-Ras appears to activate Cdc42 by recruiting its GEF. In contrast, when activated H-Ras is restricted to the PM, it does not activate Cdc42 although it can fully activate Raf-1 and induce senescence in human primary cells. Intriguingly, the PM-restricted oncogenic H-Ras can transform NIH 3T3 cells only weakly by itself, but when constitutively active Cdc42 is coexpressed, together they transform cells much more efficiently than either one alone. These results suggest that H-Ras can act via Cdc42 on the endomembrane to transform cells and that this and other compartment-specific pathways need to operate in a concerted manner to efficiently transform cells.
MATERIALS AND METHODS
Cells and cell culture.
NIH 3T3 and BJ cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured according to ATCC protocols. HTC75 and 293FT (Invitrogen, Carlsbad, CA) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 2 mM l-glutamine. All transfection experiments were performed with Lipofectamine 2000 (Invitrogen). To create NIH 3T3 cells stably expressing both Cdc42(12V) and PM-Ras(61L), NIH 3T3 cells were transfected with either pcDNA-MYC-Cdc42(12V) or vector control and then grown in medium containing G418 (500 μg/ml) for 3 days. The selected G418-resistant cells were then infected twice with either vector control or pLenti4-PM-Ras(61L)-puro and further selected by puromycin (1 μg/ml). The resulting G418 and puromycin dually resistant cells were pooled. Western blotting showed that levels of Cdc42(12V) and PM-Ras(61L) in cells that expressed both proteins were comparable to those of cells in which only one of the proteins was expressed (data not shown).
Plasmid construction.
The C-terminal PM localization signal from Rit (Rit-C) (30) was fused to the 3′ coding sequence of H-Ras(61L,186S) by two rounds of PCR using pcDNA-H-Ras(61L,186S) (5) and pEGFP-Rit (where EGFP is enhanced green fluorescent protein) (30) as templates and the following primers (5′-3′): the primer pair ATGACGGAATATAAGCTGGTGGTGGTGGGC and GTATGCAGCAGATGTCTCAAAAAAGGGACAGGAGAGCACAGACTTGCAGCTCATGCAGCC and the pair GGCTGCATGAGCTGCAAGTCTGTGCTCTCCTGTCCCTTTTTTGAGACATCTGCTGCATAC and TCAAGTTACTGAATCTTTCTTCTTCCGG. The resulting DNA fragment was subcloned into pcDNA at the HindIII and KpnI sites to generate pcDNA-PM-Ras(61L). To generate pcDNA-MYC-Cdc42, a DNA fragment was released by EcoRV and XbaI from pUM-Cdc42(12V) (38) to replace the same fragment in pcDNA-Cdc42(28L) (23). To construct pcDNA-Cdc42(17N), a DNA fragment encoding Cdc42(17N) was released from pUM-Cdc42(17N) (38) by EcoRI and XbaI and cloned into pcDNA. The DNA fragments encoding the H-Ras(12V), H-Ras(61L), EM-H-Ras(61L), PM-Ras(61L), Cdc42, Cdc42(17N), Raf-1 containing just the Ras binding domain (RafRBD), Raf-1, and RIN1 were amplified by PCR (Accuzyme, Bioline, Taunton, MA) using pM4-IRES-GFP-H-Ras(12V) (where IRES is internal ribosome entry site) (M. Thai and J. Colicelli, unpublished results), pcDNA-H-Ras(61L) (5), pcDNA-H-Ras(61L,181S,184S) (5), pcDNA-PM-Ras(61L), pcDNA-MYC-Cdc42, pcDNA-Cdc42(17N), pEYFP-RafRBD (5), pDONR223-Raf-1 (41), and pKS-RIN1 (11), respectively, as templates. All PCR forward primers start with the CACC sequence right before the start codon, allowing them to be ligated to linearized pENTR-Direct-TOPO by the Directional TOPO cloning system (Invitrogen). The resulting vectors were used as entry vectors to N-terminally tag proteins with GFP, mCherry, yellow fluorescent protein (YFP), cyan fluorescent protein (CFP), or FLAG by the Gateway system (Invitrogen) using pCL-GFP-DEST, pCL-mCherry-DEST, pCL-YFP-DEST, pCL-CFP-DEST, or pCL-FLAG-DEST, respectively, as the destination vectors. To express various H-Ras proteins in a lentiviral vector, pLenti4-DEST (Invitrogen) was used as the destination vector. The short hairpin RNA (shRNA) vectors that silence CDC42 expression and their nontargeting control (pLKO-non-target) are from Sigma (St. Louis, MO), and clones 1 and 2 used in this study contain the following sequences (5′-3′): CGGAATATGTACCGACTGTTT and CCTGATATCCTACACAACAACC. The pcDNA-MYC-Cdc42* construct was created by site-directed mutagenesis (Stratagene, La Jolla, CA) on pcDNA-MYC-Cdc42 using the following primer set (5′-3′): GTTGGTAAAACATGTCTACTTATCTCATATACGACTAACAAATTTCCATCG and CGATGGAAATTTGTTAGTCGTATATGAGATAAGTAGACATGTTTTACC AAC. This vector expresses Cdc42*, which is refractory to shRNA 2. To mutate the cysteine in the CAAX box of LCK-H-Ras(12V,181/4S) (26), site-directed mutagenesis was performed on pEF-LCK-H-Ras(12V,181/4S) using the primer set (5′-3′) AGCATGAGCAGCAAGAGTGTGCTCTCCTGACGC and GCGTCAGGAGAGCACACTCTTGCTGCTCATGCT to create pEF-LCK-H-Ras(12V,181/4/6S). pRK5-FLAG-ITSN1-L and pEGFP-ITSN1-L were kindly provided by Peter McPherson (McGill University). Src(527F) (46) was fused at the C terminus with Rit-C by two rounds of PCR and subcloned to pMSCV (where MSCV is mouse stem cell virus) (Clonetech, Mountain View, CA) at the BglII and EcoRI sites. The vector that expresses the shRNA (5′-3′) CCAGCAGAATGATGAAAAGCAA against Dbl was obtained from OpenBiosystem. All PCR and mutagenized DNAs were confirmed by sequencing.
Virus production and infection.
Viral vectors were produced in 293FT cells by cotransfecting packaging vectors (pAmpho for retrovirus, and pLP1, pLP2, and pVSVG [where VSVG is vesicular stomatitis virus G protein] for lentivirus) together with appropriate vectors to express various proteins. The supernatant containing the virus was harvested and filtered 48 h after transfection. The viral infection was carried out with 4 (retrovirus) or 6 (lentivirus) μg/ml of Polybrene (Calbiochem, San Diego, CA) for 16 h, and then the cells were fed with fresh medium for another 24 h before antibiotic selection. The titer of the virus used in the soft agar assay was determined following Invitrogen protocols.
Immunoblotting.
Proteins were separated by SDS-PAGE and transferred to the nitrocellulose membrane (Bio-Rad, Hercules, CA). To detect Ras, the pan-reactive Ras10 antibody (Calbiochem) was used (1:300). The antibodies that recognize FLAG (M2 clone; 1:1,000) and GFP (1:2,000) were from Sigma and Fitzgerald (Concord, MA). Antibodies against Na+/K+ ATPase (1:500) and caveolin (1:500) were from BD Biosciences (San Jose, CA), while Cdc42 (1:500) and p53 (1:1,000) antibodies were from Santa Cruz (Santa Cruz, CA). The MYC tag was recognized by the 9E10 monoclonal antibody (hybridoma cell culture medium; 1:10). Antibodies against p42/44 Erk kinase, phospho-p42/44 Erk kinase, p38 MAP kinase, and phospho-p38 MAP kinase were all purchased from Cell Signaling (Danvers, MA). The fluorescein-conjugated secondary antibodies (1:10,000) were purchased from Li-COR Biosciences (Lincoln, NE), and the protein levels were quantified by the Odyssey Infrared Imaging System (Li-COR Biosciences).
Statistics.
After quantifications, values are shown as averages ± standard errors of the means (SEM). Unpaired student t tests were performed to obtain P values.
Cell transformation assays.
To perform the focus formation assay, NIH 3T3 cells at 60% confluence (in a 60-mm dish) were transfected and split into four 60-mm dishes 48 h posttransfection. One dish of cells was lysed with SDS sample buffer for protein expression analysis, and the rest were grown for 14 more days before being stained by crystal violet (0.5% in 20% ethanol) to visualize foci. To compensate for low expression of PM-Ras(61L), 10 times more vector DNA was used. To carry out the soft-agar colony formation assay, NIH 3T3 cells were transfected with vectors to express various H-Ras proteins and selected in the presence of G418 (500 μg/ml). For each vector, a total of 10 clones (clones 1 to 10) were confirmed to express H-Ras proteins at similar levels, and three of them were used for this experiment. These cells were infected by pCL-FLAG-Cdc42(17N) with increasing dosages (10, 20, and 40 multiplicities of infection). Forty-eight hours after infection, they were trypsinized, and growth medium was added immediately when cells began to detach. Approximately 5 × 104 cells were mixed with SeaPlaque agar such that the final agar concentration was 0.35%. This mixture was loaded into a 60-mm dish containing 0.7% SeaPlaque agar. For the study involving coexpression of PM-Ras(61L) and Cdc42(12V), 1 × 104 cells were used and seeded in six-well plates. These cells were cultured for 14 to 28 days and fed every 4 days with 250 μl of the same medium as described above except that it contained 30% calf serum. They were finally incubated with 0.1% (wt/vol) 3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide (MTT; Sigma) in PBS to visualize colonies.
The BiFC assay.
HTC75 cells were chosen for the bimolecular fluorescence complementation (BiFC) assay for their low background fluorescence in fluorescence-activated cell sorting (FACS) (our unpublished results). We fused proteins of interest to the C and N termini of YFP fragments (Yc and Yn, respectively). While such fusion proteins worked well in this assay, we became aware that a mutant version of YFP called Venus is much brighter than the wild-type form in part because of mutations in the N terminus (28). We thus used the N terminus from Venus (amino acids 1 to 155) for this study, while Yc contains amino acids 156 to 239 from wild-type YFP. To express them, pBabe-Yn-DEST-Neo and pBabe-Yc-DEST-Puro (4) were used as destination vectors for the Gateway cloning system. The DNA fragments encoding H-RasΔE(12V) and EM-H-RasΔE(12V) were generated by two rounds of site-directed mutagenesis (Stratagene) using either pENTR-H-Ras(12V) or pENTR-EM-H-Ras(12V) as a template and the primer set (5′ to 3′) CCATTTTGTGGACGAAGCCGCCGCCGCTGTAGAGGATTCCTACCGG and CCGGTAGGAATCCTCTACAGCGGCGGCGGCTTCGTCCACAAAATGG and the set AATGACCACCTGCTTCCGGGCGGCAGCCGCTGCAGCGGCGGCGGCTT and AAGCCGCCGCCGCTGCAGCGGCTGCCGCCCGGAAGCAGGTGGTCATT to mutate amino acids 32 to 40 in the effector loop of H-Ras to alanine. HTC75 cells were infected with vectors encoding Yn-tagged H-Ras proteins and then selected with G418 (500 μg/ml). For each vector, three clones of cells expressing H-Ras proteins at similar levels were pooled. These cells were then infected by retroviral vectors expressing Yc-tagged proteins and selected in puromycin (1 μg/ml) medium. The samples were examined by either FACS or confocal microscopy.
Effector pulldown assay for Cdc42 activation.
The BL21(DE3) Escherichia coli strain was transformed using pGEX fused to Cdc42/Rac interactive binding (CRIB) domain of PAK1 (pGEX-PAKCRIB) (25), and glutathione S-transferase (GST)-PAKCRIB expression was induced by isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were washed with phosphate-buffered saline (PBS) and then lysed with PBS containing Triton X-100 (0.1%) and lysozyme (1 mg/ml). The lysate was cleared by centrifugation. GST-PAKCRIB beads were prepared using the glutathione resin (Clontech, Mountain View, CA), and the final concentration of GST-PAKCRIB was 200 ng/μl. Mammalian cells were lysed in 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, 1 mM dithiothreitol (DTT), and 5% glycerol, and the resulting lysate (500 μg of protein) was mixed with 100 μl of GST-PAKCRIB resin for 1 h at 4°C with rocking. The resin was washed three times with lysis buffer, and the bound Cdc42 was subsequently eluted by 50 μl of 2× SDS sample buffer and finally resolved by 15% SDS-PAGE.
IP.
Cells were lysed in immunoprecipitation (IP) buffer (15), supplemented with protease inhibitors (Roche Applied Science, Indianapolis, IN). Cell lysates were precleared with mouse IgG-conjugated beads (A0919; Sigma) for 4 h with agitation at 4°C. The cleared lysates were incubated with either Ras10 antibody or anti-FLAG antibody-conjugated beads (M2 clone; Sigma) overnight with agitation at 4°C. Protein A-agarose beads (Roche Applied Science) were added to samples treated with the Ras10 antibody or mouse IgG (at 4°C with agitation for 4 more hours). The beads were then washed four times with Tris-buffered saline (TBS) buffer, and the bound materials were eluted with 50 μl of 2× SDS sample buffer and resolved by 15% SDS-PAGE. Epidermal growth factor (EGF)/serum-stimulated cells were similarly lysed in the same buffer supplemented with additional phosphatase inhibitors (1 mM NaVO3 and 1 mM NaF).
Immunostaining.
Cells were seeded onto fibronectin-coated coverslips (BD) at 24 h posttransfection and were fixed 16 h later with 4% paraformaldehyde (20 min at room temperature). The fixed samples were sequentially incubated with 50 mM ammonium chloride-PBS (10 min at room temperature), permeabilized in PBS containing 0.1% Triton X-100 and 200 mM glycine (10 min at 4°C), and blocked with 3% bovine serum albumin (1 h at room temperature). All samples were incubated with the Ras10 antibody overnight at 4°C and then with the Alexa Fluor 488-conjugated secondary antibody (Invitrogen) for 1 h at room temperature. All samples were mounted using mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Lab, Burlingame, CA).
FACS and microscopy.
FACS was performed using an EPICS XL-MCL instrument (Beckman-Coulter, Fullerton, CA), and results were analyzed by the WinMDI software. For live-cell microscopy, cells were grown on glass-bottom plates (MatTek Corp., Ashland, MA). To perform deconvolution microscopy, cells were imaged with an Olympus IX70 microscope equipped with 60×/1.4 oil objective and a Retiga 1300R camera (Q-Imaging, Surrey, British Columbia, Canada). Stacks of 14 images at 0.5-μm intervals were deconvolved by Slidebook (Intelligent Imaging Innovations, Denver, CO). Confocal microscopy was performed on a Leica TCS SP5 microscope with a 63×/1.4 oil objective. To image the BiFC samples, its Ar and He/Ne lasers were operated at 30 and 100% power, respectively. CFP (458 nm) was detected with 48% of available Ar laser power, while YFP (514 nm) was imaged with 52% of the available He/Ne laser power. The samples were scanned at 700 Hz, signals from two consecutive line scannings were accumulated, and the whole frame was averaged once to reduce background. For regular confocal experiments, the 405-nm and He/Ne laser were turned on at full power while the Ar laser was set at 30% power. To visualize DAPI and YFP, 40% 405-nm and 52% He/Ne available laser power was used. To image CFP and Alexa Fluor 488, 48% and 30% Ar available laser power was used. All confocal images were analyzed by the Leica LAS AF software. To quantify the efficiency of protein localization to the PM, a region of interest (ROI) was first created to measure the fluorescent intensity of the whole cell (ROIw). A second ROI, was created to measure the signal in the cytoplasm (ROIc). The signal at the PM is defined as ROIw − ROIc.
Sucrose gradient cell fractionation.
NIH 3T3 cells were scraped and washed with PBS and resuspended and lysed in 0.5 M Na2CO3 as described previously (32). The cell lysate was centrifuged at ≈220,000 × g in a 5 to 45% sucrose gradient. Thirteen fractions of 400 μl each were collected, and an equal volume of each fraction was examined by Western blotting.
Raf-1 recruitment assay.
When CFP-Raf-1 was examined, various H-Ras proteins were YFP tagged to allow identification of cells that also express the tested H-Ras proteins. At 24 to 36 h after transfection, cells were serum starved for 1 h before imaging. Only cells that expressed both YFP and CFP were analyzed. The efficiency of Raf-1 recruitment to the PM was determined by measuring the portion of CFP-Raf-1 at the PM over the total CFP signal in the cell, as described above in “FACS and microscopy.” This study also examined various H-Ras proteins that were tagged by LCK using YFP-RafRBD. These LCK-tagged H-Ras proteins may not be further tagged without affecting proper localization because they already contain a myristoylation signal at the N terminus and a CAAX motif at the C terminus. Thus, in the doubly transfected cells, we could observe only YFP signal from YFP-RafRBD, which occurred in 50 to 60% of these cells. When cells were cotransfected by the vector control, nearly all YFP-positive cells showed diffused YFP signal. In contrast, when different LCK-tagged H-Ras mutants were expressed, about 20% of the YFP-positive cells displayed nondiffused YFP. In comparison, 40 to 50% of YFP-positive cells showed nondiffused YFP when unrestricted H-Ras(61L) was expressed. Nine to 20 such cells from each sample were chosen to further analyze whether YFP-RafRBD was preferentially recruited to the PM or to the internal membranes in the cytoplasm, as described above.
Raf-1 kinase activity measurement.
A Raf-1 kinase experiment was performed as described previously (40). Briefly, transfected 293FT cells were serum starved and lysed, and the lysate was centrifuged at 100,000 × g to yield the supernatant (S100) and pellet (P100). The pellet was resuspended in the lysis buffer at the same volume as the supernatant to yield the P100 fraction. The presence of Raf-1 in these fractions was analyzed by Western blotting. Ten micrograms of protein from the P100 fraction was taken to measure Raf-1 activity using a MEK1/Erk2-coupled in vitro kinase assay kit from Millipore (Billerica, MA). The phosphorylated Erk2 was detected by Western blotting.
Yeast two-hybrid assay.
A yeast two-hybrid assay was performed as previously described using the L40 reporter strain (2). The DNAs encoding H-Ras(61L), EM-H-Ras(61L), and PM-Ras(61L) were cloned to pBluescript at the BamHI and SacI sites by PCR. A BglII linker was added to the SacI site of the resulting vectors, and then the DNA fragments encoding various H-Ras mutants were released and subcloned to pVJL11 at the BamHI site. Plasmids pGAD424-AF6 (49), pGADGE-RalGDS (53), and pGAD-Raf-1 (52) were kind gifts from Michael White (University of Texas Southwestern Medical School). In these pGAD vectors, the cDNAs encoding the tested Ras effectors were fused with the coding sequences of the GAL4 activation domain and a nuclear localization signal from simian virus 40 (SV40).
Senescence assay.
BJ cells between population doublings 23 to 33 were infected with the described lentiviral vectors to express various H-Ras mutants. The virus was either used straight for infection [vector control, H-Ras(61L), and EM-H-Ras(61L)] or concentrated by an Amicon Ultra-15 spin column (Millipore) to compensate for the low expression level of PM-Ras(61L). Infected cells were divided into two groups at 36 h postinfection. Cells in group 1 were serum starved for 20 h and then lysed with IP buffer supplemented with protease and phosphatase inhibitors as described previously (15) and analyzed by Western blotting. Cells in group 2 were selected with zeocin (100 μg/ml) for 12 days and then stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and scored microscopically (15).
Tumorigenicity assay in nude mice.
Female nu/nu mice (Harlan, Houston, TX) at 6 to 7 weeks of age were divided into three groups of 8 animals each. The three selected clones of cells were pooled and allowed to grow to approximately 80% confluence. They were then scraped and washed with PBS. Cell pellets were resuspended in Hanks balanced salt solution (Invitrogen) at a density of 1 × 106/ml, and 100 μl of these cells was injected subcutaneously into the right flank of each animal. Tumor volumes were measured 10 days after injection (44).
RESULTS
Inactivating Cdc42 blocks cell transformation induced by endomembrane-restricted oncogenic H-Ras.
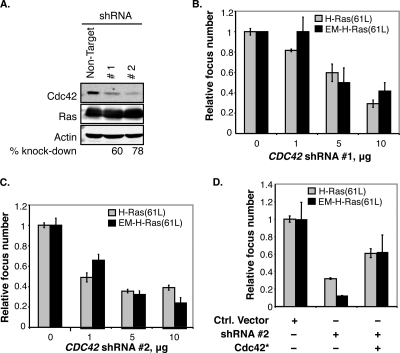
To determine whether endomembrane-restricted H-Ras requires Cdc42 to transform cells, we sought to block Cdc42 activity and then measure cell transformation as induced by endomembrane-restricted H-Ras. We first screened commercially available shRNAs and found two clones that are particularly efficient in silencing Cdc42 expression without affecting expression of actin, H-Ras, and other Rho GTPases (RhoA and Rac) (Fig. 1A and data not shown). These two shRNAs were expressed in NIH 3T3 cell lines that stably express a constitutively active H-Ras [H-Ras(61L,181S,184S)] whose two cysteine residues for palmitoylation (5) have been mutated to serine to prevent PM association (see Fig. S1 in the supplemental material). We renamed it EM-H-Ras(61L) to denote its restriction to the endomembrane. Our data show that when Cdc42 protein levels were reduced by shRNA, cell transformation (Fig. 1B and C show data for focus formation; data for colony formation in soft agar are not shown) induced by EM-H-Ras(61L) was blocked. We note that the efficiencies of shRNAs to block EM-H-Ras(61L)-induced transformation correlated with those of Cdc42 knockdown; further, overexpressing either normal CDC42 or a mutant CDC42, CDC42*, that is refractory to the CDC42 shRNA could restore cell transformation (Fig. 1D and data not shown). These results suggest that these shRNAs block Cdc42 expression with high specificity. To exclude the possibility that CDC42 shRNA blocks cell growth indiscriminately, we determined that cells transfected by the shRNA vector, which contains a puromycin resistance marker, could efficiently form colonies in puromycin (data not shown). We similarly examined a dominant negative Cdc42 [Cdc42(17N)] and obtained the same results (data not shown).
FIG. 1.
Cdc42 inactivation blocks cell transformation induced by endomembrane-restricted oncogenic H-Ras. (A) NIH 3T3 cells were transiently cotransfected with pcDNA-H-Ras(61L) plus a vector carrying shRNA 1 or 2 against Cdc42 or a nontarget shRNA. Cells were lysed with SDS sample buffer, and cell lysates were examined by Western blotting to determine the efficiency of the knockdown and its effects on Ras expression. Actin was the loading control. (B and C) NIH 3T3 cells were cotransfected with vectors expressing either H-Ras(61L) or EM-H-Ras(61L) and the indicated amount of Cdc42 shRNA 1 or 2. The mean numbers of foci were calculated from cells seeded in triplicate (n = 3), and the number of foci from cells expressing the nontarget shRNA was taken as 1. (D) NIH 3T3 cells were transfected with vectors expressing either H-Ras(61L) or EM-H-Ras(61L) together with either the vector control (−) or a vector expressing Cdc42 shRNA 2 and with or without Cdc42*, which is refractory to the shRNA knockdown (data not shown). Focus formation was assayed as described for panel B. A normal CDC42 cDNA was similarly tested, which restored Cdc42 to endogenous levels, and the same rescue results were obtained (data not shown).
To determine whether the “normal” unrestricted H-Ras(61L) also required Cdc42 for cell transformation, we repeated these experiments and found that cell transformation induced by the unrestricted H-Ras(61L) protein was also inhibited by Cdc42(17N) and Cdc42 shRNAs (Fig. 1). These data collectively support the idea that endomembrane-localized H-Ras acts through Cdc42 to transform cells.
Activated H-Ras forms a complex with Cdc42.
To further investigate how Ras interacts with Cdc42, we ascertained whether H-Ras and Cdc42 formed a complex. We first performed colocalization experiments examining ectopically expressed epitope-tagged proteins by confocal microscopy. Our data show that these two proteins colocalize in a vesicular structure near the nucleus (see Fig. S2 in the supplemental material). We then employed the bimolecular fluorescence complementation (BiFC) method, which takes advantage of the fact that fusing the N- and C-terminal YFP fragments (Yn and Yc, respectively) to two proteins that form a complex can restore the YFP signal, detectable by both FACS and fluorescence microscopy (17). This method can detect transient and weak protein-protein interactions because once a protein complex is formed, it is very stable as the binding is secured by not only the binding between the two proteins of interest but also the very strong H-bonds formed between the N- and C-terminal regions of YFP (12). Another obvious advantage of this system is that it allows visualization of where the binding takes place in live, but not fixed, cells.
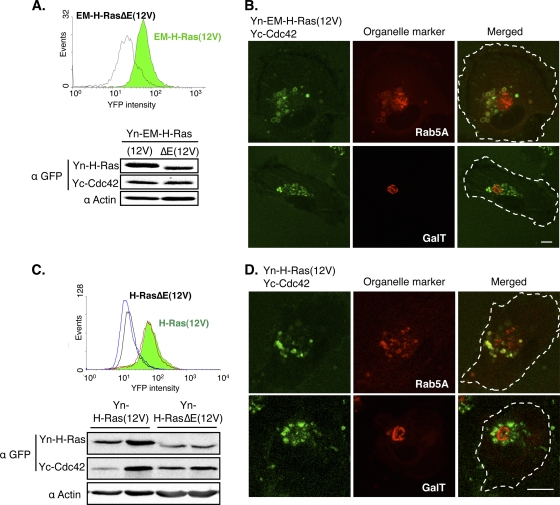
During the course of studying Ras and effector binding, we found that another constitutively active allele of H-Ras, H-Ras(12V), bound more strongly to several effectors than H-Ras(61L) (data not shown); thus, H-Ras(12V) was chosen over H-Ras(61L) for this assay. In addition to H-Ras(12V), we created a mutant Ras control, called H-RasΔE(12V), whose amino acids in the effector binding region (residues 32 to 40) were all changed to alanine. We showed that Yn-H-Ras(12V), but not Yn-H-RasΔE(12V), bound the well-known Ras effectors Yc-Raf-1 and RIN1-Yc (see Fig. S3 in the supplemental material; also data not shown). Having confirmed that the BiFC method can detect protein binding to Ras, Cdc42 was similarly analyzed by FACS. As shown in Fig. 2A, Yc-Cdc42 bound Yn-EM-H-Ras(12V) while its binding to the Yn-EM-H-RasΔE(12V) control was substantially reduced. Furthermore, as shown by microscopy (Fig. 2B), consistent with the function of Cdc42 in regulating endocytic transport (20), the binding between EM-H-Ras(12V) and Cdc42 can be readily detected on endosomes but not on the Golgi apparatus. We have similarly examined H-Ras(12V) and the effector binding mutant control [H-RasΔE(12V)] and obtained similar results (Fig. 2C and D). These data suggest that H-Ras forms a complex with Cdc42, and this binding is readily detectable on endosomes and dependent on the effector loop of H-Ras.
FIG. 2.
H-Ras forms a complex with Cdc42 on endosomes. (A) A representative clone of HTC75 cells stably coexpressing Yc-Cdc42 and either Yn-EM-H-Ras(12V) (green) or Yn-EM-H-RasΔE(12V) (white) was analyzed by FACS (top). The cells were lysed, and the protein levels were examined by Western blotting using an anti-GFP (α-GFP) antibody, which cross-reacts with YFP (bottom). (B) The same cells as in panel A were transfected with vectors expressing either CFP-tagged Rab5A or GalT, which mark endosomes and the Golgi compartment, respectively, and examined by confocal microscopy. YFP and CFP signals were pseudocolored green and red, and the outline of the cell was marked by a dotted line in the merged image. Scale bar, 10 μm. (C) Two clones each of HTC75 cells stably coexpressing Yc-Cdc42 and either Yn-H-Ras(12V) (green) or Yn-H-RasΔE(12V) (white) were analyzed by FACS, and protein expression levels were examined by Western blotting as described for panel A. (D) Cells in panel C were transfected with vectors expressing organelle markers and examined as described for panel B.
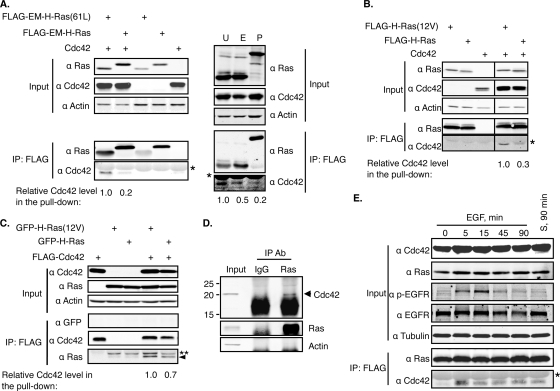
To validate the binding biochemically and to determine whether it is dependent on the guanine nucleotide binding state of Ras, we ectopically expressed a FLAG-tagged nononcogenic or oncogenic constitutively GTP-bound form of EM-H-Ras together with Cdc42 and performed coimmunoprecipitation experiments. As shown in Fig. 3A, the oncogenic EM-H-Ras could bring down Cdc42 approximately 5-fold more efficiently than the nononcogenic form. The same coimmunoprecipitation experiment was performed with the unrestricted oncogenic H-Ras, and the same results were obtained (Fig. 3B). We performed the reverse coimmunoprecipitation experiments and found that FLAG-Cdc42 also pulled down oncogenic H-Ras more efficiently (Fig. 3C). To determine whether the binding between endogenous Ras and Cdc42 could be readily detected, we immunoprecipitated Ras proteins from HTC75 cells, which were derived from HT1080 cells that contain an oncogenic N-RAS, and detected coimmunoprecipitation of endogenous Cdc42 (Fig. 3D). We conclude from these data that Cdc42 preferentially binds H-Ras-GTP.
FIG. 3.
Cdc42 preferentially binds H-Ras-GTP. (A) 293FT cells were cotransfected to express FLAG-tagged EM-H-Ras or EM-H-Ras(61L) together with Cdc42 (left). The FLAG-tagged H-Ras proteins were immunoprecipitated, and the resulting samples, together with the cell lysate (input), were examined by Western blotting. The asterisk denotes the IgG light chain. The levels of immunoprecipitated H-Ras were used to normalize levels of coprecipitated Cdc42. The normalized level of Cdc42 in cells that ectopically expressed the FLAG-EM-H-Ras(61L) was set to 1. On the right, the same cells were also cotransfected to express FLAG-tagged unrestricted H-Ras(61L) (U) or PM-Ras(61L) (P), in addition to the FLAG-tagged EM-H-Ras(61L) (E), together with Cdc42. The coimmunoprecipitation was performed as described above. (B) 293FT cells were similarly transfected to express FLAG-tagged unrestricted wild-type or oncogenic (12V) H-Ras, together with Cdc42, and the coimmunoprecipitation experiment was performed as described for panel A. Note that images were cropped from different lanes on the same Western blot membrane. (C) 293FT cells were transfected to express FLAG-tagged Cdc42 together with GFP-tagged H-Ras or oncogenic H-Ras(12V), and FLAG-tagged Cdc42 proteins were immunoprecipitated as described for panel A. The level of immunoprecipitated Cdc42 was used to normalize coprecipitated H-Ras proteins. The normalized level of H-Ras(12V) was set to 1. The double asterisks denote the nonprenylated H-Ras, and the arrowhead marks the immunoprecipitated prenylated H-Ras (19). (D) Endogenous Ras proteins of HTC75 cells were immunoprecipitated by the Ras10 antibody, and mouse IgG was used as the antibody control. The cell lysates (input) and immunoprecipitated samples were examined by Western blotting. (E) 293FT cells were transfected to express FLAG-tagged H-Ras and Cdc42 for 36 h. These cells were then serum starved for 16 h before being stimulated with either EGF (100 ng/ml) for the indicated time or serum (S; 10%) for 90 min. EGF or serum stimulation is expected to induce EGFR phosphorylation, which we confirmed using Western blotting. H-Ras and Cdc42 coimmunoprecipitation was examined as described for panel A. α, anti; Ab, antibody.
H-Ras and Cdc42 interaction is stimulated by EGF and serum.
Since GTP-bound H-Ras binds Cdc42 more efficiently, we asked whether this interaction was stimulated by growth factors that activate Ras proteins. To this end, nononcogenic H-Ras and Cdc42 were ectopically coexpressed, and the cells were then serum starved before being stimulated by either epidermal growth factor (EGF) or serum. As shown in Fig. 3E, the binding between H-Ras and Cdc42 was readily induced 5 min after EGF addition. Similar results were obtained with cells that were stimulated by serum.
Active H-Ras can activate Cdc42 by recruiting and activating Cdc42 GEFs.
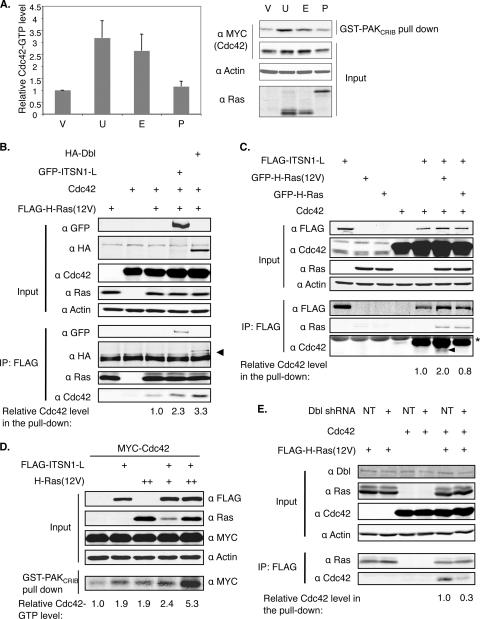
To determine whether the binding of H-Ras to Cdc42 could lead to Cdc42 activation, we carried out an effector pulldown assay using the CRIB (Cdc42/Rac interactive binding) domain from the Cdc42 effector PAK1, which preferentially binds Cdc42 in a GTP-bound state. As shown in Fig. 4A, when EM-H-Ras(61L) or the unrestricted H-Ras(61L) was expressed, approximately 3-fold more Cdc42 was pulled down than from control cells. These data suggest that the binding of activated H-Ras induces Cdc42 activation.
FIG. 4.
Constitutively active H-Ras can efficiently activate Cdc42 by recruiting a Cdc42 GEF. (A) 293FT cells were cotransfected with a vector control (V) or vectors expressing various H-Ras mutants (U, unrestricted; E, EM-restricted; and P, PM-restricted) together with MYC-tagged Cdc42 for 36 h and then serum starved for 16 h before being lysed. The levels of Cdc42 in the lysates as well as in the GST-PAKCRIB pulldown were analyzed by Western blotting. The portion of Cdc42 presumed to be GTP bound was obtained by dividing the latter by the former. The Cdc42-GTP level in cells transfected with the vector control was defined as 1 (n = 3 separate experiments). (B and C) 293FT cells were transfected to express the indicated proteins. FLAG-tagged H-Ras(12V) (B) or ITSN1-L (C) was immunoprecipitated as described in the legend of Fig. 3A. Note that the asterisk denotes the IgG light chain. In panel B, the arrowhead denotes the coimmunoprecipitated Dbl, and in panel C, endogenous Cdc42 proteins were marked by an arrowhead. (D) 293FT cells were examined as described for panel A, and the Cdc42-GTP level in cells expressing MYC-Cdc42 alone was set to 1. This experiment was performed twice and yielded the same results. (E) 293FT cells were transfected to express either nontarget shRNA (NT) or shRNA against Dbl, together with FLAG-tagged H-Ras(12V) and Cdc42. FLAG-tagged H-Ras(12V) was immunoprecipitated, and relative Cdc42 levels in the pulldown were quantified as described in the legend of Fig. 3A.
Next, we ascertained how Ras activates Cdc42. In S. pombe, we have shown that Ras1 activates Cdc42 by recruiting a Cdc42 GEF, called Scd1/Ral1 (2). In mammalian cells, intersectin 1-L (ITSN1-L) is a Cdc42 GEF (13, 57), and a shorter version of this protein has been shown to bind Ras at the endomembrane by fluorescence resonance energy transfer (FRET) (27). We thus investigated as a proof of principle whether H-Ras in the GTP-bound state can form a complex with both Cdc42 and ITSN1-L. We transiently transfected cells to express H-Ras(12V), Cdc42, or ITSN1-L either alone or in various combinations. The data show that immunoprecipitating H-Ras(12V) brought down ITSN1-L and Cdc42 (Fig. 4B). Note that approximately 2-fold more Cdc42 was coimmunoprecipitated with H-Ras(12V) when ITSN1-L was also coexpressed, supporting the idea that the binding between activated H-Ras and Cdc42 is mediated by a Cdc42 GEF, such as ITSN1-L. In S. pombe, when Ras1 is overexpressed, the binding between Scd1 and Cdc42 is enhanced (2). To examine whether H-Ras can stimulate ITSN1-L and Cdc42 binding, we analyzed how much Cdc42 was coimmunoprecipitated with ITSN1-L as a function of Ras. As shown in Fig. 4C, the coprecipitation between ITSN1-L and Cdc42 was indeed enhanced by oncogenic H-Ras but not by nononcogenic H-Ras. We note that in this experiment, we were also able to detect the presence of endogenous Cdc42 in the complex. The combination of unrestricted H-Ras(12V) (Fig. 4D) or EM-H-Ras(61L) (data not shown) with ITSN1-L also acted synergistically to activate Cdc42, as measured by the same CRIB pulldown assay.
While the ITSN1-L experiments support the idea that Ras can activate Cdc42 by acting via its GEFs, ITSN1-L is expressed mostly in cells of neuronal original (14) and is not expressed in many cells used in this study (e.g., 293FT or HTC75 cells) (data not shown). Thus, the binding between H-Ras and Cdc42 may be mediated by other GEFs, such as Dbl, which is expressed in a wide range of cell lines. Our data in Fig. 4B showed that when Dbl was overexpressed, the binding between oncogenic H-Ras and Cdc42 was also enhanced (Fig. 4B); conversely, when Dbl expression was reduced to half by shRNA, the binding was substantially weakened (Fig. 4E). We conclude from these results that the interaction between H-Ras and Cdc42 can be mediated by many Cdc42 GEFs.
Restrict H-Ras to the PM.
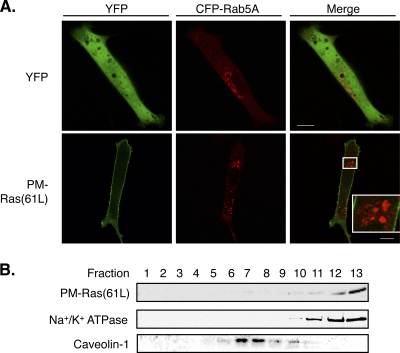
To further investigate the idea that the described H-Ras-Cdc42 interaction occurs selectively on the endomembrane, we targeted H-Ras to the PM as a control. Since endomembrane-restricted oncogenic H-Ras proteins transform cells very efficiently, it is critical to target H-Ras to the PM without association with internal membranes, a criterion that is hard to meet by existing methods. For example, a transmembrane domain from CD8 was used to target proteins to the PM (5, 26), but substantial amounts of the fusion proteins appear to localize also to the endomembrane (6). Likewise, the myristoylation and palmitoylation signals in LCK have been used for PM targeting, but Zlatkine et al. (58) reported that an LCK-tagged soluble protein, chloramphenicol acetyltransferase, associates not only with the PM but also with the endomembrane, an observation that we have confirmed (see below). Therefore, in this and in our previous S. pombe study, we employed a PM-targeting sequence from the C terminus of the Rit GTPase (called Rit-C), which is a small GTPase without a CAAX motif that localizes to the PM without detectable association with the endomembrane (21, 30). When S. pombe Ras1 is tagged with the same sequence, the resulting protein selectively activates the MAP kinase pathway on the PM but not the Scd1-Cdc42 pathway on the endomembrane (30). To tag H-Ras with Rit-C, we first mutated the cysteine in the CAAX box of H-Ras(61L) to serine to prevent farnesylation (and nonspecific binding to internal membranes). This mutant H-Ras was then tagged at its C terminus by Rit-C. Since such modification may alter the activity of the isoform-specific HVR, we named the resulting protein with a more generic term, PM-Ras(61L) (see Fig. S1 in the supplemental material). We confirmed that PM-Ras(61L) localizes to the PM by both fluorescent protein tagging and immunostaining using Ras-specific antibody (Fig. 5A and data not shown). The signal of PM-Ras(61L) in the interior of the cell was barely detectable and did not appear to colocalize with the markers of endosomes (Fig. 5A), Golgi network (see Fig. S4A in the supplemental material), and endoplasmic reticulum (ER) (see Fig. S4B). Activated H-Ras is known to associate more with bulk membrane than with lipid rafts on the PM (32). To examine this biochemically, we transiently expressed PM-Ras(61L), and the cell lysates were fractionated by sucrose gradient centrifugation in the presence of Na2CO3, which prevents extraction of membrane-bound Ras proteins. Our data show that, indeed, PM-Ras(61L) primarily cofractionated with the bulk membrane marker Na+/K+ ATPase but not with the lipid raft marker caveolin-1 (Fig. 5B). We similarly examined cells that coexpressed both PM-Ras(61L) and unrestricted H-Ras(61L) and found that they cofractionated in the bulk membrane fractions (see Fig. S4C in the supplemental material). These data suggest that PM-Ras(61L) localizes to the PM in a manner that is indistinguishable from that of unrestricted H-Ras(61L).
FIG. 5.
Targeting H-Ras to the PM. (A) NIH 3T3 cells were transfected to express either YFP or YFP-tagged PM-Ras(61L), together with CFP-tagged Rab5A, an endosome marker, and examined by confocal microscopy. Confocal images from the same focal plane are shown with YFP and CFP signals pseudocolored green and red, respectively. We note that the YFP-PM-Ras signal in the interior of the cell was barely detectable and did not colocalize with that of CFP-Rab5A. Scale bar, 10 μm. (B) Cell lysates of NIH 3T3 cells containing PM-Ras(61L) were fractionated by sucrose gradient ultracentrifugation. Western blotting was performed to detect indicated proteins. Na+/K+ ATPase and caveolin-1 are bulk membrane and lipid raft markers, respectively.
PM-restricted active Ras efficiently activates Raf but not Cdc42.
As shown above, endomembrane-restricted H-Ras in the GTP-bound state can efficiently activate Cdc42. We thus further investigated whether this activation was cell compartment specific. As shown in Fig. 4A, when PM-Ras(61L) was expressed, the level of Cdc42-GTP was not significantly different from that of cells carrying the vector control, suggesting that PM-Ras(61L) does not efficiently activate Cdc42. The lack of induction of Cdc42 activation by PM-Ras appears to be partly caused by a reduction of Cdc42 binding (Fig. 3A).
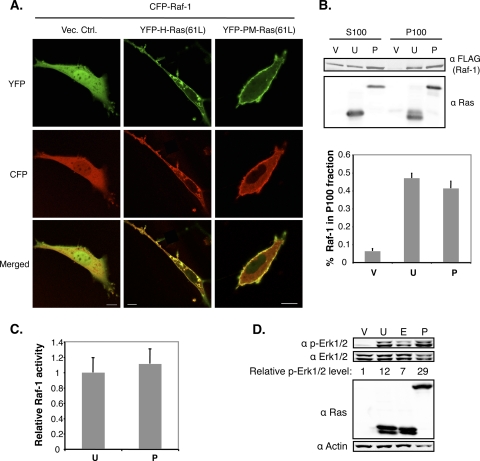
As a control, we examined whether PM-Ras(61L) can activate Raf, an event known to occur on the PM. We first examined whether PM-Ras(61L) could recruit Raf-1 to the PM by confocal microscopy. To this end, we coexpressed YFP-tagged PM-Ras(61L) or H-Ras(61L) as a control together with CFP-tagged Raf-1 and measured the portion of CFP-Raf-1 on the PM. Our data show that PM-Ras(61L) recruited Raf-1 to the PM as efficiently as the unrestricted H-Ras(61L) (37% ± 3% versus 30% ± 6%) (Fig. 6A). A truncated Raf-1 containing just the Ras binding domain (RBD) was similarly examined and yielded the same results (see Fig. S5A in the supplemental material).
FIG. 6.
PM-targeted active Ras efficiently activates Raf. (A) NIH 3T3 cells were cotransfected to express CFP-Raf-1, together with either YFP-tagged H-Ras(61L) or PM-Ras(61L). Thirty-six hours after transfection, cells positive for both YFP and CFP were analyzed by confocal microscopy. YFP and CFP signals were pseudocolored green and red. Scale bar, 10 μm. The efficiency of CFP-Raf-1 recruitment to the PM in these cells was determined by measuring the CFP-Raf-1 signal at the PM over the total CFP signal in the cell. The proportions of Raf-1 at the PM in cells carrying H-Ras(61L) or PM-Ras(61L) were 30% ± 6% (n = 4 cells) or 37% ± 3% (n = 9 cells), respectively. (B) The 293FT cells were transfected to express various H-Ras mutants for 36 h and then serum starved for 16 h before being lysed. These samples were similarly labeled as V, U, and P as in Fig. 4A. The resulting cell lysates were fractionated into the S100 and P100 fractions after centrifugation. An equal volume of each fraction was examined by Western blotting using the indicated antibodies. The proportions of Raf-1 in the P100 fractions are shown in the graph (n = 7 separate experiments). (C) To measure Raf-1 kinase activity, an equal amount of protein from the P100 fraction of each transfection was added to the recombinant substrate MEK1 which, when activated, will phosphorylate recombinant Erk2, detectable by Western blotting. The Raf-1 activity was defined by dividing the portion of Erk2 that is phosphorylated by the Raf-1 protein level. The results are presented in a graph, with the Raf-1 activity induced by H-Ras(61L) set to 1 (n = 3 separate experiments). (D) 293FT cells were transfected to express various H-Ras mutants for 36 h and then serum starved for 16 h before being lysed. These samples were labeled as V, U, E, and P as in Fig. 4A. Cell lysates were examined by Western blotting. The portion of Erk1/2 that is phosphorylated in cells carrying just the vector control was set to 1.
To directly measure Raf kinase activity, we coexpressed PM-Ras(61L) or H-Ras(61L) with Raf-1 and fractionated cell lysates by ultracentrifugation to obtain the soluble (S100) and membrane (P100) fractions. The latter were further analyzed by an in vitro MEK/Erk kinase assay system. Consistent with the microscopy data above, PM-Ras(61L) recruited Raf-1 to the membrane fraction as efficiently as the unrestricted H-Ras(61L) (Fig. 6B). Furthermore, PM-Ras(61L) activated Raf-1 kinase activity as well as H-Ras(61L) (Fig. 6C). Consistent with the possibility that Raf and its targets in the cell are activated, we examined the activation of endogenous Erk proteins in cells expressing PM-Ras(61L) or unrestricted H-Ras(61L) and found that they were similarly phosphorylated in both cells (Fig. 6D). In further confirmation that tagging H-Ras by Rit-C did not fundamentally change H-Ras and effector interactions, we performed a yeast two-hybrid assay and found that PM-Ras(61L) bound well to Raf-1, RalGDS, and AF6 in a manner that was indistinguishable from that of the unrestricted H-Ras(61L) (see Fig. S5B in the supplemental material; also data not shown). We conclude from these data that PM-restricted active H-Ras does not efficiently activate Cdc42 although it recruits Raf to the PM efficiently, leading to activation of Raf and Erk1/Erk2 (Erk1/2).
PM-restricted constitutively active Ras can efficiently induce senescence in human primary fibroblast cells.
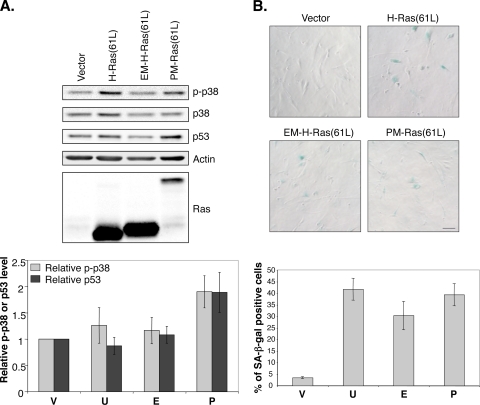
Constitutively active Ras proteins have been shown to induce senescence in human primary cells, such as BJ foreskin fibroblast cells (51). Ras presumably does so by activating the p38 protein kinase via the Raf/MEK/MAP kinase pathway (51). Since the PM-restricted Ras(61L) appears to selectively activate Raf, we investigated by Western blotting whether PM-Ras(61L) efficiently induced p38 phosphorylation. As shown in Fig. 7A, PM-Ras(61L), like H-Ras(61L), efficiently induced p38 phosphorylation; furthermore, these cells also appeared enlarged and flat and expressed senescence-associated β-Gal activity (Fig. 7B, SA-β-Gal). Ras-induced senescence also correlated with elevated p53 levels (42), which was also induced by PM-Ras(61L) (Fig. 7A). These data together with the Raf activation data strongly suggest that PM-restricted active Ras can fully activate Ras pathways, such as the Raf pathway, that are operative on the PM.
FIG. 7.
PM-restricted activated Ras can efficiently induce senescence in human primary fibroblast cells. (A) BJ cells infected by lentiviral vectors expressing various H-Ras mutants were serum starved and then lysed to be examined by Western blotting (top). The samples were labeled as V, U, E, and P as in Fig. 4A. Phosphorylated p38 and p53 levels in cells carrying the vector control were set to 1, and the results are shown in the graph at the bottom (n = 3 separate experiments). (B) BJ cells infected as described for panel A were selected by zeocin and examined by microscopy after being stained for β-Gal activity (top). Scale bar, 40 μm. The samples were similarly labeled as V, U, E, and P (see Fig. 4A). For each sample, more than 300 cells in each experiment were scored, and the percentage (n = 5 separate experiments) of senescence-associated β-Gal (SA-β-Gal)-positive cells is shown at the bottom.
Transiently expressed PM-targeted oncogenic Ras does not efficiently transform cells.
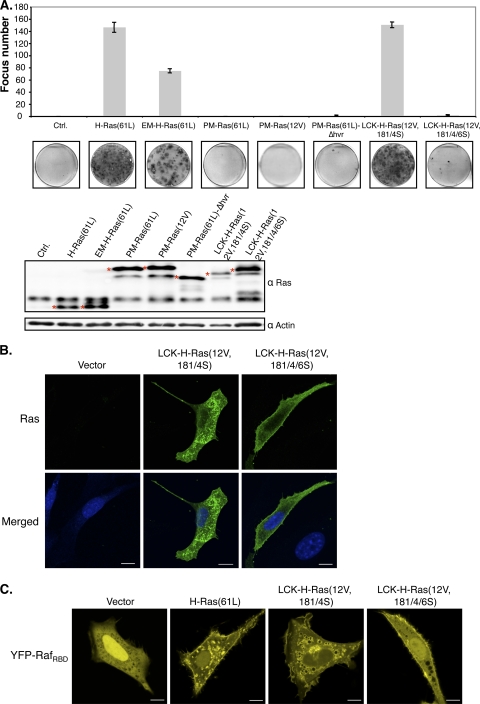
To assess the ability of PM-restricted Ras to transform NIH 3T3 cells, we transiently expressed PM-Ras(61L) and measured focus formation. Surprisingly, our data showed that PM-Ras(61L) did not efficiently induce foci (Fig. 8A). To exclude the possibility that PM-Ras(61L) was cleared from cells faster than normal, we compared its level to that of ectopically expressed GFP and found that they both declined at about the same rate over a 5-day period (data not shown). As mentioned above, we found that H-Ras(12V) bound Ras effectors more strongly than H-Ras(61L); thus, we created a PM-targeted mutant with the 12V mutation [PM-Ras(12V)], but it too failed to efficiently induce foci (Fig. 8A). We also removed the entire HVR before fusing H-Ras(61L) to Rit-C (see Fig. S1 in the supplemental material) and again found that the resulting protein failed to readily transform cells [Fig. 8A, PM-Ras(61L)-Δhvr]; thus, the extra length at the C terminus afforded by Rit-C was not the major factor inactivating cell transformation. The Rit-C tag was also fused to H-Ras(61L) with the normal CAAX motif, and the resulting protein was still inefficient in transforming these cells (data not shown). To exclude the possibility that tagging with Rit-C fundamentally inactivates the ability of oncoproteins to transform, we similarly tagged oncogenic Src [Src(527F)] and found that Src(527F)-Rit-C transformed NIH 3T3 cells efficiently (see Fig. S6 in the supplemental material).
FIG. 8.
PM-targeted oncogenic Ras does not efficiently transform NIH 3T3 cells. (A) NIH 3T3 cells were transfected to express the indicated H-Ras mutants and examined by crystal violet staining for focus formation. The numbers of foci were counted and graphed (top), and a representative dish of cells from each transfection is shown (bottom). Transfected cells were also analyzed for Ras protein expression levels (marked by asterisks) by Western blotting. (B) NIH 3T3 cells transfected with a control vector or vectors expressing LCK-H-Ras(12V,181/4S) or LCK-H-Ras(12V,181/4/6S) for 36 h were fixed and immunostained with an anti-Ras antibody. Their DNA was counterstained by DAPI. These samples were examined by confocal microscopy, and signals corresponding to Ras and DNA were pseudocolored green and blue, respectively. Scale bar, 10 μm. The proportions of LCK-H-Ras(12V,181/4S) and LCK-H-Ras(12V,181/4/6S) on the PM, determined as described in the legend of Fig. 6A, were 29% ± 4% (n = 8 cells) and 42% ± 4% (n = 8 cells), respectively. This difference is statistically significant (P = 0.04). (C) NIH 3T3 cells were cotransfected with vectors to express YFP-RafRBD and the indicated H-Ras mutants and examined by confocal microscopy 36 h posttransfection. Scale bar, 10 μm. The efficiencies of YFP-RafRBD recruitment to the PM by H-Ras(61L), LCK-H-Ras(12V,181/4S), and LCK-H-Ras(12V,181/4/6S), determined as described in the legend of Fig. 6A, were 27% ± 4% (n = 9 cells), 21% ± 3% (n = 14 cells), and 33% ± 2% (n = 20 cells), respectively. The difference in the last two values is statistically significant (P = 0.004).
Matallanas et al. have recently similarly investigated whether PM-restricted Ras can transform cells (26). One of the methods they used to restrict H-Ras to the PM was the aforementioned myristoylation and palmitoylation signals from LCK. However, this method has been previously demonstrated to cause a fusion protein to localize substantially to the endomembrane as well as the PM. Indeed, we confirmed that the reported LCK-H-Ras(12V,181/4S) protein showed significant localization in the endomembrane (Fig. 8B). We mutated the CAAX box in this protein and found that the resulting protein [LCK-H-Ras(12V,181/4/6S)] (see Fig. S1 in the supplemental material) now showed more pronounced PM localization. Curiously, while the original LCK-tagged H-Ras protein could efficiently transform cells as reported, the mutated form could no longer do so (Fig. 8A). To ascertain that the LCK-H-Ras(12V,181/4/6S), like our PM-Ras(61L), can efficiently bind Ras effectors on the PM, we performed the same Raf recruitment assay as described earlier. Our data showed that the LCK-H-Ras(12V,181/4/6S) recruited RafRBD to the PM more efficiently than the original LCK-tagged H-Ras protein, which also recruited RafRBD to the endomembrane (Fig. 8C), an observation that is consistent with our data that it localizes to the endomembrane, as well as the PM. Raf recruitment to the PM is expected to induce Erk1/2 activation, and, indeed, our data showed that LCK-H-Ras(12V,181/4/6S) also activated Erk1/2 in the cytosol as efficiently as the original LCK-tagged H-Ras (see Fig. S7). These observations agree with our study of the Rit-C-tagged H-Ras protein in that the efficiency in PM targeting inversely correlated with the ability to transform cells.
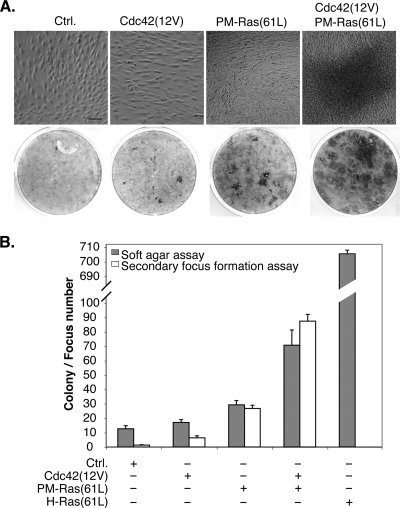
PM-restricted oncogenic Ras synergizes with activated Cdc42 to transform cells.
PM-restricted oncogenic Ras proteins do not efficiently transform cells in the focus formation assay, despite the fact that they can efficiently interact with Raf. Interestingly, several groups have reported that overexpressing activated Raf mutants alone can only weakly transform cells (18, 34, 35, 37, 54), and at least one such Raf mutant can synergize with activated Cdc42 for cell transformation (54). It is possible that to efficiently transform cells, multiple compartment-specific Ras pathways must be activated in a concerted fashion. We thus investigated whether PM-Ras(61L) can synergize with a constitutively active Cdc42 [Cdc42(12V)], which is also very weak in transforming cells when expressed alone (50), to efficiently transform cells. We created stable cell lines that expressed Cdc42(12V) either alone or together with PM-Ras(61L). These cells were allowed to grow beyond confluence in order to measure the ability to overcome contact inhibition, which can be readily detected by microscopy or by crystal violet staining. As shown in Fig. 9A, the cells carrying a vector control could grow to confluence only as a monolayer; similarly, the cells expressing just Cdc42(12V), as expected, did not grow substantially beyond confluence. In contrast, cells expressing just PM-Ras(61L) appeared to be weakly transformed and grew to a slightly higher density than cells carrying the vector control or Cdc42(12V). Importantly, when PM-Ras(61L) was coexpressed with Cdc42(12V), the resulting cells were transformed to a much higher degree and formed dense layers (Fig. 9A). When these cells were diluted with untransfected cells and replated, secondary foci could be readily detected in cells that carried both PM-Ras(61L) and Cdc42(12V); furthermore, a similar synergy was detected when they were seeded in soft agar for colony formation (Fig. 9B). These data are consistent with the model that activation of Ras pathways restricted to the PM can act in a synergistic manner with activated Cdc42 on the endomembrane to promote cell transformation.
FIG. 9.
PM-restricted activated Ras synergizes with activated Cdc42 to transform cells. (A) NIH 3T3 cells stably expressing Cdc42(12V) or PM-Ras(61L) either alone or in combination were cultured to saturation and photographed after 10 days (top). Scale bar, 40 μm. These cells were then stained with crystal violet to show dense layers of cells (bottom). This experiment was performed at least twice and yielded the same results. (B) Approximately 1 × 104 cells as described in panel A were seeded in soft agar in triplicate, and colony numbers (n = 3) were counted 28 days later after MTT staining. For secondary focus formation, 1 × 103 cells described in panel A were mixed with 3 × 105 untransfected NIH 3T3 cells and seeded in six-well plates in triplicate. These samples were cultured for 7 days, and the numbers of foci (n = 3) were scored after crystal violet staining.
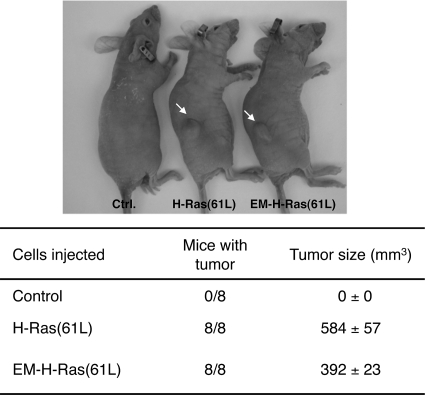
Endomembrane-restricted oncogenic H-Ras can induce tumor formation.
Lastly, while endomembrane-restricted oncogenic H-Ras can induce cell transformation in vitro, whether it can induce tumor formation in vivo has not been tested. We thus subcutaneously injected NIH 3T3 cells that stably expressed EM-H-Ras(61L) into immunodeficient mice. As shown in Fig. 10, in less than 10 days all injected mice produced tumors that were similar in size to those caused by unrestricted H-Ras(61L); in contrast, injecting cells carrying the vector control produced no detectable tumors.
FIG. 10.
Endomembrane-restricted oncogenic H-Ras efficiently induces tumor formation in nude mice. NIH 3T3 cells stably carrying either the vector control or the vector that expresses H-Ras(61L) or EM-H-Ras(61L) were injected subcutaneously into the flank region of immunodeficient mice (top), with eight mice per strain of cells. The tumor volume at 10 days postinjection is tabulated at the bottom.
DISCUSSION
One leading model suggests that Ras proteins can perform different functions by signaling from different cell compartments. However, compartment-specific Ras pathways have not been thoroughly defined. In this study, we illustrate that upon growth factor stimulation, H-Ras can activate Cdc42 on the endomembrane, an activity that is critical for cell transformation. In contrast, when H-Ras is restricted to the PM, it does not efficiently activate Cdc42 although it can efficiently activate Raf-1 and induce senescence in human primary cells. Surprisingly, while oncogenic H-Ras that is restricted to the endomembrane can very efficiently transform NIH 3T3 cells, PM-restricted oncogenic Ras can do so only weakly. However, when constitutively active forms of PM-restricted Ras and Cdc42 are coexpressed, they work in a synergistic fashion to efficiently transform cells. We conclude from these results that Cdc42 is a downstream target of H-Ras on the endomembrane, and this H-Ras-Cdc42 interaction is critical for cell transformation. However, to fully transform cells, multiple compartment-specific Ras pathways may need to work in a concerted fashion.
While H-Ras can be detected in the same protein complex with Cdc42, this interaction may not be direct. Since such binding leads to Cdc42 activation, it seems highly probable that Cdc42 GEFs must be in the same complex. Our data agree with this prediction as both DBL and ITSN1-L appear to be recruited by H-Ras to stimulate Cdc42. The H-Ras-ITSN1-L/Dbl-Cdc42 pathway in mammalian cells resembles the Ras1-Scd1-Cdc42 pathway in S. pombe, suggesting that this mode of interaction may be evolutionarily conserved.
Our BiFC data suggest that H-Ras and Cdc42 interact on endosomes. In keeping with this possibility, Ras proteins and Cdc42 have all been shown to localize to endosomes. Furthermore, Cdc42 is well known to regulate intracellular trafficking such as endocytosis (13, 29), and the Ras effector, RIN1, a GEF for Rab5, is a key regulator for endocytosis (45, 47). RIN1 and EGF receptor (EGFR) have been shown to colocalize in endosomes, and we have shown that H-Ras and RIN1 can bind in endosomes (see Fig. S2B and C in the supplemental material). These results suggest that the endosome is an authentic compartment for both H-Ras and Cdc42 signaling although it is unclear whether these two molecules depend on one another to localize to the endosome. Finally, ubiquitylation is an important step allowing proteins to be transported through the endosome compartments. Like H-Ras, N-Ras is concentrated on the endomembrane; furthermore, as with H-Ras, N-Ras can be ubiquitylated (15). Although at steady state K-Ras is not concentrated at the endomembrane, a recent study by Lu et al. has shown that active K-Ras proteins are present on the endosome (24). These results suggest that Cdc42 may also interact with N-Ras and K-Ras on endosomes. Consistent with this idea, we found that that both N- and K-Ras can also form a complex with Cdc42 as determined by coimmunoprecipitation (see Fig. S8 in the supplemental material).
To stringently test the compartmentalized Ras signaling model, it is central to seek a method that can target Ras proteins to the PM without significant “mislocalization” to the endomembrane, where Ras proteins transform cells very efficiently. The data from this and other studies, as discussed earlier, suggest that PM-targeting using CD8 and LCK may be inadequate because the resulting fusion proteins localize to not only the PM but also the endomembrane. We also tested the polylysine region from K-Ras4B in S. pombe and found that the resulting fusion Ras1 protein still activates both Ras1 pathways in S. pombe because it can be found on both the PM and the endomembrane (B. Onken and E. Chang, unpublished data). Therefore, in comparison, the C terminus of Rit is a good PM targeting signal in both yeast and mammalian cells because it can restrict H-Ras to the PM without substantial mislocalization to the endomembrane.
The fact that Ras targeted to the PM cannot efficiently transform NIH 3T3 cells is counterintuitive at first glance. One obvious interpretation of this is that the Rit-C tagging itself somehow fundamentally weakens the ability of H-Ras to stimulate effector pathways on the PM because electrostatic interactions in the HVR have been elegantly demonstrated to influence how different Ras proteins associate with microdomains (called nanoclusters) in the plasma membrane, which in turn can impact on the interaction with effectors (9). Our data do not support this possibility, however, since PM-Ras(61L) evidently recruits Raf to the PM and activates the Raf-Erk pathway as efficiently as the unrestricted H-Ras(61L). Furthermore, PM-Ras(61L) induces senescence very efficiently in human primary cells. In contrast, since PM-Ras(61L) synergizes with activated Cdc42 to more efficiently transform cells, these data support an alternative interpretation that multiple compartment-specific pathways, which include the endomembrane Cdc42 pathway, must be activated together for efficient cell transformation. This conclusion is consistent with several reports in which Ras targets, such as Raf and Cdc42, can work in concert to transform cells (18, 34, 35, 37, 54). One obvious caveat of restricting H-Ras to the PM by the use of the Rit protein is that the resulting H-Ras can no longer undergo the natural palmitoylation and depalmitoylation cycle, which is thought to control PM targeting and internalization of H-Ras. It is possible that some Ras-effector complexes may need to be internalized from the PM for efficient transformation, but this process is now blocked.
While oncogenic K-Ras4B can transform NIH 3T3 cells, when it was examined side by side with oncogenic H-Ras in transient transfection assays, we along with others (22) have found that oncogenic H-Ras transforms cells much more efficiently than oncogenic K-Ras4B. Li et al. (22) further investigated the potential cause of this difference in transformation efficiency, and they found that H-Ras appears to selectively act through the phosphatidylinositol 3-kinase (PI3K)-AKT pathway while K-Ras activates the Raf-MEK kinase pathway (22). These data agree with those in our study that activation of the Raf-Erk pathway by oncogenic PM-Ras can only weakly transform NIH 3T3 cells and that oncogenic K-Ras4B is the least efficient among the three Ras proteins tested in the binding to Cdc42.
We caution that cell transformation induced by the Cdc42(12V) and PM-Ras(61L) pair is weaker than that induced by normal unrestricted H-Ras(61L). One interpretation of this is that, besides Cdc42, there may be additional endomembrane effectors that need to be activated for more efficient transformation. Intriguingly, Tsutsumi et al. have recently examined the binding between H-Ras and the catalytic subunit of PI3K and found that this also occurs in endosomes (48). It is possible that some fractions of PI3K may be activated by Ras in the endomembrane but not the PM, as generally assumed, during transformation. Our model may also explain why cell transformation induced by EM-H-Ras is about 30 to 50% weaker than that induced by unrestricted H-Ras because the former does not efficiently activate effectors on the PM. As an alternative, it is also possible that the tested EM-H-Ras may not localize properly to all the endomembrane compartments visited by normal H-Ras. One major difference between the normal and the EM-restricted H-Ras is the lack of a palmitoylation site in the latter, which has long been thought to influence only PM targeting. However, in Rocks et al. (36), when a modified form of H-Ras that is resistant to depalmitoylation was injected to the cell, it associated nonspecifically with internal membranes without being concentrated at the Golgi compartment (36), suggesting that proper cycling between palmitoylation and depalmitoylation is important for proper Ras localization to the endomembrane as well. The difference in localization to the endomembrane compartments may also partly explain the observation that the unrestricted H-Ras binds Cdc42 more efficiently than EM-H-Ras does.
It has been well documented that Ras proteins can regulate different pathways in different cells (43). The Ras pathways in NIH 3T3 cells have long been used as a model system to study Ras signaling. As such, we caution that while activating PM-restricted Ras pathways can only weakly transform these cells, activating these Ras pathways may be sufficient for tumorigenesis in other types of cells. Furthermore, in addition to transformation, Ras proteins also control a wide range of activities, and it is unclear to what extent these activities are regulated in a cell compartment-specific manner. In fact, in this study we show that PM-restricted Ras appears to more efficiently induce senescence in human primary cells. Despite these caveats, our ability to evaluate cancer risk may be improved when we can detect where in the cell activated Ras proteins reside.
Supplementary Material
Acknowledgments
We thank Michael White, Marc Symons, Piero Crespo, John Colicelli, Danny Manor, Mark Philips, Peter McPherson, and Zhou Songyang for kindly providing reagents and the BCM Cytometry and Cell Sorting Core for superb help. We appreciate John Hancock and Sarah Plowman for technical support. We thank Gary Chamness for thoughtfully reading the manuscript and the Chang lab for discussion and technical help.
R.S., J.H., and the Pathology Core at the Lester and Sue Smith Breast Center are supported by a SPORE NIH grant (P50-CA58183), and E.C.C. is supported by grants from the NIH (CA90464, CA107187, GM81627, and P50-CA58183).
Footnotes
Published ahead of print on 28 December 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Cerione, R. A. 2004. Cdc42: new roads to travel. Trends Cell Biol. 14:127-132. [DOI] [PubMed] [Google Scholar]
- 2.Chang, E. C., et al. 1994. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 79:131-141. [DOI] [PubMed] [Google Scholar]
- 3.Chang, E. C., and M. R. Philips. 2006. Spatial segregation of Ras signaling: new evidence from fission yeast. Cell Cycle 5:1936-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, L. Y., D. Liu, and Z. Songyang. 2007. Telomere maintenance through spatial control of telomeric proteins. Mol. Cell. Biol. 27:5898-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu, V. K., et al. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, V. K., et al. 2004. Carboxyl methylation of Ras regulates membrane targeting and effector engagement. J. Biol. Chem. 279:7346-7352. [DOI] [PubMed] [Google Scholar]
- 7.D'Cruz, C. M., et al. 2001. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat. Med. 7:235-239. [DOI] [PubMed] [Google Scholar]
- 8.Hancock, J. F., A. I. Magee, J. E. Childs, and C. J. Marshall. 1989. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57:1167-1177. [DOI] [PubMed] [Google Scholar]
- 9.Henis, Y. I., J. F. Hancock, and I. A. Prior. 2009. Ras acylation, compartmentalization and signaling nanoclusters (review). Mol. Membr. Biol. 26:80-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoadley, K. A., et al. 2007. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics 8:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, H., et al. 2008. Integration of transforming growth factor beta and RAS signaling silences a RAB5 guanine nucleotide exchange factor and enhances growth factor-directed cell migration. Mol. Cell. Biol. 28:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, J. R., T. D. Craggs, J. Christodoulou, and S. E. Jackson. 2007. Stable intermediate states and high energy barriers in the unfolding of GFP. J. Mol. Biol. 370:356-371. [DOI] [PubMed] [Google Scholar]
- 13.Hussain, N. K., et al. 2001. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3:927-932. [DOI] [PubMed] [Google Scholar]
- 14.Hussain, N. K., et al. 1999. Splice variants of intersectin are components of the endocytic machinery in neurons and nonneuronal cells. J. Biol. Chem. 274:15671-15677. [DOI] [PubMed] [Google Scholar]
- 15.Jura, N., E. Scotto-Lavino, A. Sobczyk, and D. Bar-Sagi. 2006. Differential modification of Ras proteins by ubiquitination. Mol. Cell 21:679-687. [DOI] [PubMed] [Google Scholar]
- 16.Karnoub, A. E., and R. A. Weinberg. 2008. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9:517-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerppola, T. K. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khosravi-Far, R., et al. 1996. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol. Cell. Biol. 16:3923-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khwaja, A., M. E. Dockrell, B. M. Hendry, and C. C. Sharpe. 2006. Prenylation is not necessary for endogenous Ras activation in non-malignant cells. J. Cell. Biochem. 97:412-422. [DOI] [PubMed] [Google Scholar]
- 20.Kroschewski, R., A. Hall, and I. Mellman. 1999. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol. 1:8-13. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. H., N. G. Della, C. E. Chew, and D. J. Zack. 1996. Rin, a neuron-specific and calmodulin-binding small G-protein, and Rit define a novel subfamily of ras proteins. J. Neurosci. 16:6784-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, W., T. Zhu, and K. L. Guan. 2004. Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J. Biol. Chem. 279:37398-37406. [DOI] [PubMed] [Google Scholar]
- 23.Lin, R., S. Bagrodia, R. Cerione, and D. Manor. 1997. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 7:794-797. [DOI] [PubMed] [Google Scholar]
- 24.Lu, A., et al. 2009. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J. Cell Biol. 184:863-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manser, E., et al. 1998. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell 1:183-192. [DOI] [PubMed] [Google Scholar]
- 26.Matallanas, D., et al. 2006. Distinct utilization of effectors and biological outcomes resulting from site-specific Ras activation: Ras functions in lipid rafts and Golgi complex are dispensable for proliferation and transformation. Mol. Cell. Biol. 26:100-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohney, R. P., et al. 2003. Intersectin activates Ras but stimulates transcription through an independent pathway involving JNK. J. Biol. Chem. 278:47038-47045. [DOI] [PubMed] [Google Scholar]
- 28.Nagai, T., et al. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, M., S. Schoch, and T. C. Sudhof. 1999. EHSH1/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. A protein connection between exocytosis and endocytosis? J. Biol. Chem. 274:18446-18454. [DOI] [PubMed] [Google Scholar]
- 30.Onken, B., H. Wiener, R. M. Philips, and E. C. Chang. 2006. Compartmentalized signaling of Ras in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 103:9045-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podsypanina, K., Y. Li, and H. E. Varmus. 2004. Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype. BMC Med. 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prior, I. A., et al. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3:368-375. [DOI] [PubMed] [Google Scholar]
- 33.Qiu, R. G., A. Abo, F. McCormick, and M. Symons. 1997. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol. Cell. Biol. 17:3449-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu, R. G., J. Chen, D. Kirn, F. McCormick, and M. Symons. 1995. An essential role for Rac in Ras transformation. Nature 374:457-459. [DOI] [PubMed] [Google Scholar]
- 35.Qiu, R. G., J. Chen, F. McCormick, and M. Symons. 1995. A role for Rho in Ras transformation. Proc. Natl. Acad. Sci. U. S. A. 92:11781-11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocks, O., et al. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307:1746-1752. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Viciana, P., et al. 1997. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89:457-467. [DOI] [PubMed] [Google Scholar]
- 38.Rojas, R., W. G. Ruiz, S. M. Leung, T. S. Jou, and G. Apodaca. 2001. Cdc42-dependent modulation of tight junctions and membrane protein traffic in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 12:2257-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roux, P., C. Gauthier-Rouviere, S. Doucet-Brutin, and P. Fort. 1997. The small GTPases Cdc42Hs, Rac1 and RhoG delineate Raf-independent pathways that cooperate to transform NIH 3T3 cells. Curr. Biol. 7:629-637. [DOI] [PubMed] [Google Scholar]
- 40.Roy, S., A. Lane, J. Yan, R. McPherson, and J. F. Hancock. 1997. Activity of plasma membrane-recruited Raf-1 is regulated by Ras via the Raf zinc finger. J. Biol. Chem. 272:20139-20145. [DOI] [PubMed] [Google Scholar]
- 41.Rual, J. F., et al. 2004. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14:2128-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 43.Shields, J. M., K. Pruitt, A. McFall, A. Shaub, and C. J. Der. 2000. Understanding Ras: “it ain't over ′til it's over.” Trends Cell Biol. 10:147-154. [DOI] [PubMed] [Google Scholar]
- 44.Shin, S. 1979. Use of nude mice for tumorigenicity testing and mass propagation. Methods Enzymol. 58:370-379. [DOI] [PubMed] [Google Scholar]
- 45.Somsel Rodman, J., and A. Wandinger-Ness. 2000. Rab GTPases coordinate endocytosis. J. Cell Sci. 113:183-192. [DOI] [PubMed] [Google Scholar]
- 46.Songyang, Z., Y. Yamanashi, D. Liu, and D. Baltimore. 2001. Domain-dependent function of the rasGAP-binding protein p62Dok in cell signaling. J. Biol. Chem. 276:2459-2465. [DOI] [PubMed] [Google Scholar]
- 47.Tall, G. G., M. A. Barbieri, P. D. Stahl, and B. F. Horazdovsky. 2001. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1:73-82. [DOI] [PubMed] [Google Scholar]
- 48.Tsutsumi, K., Y. Fujioka, M. Tsuda, H. Kawaguchi, and Y. Ohba. 2009. Visualization of Ras-PI3K interaction in the endosome using BiFC. Cell Signal. 21:1672-1679. [DOI] [PubMed] [Google Scholar]
- 49.Van Aelst, L., M. A. White, and M. H. Wigler. 1994. Ras partners. Cold Spring Harb. Symp. Quant Biol. 59:181-186. [DOI] [PubMed] [Google Scholar]
- 50.Vanni, C., et al. 2005. Constitutively active Cdc42 mutant confers growth disadvantage in cell transformation. Cell Cycle 4:1675-1682. [DOI] [PubMed] [Google Scholar]
- 51.Wang, W., et al. 2002. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol. Cell. Biol. 22:3389-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, M. A., et al. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 53.White, M. A., T. Vale, J. H. Camonis, E. Schaefer, and M. H. Wigler. 1996. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J. Biol. Chem. 271:16439-16442. [DOI] [PubMed] [Google Scholar]
- 54.Whitehead, I. P., K. Abe, J. L. Gorski, and C. J. Der. 1998. CDC42 and FGD1 cause distinct signaling and transforming activities. Mol. Cell. Biol. 18:4689-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright, L. P., and M. R. Philips. 2006. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47:883-891. [DOI] [PubMed] [Google Scholar]
- 56.Wu, W. J., J. W. Erickson, R. Lin, and R. A. Cerione. 2000. The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature 405:800-804. [DOI] [PubMed] [Google Scholar]
- 57.Zamanian, J. L., and R. B. Kelly. 2003. Intersectin 1L guanine nucleotide exchange activity is regulated by adjacent src homology 3 domains that are also involved in endocytosis. Mol. Biol. Cell 14:1624-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zlatkine, P., B. Mehul, and A. I. Magee. 1997. Retargeting of cytosolic proteins to the plasma membrane by the Lck protein tyrosine kinase dual acylation motif. J. Cell Sci. 110:673-679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.