Abstract
Nuclear factor κB (NF-κB) signaling controls a wide range of cellular functions such as tumor progression and invasion by inducing gene expression. Upon stimulation, NF-κB is translocated to the nucleus and binds to its target gene promoters to activate transcription by recruiting transcription coactivators. Although significant progress has been made in understanding NF-κB-mediated transactivation, little is known about how NF-κB is recruited to its target gene promoters. Here, we report that transducin β-like protein 1 (TBL1) controls the expression of NF-κB target genes by directly binding with NF-κB and facilitating its recruitment to target gene promoters. Tumor necrosis factor alpha stimulation triggered the formation of an NF-κB and TBL1 complex and subsequent target gene promoter binding. Knockdown of TBL1 impaired the recruitment of NF-κB to its target gene promoters. Interestingly, analysis of the Oncomine database revealed that TBL1 mRNA levels were significantly higher in invasive breast cancer tissues than in breast adenocarcinoma tissue. Consistently, TBL1 knockdown significantly reduced the invasive potential of breast cancer cells by inhibiting NF-κB. Our results reveal a new mechanism for the regulation of NF-κB activation, with important implications for the development of novel strategies for cancer therapy by targeting NF-κB.
NF-κB, a pleiotropic transcription factor, activates a variety of gene transcription factors, including cytokines, angiogenesis modifiers, cell adhesion molecules, and antiapoptotic factors (29, 34, 45-48). The classical form of NF-κB is a heterodimer consisting of p65/RelA and p50 subunits. In most unstimulated normal cells, NF-κB is retained in the cytoplasm by IκB inhibitory proteins. The stimulation of cells with tumor necrosis factor alpha (TNF-α), interleukin-1, or various bacterial products activates the IκB kinase (IKK) complex, leading to the phosphorylation of IκB family proteins. Phosphorylated IκB is polyubiquitinated by ubiquitin ligase and subsequently degraded by 26S proteasome machinery, resulting in translocation of NF-κB to the nucleus to induce gene expression (31, 32, 45-48). Growing evidence suggests that aberrant activation of NF-κB is a common feature of cancer cells (37, 38). NF-κB has been shown to promote tumor invasion and metastasis by regulating the expression of chemokines, cellular adhesion molecules, and matrix metalloproteinases (2, 9, 30, 31, 33). Previously we demonstrated that constitutive activation of NF-κB promotes osteolytic breast cancer bone metastasis (33). Improved understanding of the molecular mechanisms that control NF-κB-mediated gene regulation may lead to development of novel strategies to target NF-κB signaling for inhibition of cancer progression and metastasis.
When NF-κB moves into the nucleus, it interacts with the transcription coactivator CBP/p300 to activate transcription. While many studies have shown that posttranscriptional modification of p65 affects how it interacts with coactivators and corepressors, it is unknown how p65 is recruited to the NF-κB target gene promoter to activate gene transcription. In general, transcriptional activation by liganded nuclear receptors and other regulated transcription factors requires the dismissal of a corepressor and subsequent recruitment of coactivators with intrinsic enzyme activities (36). Transducin β-like protein 1 (TBL1) and TBL1-related protein (TBLR1), two highly related F-box and WD-40 domain-containing proteins, are the intrinsic components of the NCoR corepressor complex (11, 26, 42, 49, 51). TBL1 and TBLR1 have been found to be important in ligand-induced activation by serving as specific adaptors for the recruitment of the ubiquitin-conjugating/19S proteasome complex, which mediates the exchange of corepressors for coactivators (35). TBL1 is also involved in p53-mediated degradation of β-catenin induced by DNA-damaging agents (28). Recently, TBL1 was reported to interact with CtBP and mediate its ubiquitylation and degradation, thus leading to a dismissal of corepressor complexes during gene activation (36).
Aside from serving a crucial role in regulated gene expression, our recent findings revealed unique functions of TBL1 in canonical Wnt signaling. We found that TBL1 plays an important role in the recruitment of β-catenin to the Wnt target gene promoter to activate transcription (25). Interestingly, TBL1 has also been found to play a role in NF-κB-induced gene expression (35). In this study, we investigated the role of TBL1 in NF-κB-mediated gene transcription. We found that TBL1 was required for the recruitment of p65 to the NF-κB target gene promoter. Importantly, knockdown of TBL1 impaired p65 binding to the NF-κB target gene promoter and subsequent gene transactivation. Analysis of the Oncomine database revealed that levels of TBL1 mRNA expression were increased in breast carcinoma, compared with normal breast tissues. Moreover, levels of TBL1 mRNA expression in invasive breast carcinoma were significantly higher than those in breast adenocarcinoma, implying that TBL1 has a proinvasive function in breast cancer. Consistently, we found that knockdown of TBL1 in the highly metastatic breast cancer cell line MDA-MB-231 suppressed invasion by inhibiting NF-κB activation. Taken together, our findings suggest that TBL1 controls NF-κB activation by recruiting NF-κB to its target gene promoter.
MATERIALS AND METHODS
Cell culture, plasmids, and viral infection.
Human embryonic kidney 293T, HT1080, and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (streptomycin and penicillin) at 37°C in a 5% CO2-95% air atmosphere. Human squamous cell carcinoma M4e cells were maintained in DMEM-F12 (1:1) medium supplemented with 10% heat-inactivated FBS. p65+/+ and p65−/− mouse embryonic fibroblasts (MEFs) were maintained in DMEM with 15% FBS. For transient transfections, 2 × 106 293T cells were seeded into six-well plates for 12 h and then transfected using Lipofectamine 2000 reagents according to the manufacturer's protocol (Invitrogen). The total amount of DNA in each individual well was kept constant by adding empty vectors as appropriate. To stably knock down p65 and TBL1, lentiviruses expressing p65 shRNA or TBL1 shRNA were packaged and generated in 293T cells as described previously (25, 33). MDA-MB-231, HT1080, or M4e cells were seeded in the six-well plates overnight and then infected with lentiviral particles. At 24 h after infection, the cells were selected with puromycin (1 μg/ml) for at least 1 week. Knockdown efficiency was determined by real-time reverse transcription-PCR (RT-PCR) or Western blot analysis. For small interfering RNA(siRNA)-mediated knockdown of TBL1, cells were transfected with control or TBL1 siRNA by using Oligofectamine reagents according to the manufacturer's protocol (Invitrogen).
Flag-TBL1, p65, and the fragments of these two molecules were constructed in a pQNCX2 vector by standard PCR subcloning. TBL1 and p65 shRNA constructs were prepared using the lentiviral expression vector pLKO.1. The targeting sequence used for TBL1 shRNA was 5′-AAGATGAGCATAACCAGTGAC-3′, and the sequence used for TBL1 siRNA was 5′-TCACTGGACTGGAATACCA-3′. The targeting sequences used for p65 used were the following: 5′-GTGACAAGGTGCAGAAAGA-3′ (p65 shRNA1), 5′-GTGACAAGGTGCAGAAAGA-3′ (p65 shRNA2), and 5′-ACCATCAACTATGATGAGTT-3′ (p65 shRNA3).
Real-time RT-PCR.
Total RNA from cells was purified using TRIzol reagent, and cDNA was synthesized with oligo(dT) primers by using SuperScript III (Invitrogen). Real-time RT-PCR analysis was carried out with iQ SYBR green supermix (Bio-Rad) on an iCycler iQ real-time PCR detection system (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels were used as an internal control for real-time PCR. Sequences of the primer pairs used were as follows: IκBα, 5′-GATCCGCCAGGTGAAGGG-3′ and 5′-GCAATTTCTGGCTGGTTGG-3′; c-IAP2, 5′-CCATTTTGAACCTGGATA-3′ and 5′-CATTTCCACGGCAGCA-3′; GAPDH, 5′-ATCATCCCTGCCTCTACTGG-3′ and 5′-GTCAAGTCCACCACTGACAC-3′.
Coimmunoprecipitation and Western blot analysis.
Cells were lysed in 500 μl of lysis buffer for 30 min on ice. After centrifugation at 10,600 × g at 4°C, the supernatants were incubated with antibodies at 4°C overnight, followed by incubation with protein G-Sepharose (GE Healthcare) for 1 h. For nuclear coimmunoprecipitation, the nuclear extracts were prepared from HT1080, MDA-MB-231, or M4e cells, as described previously (25). Immunoprecipitates were washed three times with lysis buffer at 4°C. Proteins bound to the beads were eluted with SDS loading buffer at 98°C for 2 min and then subjected to SDS-PAGE. Western blot analysis was performed as described previously (25). All antibodies used are commercially available: p65 (1:1,000; Upstate), anti-Flag (1:5,000; Sigma), antihemagglutinin (anti-HA; 1:5,000; Covance), TBL1 (1:1,000; Abcam), TBLR1 (1:5,000; Bethyl Laboratories), α-tubulin (1:10,000; Sigma-Aldrich).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP assay kit according to the manufacturer's protocol (Upstate Biotechnology). Cells were preincubated with a dimethyl 3,3′-dithiobispropionimidate-HCl (DTBP; Pierce) solution (5 mM) for 30 min on ice and then treated with formaldehyde. For each ChIP reaction mixture, 2 × 106 cells were used. The resulting precipitated DNA samples were quantified by real-time PCR. Data are expressed as the percentage of input DNA. The primer sequences used were the following: human IκBα promoter, 5′-TTGCCAGGCCACTGATTAAGAGGAA-3′ and 5′-GCTCATCAAAAAGTTCCCTGTCCGT-3′; human IκBα ORF, 5′-CCAGTCTCGGTGTACCTCCT-3′ and 5′-GGCCATTTGTGGAAGAAGTT-3′; human c-IAP2 promoter, 5′-TGCACTGGTGCTTTCCTTTTAGGA-3′ and 5′-GGGGGAAAAAGAAGCCTAAACCCT-3′); human c-IAP2 ORF, 5′-TACAATAGAATCAGTTCCCAGTGGGC-3′ and 5′-CTGGTTGTTTGCCTCAAATCACAG-3′; mouse IκBα promoter, 5′-CAGGGAGTTTCTCCGATGA-3′ and 5′-GGAATTTCCAAGCCAGTCAG-3′; mouse IκBα ORF, 5′-CTGGTGCTGAGAAGGATCAA-3′ and 5′-GCCAAGTCTTATGTAGACCAAGG-3′; mouse IP-10 promoter, 5′-ACTGGAATTACTCTTACGGC-3′ and 5′-CTAAGAGTCCGCTCCCTATG-3′; mouse IP-10 ORF, 5′-TGCACAGCAGCTACAACAAC-3′ and 5′-TAAATCCTCCAGGTGGGAAC-3′.
Oncomine data analysis.
TBL1 mRNA expression in breast cancers from three studies (38-40) was analyzed using Oncomine. Details of standardized normalization techniques and statistical calculations can be found on the Oncomine website. First, standard analyses were applied to raw microarray data by using either the robust multichip average for Affymetrix data or the Loess normalization for cDNA arrays. To scale the data and allow comparison of multiple independent studies, Z-score normalization was applied by Oncomine. This included log2 transformation, setting the array median to 0, and standard deviation to 1. To determine whether TBL1 was differentially expressed, two-sided t tests were conducted using SPSS 13.0.
Matrigel invasion assays.
BD BioCoat Matrigel invasion chambers (24 wells) were used for tumor cell invasion assays. Cells (1 × 105) suspended in 0.5 ml serum-free medium were plated on Matrigel. In the lower chamber, 0.75 ml of medium containing 0.1% FBS was added as a chemoattractant. At 24 h after incubation, Matrigel was removed and the invaded cells were stained with the HEMA-3 kit (Fisher) and counted.
RESULTS
Requirement of TBL1 for NF-κB target gene expression.
We previously generated a TBL1 shRNA vector that is able to specifically knock down TBL1 (25). To investigate a potential role of TBL1 in NF-κB-dependent gene expression, we utilized the TBL1 shRNA vector to knock down TBL1 in the sqaumous carcinoma cell line M4e. Western blot analysis showed that over 85% of TBL1 was depleted in M4e cells expressing TBL1shRNA (M4e/TBL1sh), compared with M4e cells expressing scramble shRNA (M4e/Scrsh). In contrast, TBL1 knockdown did not affect TBLR1 or p65 expression (Fig. 1A). To determine whether the knockdown of TBL1 affected NF-κB target gene expression, both M4e/Scrsh and M4e/TBL1sh cells were treated with TNF-α for 0, 2, and 4 h. Real-time RT-PCR revealed that two well-known NF-κB target genes, IκBα and c-IAP2, were significantly inhibited in M4e/TBL1sh cells compared with M4e/Scrsh cells (Fig. 1B). To rule out off-target effects, we also transiently knocked down TBL1 using TBL1 siRNA that targeted a different TBL1 sequence. Similarly, knockdown of TBL1 by siRNA also suppressed the expression of IκBα and c-IAP2 induced by TNF-α (Fig. 1C and D). Previously, we and others reported that NF-κB is constitutively activated in the highly metastatic breast cancer cell line MDA-MB-231 (13, 30, 33). To determine whether TBL1 was required for the expression of c-IAP2 and IκBα in MDA-MB-231 cells, we also depleted TBL1 in these cells (Fig. 1E). As shown in Fig. 1F, while the basal levels of IκBα and c-IAP2 were higher in MDA-MB-231 cells expressing scramble shRNA (MDA/Scrsh) than in MDA-MB-231 cells expressing TBL1 shRNA (MDA/TBL1sh), the depletion of TBL1 significantly inhibited TNF-α-induced expression of IκBα and c-IAP2 in MDA/TBL1sh cells. Taken together, our results suggest that TBL1 plays an important role in NF-κB-mediated gene expression.
FIG. 1.
Requirement of TBL1 for NF-κB target gene expression. (A) Knockdown of TBL1 in M4e cells by shRNA. Western blotting was performed using anti-TBL1, anti-p65, anti-TBLR1, and anti-α-tubulin antibodies. (B) Knockdown of TBL1 inhibited TNF-α-stimulated NF-κB target gene expression in M4e cells. M4e/Scrsh and M4e/TBL1sh cells were stimulated with TNF-α for the indicated times. The expression levels of IκBα and c-IAP2 were determined by quantitative RT-PCR. Values are means ± standard deviations of triplicate samples from a representative experiment. (C) Knockdown of TBL1 in M4e cells by siRNA. M4e cells were transiently transfected with scramble siRNA (Scrsi) or TBL1 siRNA (TBL1si). TBL1 expression was determined by the real-time RT-PCR. (D) Knockdown of TBL1 by siRNA inhibited TNF-α-induced NF-κB target gene expression in M4e cells. The expression levels of IκBα and c-IAP2 were determined by quantitative RT-PCR. (E) Knockdown of TBL1 in MDA-MB-231 cells. Western blotting was performed using anti-TBL1, anti-p65, anti-TBLR1, and anti-α-tubulin antibodies. (F) TBL1 knockdown inhibited NF-κB target gene expression in MDA-MB-231 cells. MDA/Scrsh and MDA/TBL1sh cells were stimulated with TNF-α for the indicated times. The expression levels of IκBα and c-IAP2 were determined by quantitative RT-PCR. Values are means ± standard deviations of triplicate samples from a representative experiment.
TBL1 interacts with RelA/p65.
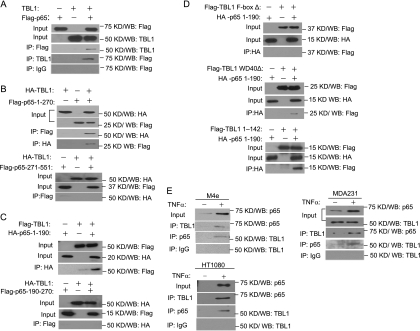
To gain mechanistic insight into the way in which TBL1 positively regulates NF-κB target genes, we examined whether TBL1 interacted with p65 by overexpressing Flag-tagged p65 and TBL1 in HEK293T cells. The whole-cell lysates were prepared and immunoprecipitated with either anti-Flag or anti-TBL1 antibodies. Western blot analysis revealed that anti-Flag antibodies could immunoprecipitate TBL1 or vice versa (Fig. 2A). p65 contains the N-terminal Rel homology domain (RHD), which mediates DNA binding, dimerization, and nuclear localization and enables association with the inhibitory protein IκB, and the C-terminal transactivation domain (TAD), which enables the recruitment of cotranscriptional regulators and basal transcriptional machinery to target genes (31). TBL1 is composed of an N-terminal F-box domain and a C-terminal WD-40 domain (25). Seeking to determine the specific domains that are involved in the interaction between p65 and TBL1, we performed extensive map analysis by overexpressing various domains of NF-κB and TBL1 in HEK293T cells. As shown in Fig. 2B, we found that HA-TBL1 was able to pull down the N-terminal RHD (amino acids 1 to 270) of p65, but not the C-terminal domain (amino acids 271 to 551) of p65. Reciprocally, p65-1-270 was able to pull down TBL1. Further deletion analysis found that the N-terminal amino acids 1 to 190 of p65 (p65-1-190) interacted with TBL1, while the fragment covering amino acids 191 to 270 did not show any binding activity (Fig. 2C). As shown in Fig. 2D, deletion of the F-box domain in TBL1 abolished the interaction between TBL1 and p65-1-190, while the deletion of the WD-40 domain did not affect the interaction between TBL1 and p65-1-190. Moreover, the TBL1-F-box could also pull down p65-1-190, suggesting that the F-box domain of TBL1 interacted with the RHD of NF-κB (Fig. 2D). To further determine whether endogenous TBL1 and p65 interacted with each other, we treated M4e or HT1080 cells with TNF-α and isolated nuclear extracts for immunoprecipitation. As shown in Fig. 2E, TNF-α induced interaction between TBL1 and p65 in both M4e and HT1080 cells. In MDA-MB-231 cells, endogenous TBL1 could be coimmunoprecipitated with p65 at basal levels, and TNF-α stimulation further enhanced the complex formation. The reciprocal coimmunoprecipitation experiment also showed that TBL was present in the p65 precipitates (Fig. 2E).
FIG. 2.
TBL1 interacts with RelA/p65. (A) TBL1 interacts with Flag-p65. 293T cells were transfected with vectors expressing Flag-p65 or TBL1 alone or in combination as indicated in the blots. (B) TBL1 interacts with the N-terminal RHD of p65. (C) TBL1 interacts with the N-terminal amino acids 1 to 190 of p65. (D) The F-box of TBL1 is required for the p65-TBL1 interaction. (E) TNF-α-induced endogenous interaction between TBL1 and p65. M4e, HT1080, or MDA-MB-231 cells were treated with TNF-α (10 ng/ml) for 30 min. The nuclear extracts were prepared and immunoprecipitated with anti-TBL1 or anti-p65 antibodies.
Simultaneous recruitment of TBL1 and p65 to NF-κB target gene promoters.
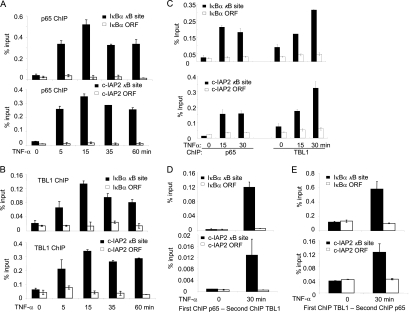
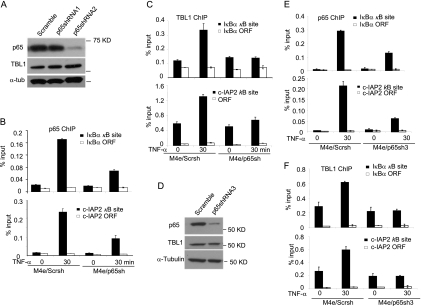
Due to the facts that TBL1 interacts with p65 in the nucleus and is required for TNF-α-induced NF-κB target gene expression, we hypothesized that TBL1 might be recruited along with NF-κB to the target gene promoter to induce gene transactivation. To examine this possibility, we performed ChIP assays to determine whether TBL1 was recruited to IκBα and c-IAP2 promoters upon TNF-α stimulation. The ChIP-enriched DNA was quantified by real-time PCR using specific primers covering κB binding sites on the IκBα and c-IAP2 gene promoters. As a strict negative control, a region located in the open reading frame (ORF) was also examined. In the absence of TNF-α stimulation, there was basal binding of TBL1 to IκBα and c-IAP2 promoters in M4e cells. TNF-α stimulation strongly enhanced both p65 and TBL1 binding to the IκBα and c-IAP2 promoters with similar kinetics (Fig. 3A and B). We also found that TNF-α induced both TBL1 and p65 binding to the promoter of IκBα and c-IAP2 in HT1080 cells (Fig. 3C). To determine whether TBL1 and p65 were colocalized on the promoters of IκBα and c-IAP2, we performed two-step ChIP or re-ChIP assays in M4e cells. The chromatin complexes isolated from control and TNF-α-stimulated cells were subjected to a first round of ChIP with anti-p65 antibodies followed by re-ChIP with anti-TBL1 antibodies. Compared with the nonstimulated cells, a significant increase in TBL1 binding to the IκBα and c-IAP2 promoters was detected as determined by real-time PCR (Fig. 3D). The reciprocal re-ChIP assay using anti-TBL1 antibody for the initial isolation of chromatin complexes and anti-p65 antibody for re-ChIP also demonstrated that anti-p65 could pull down TBL1 chromatin complexes (Fig. 3E). These results suggest the simultaneous recruitment and cooccupancy of p65 and TBL1 on NF-κB target gene promoters.
FIG. 3.
TNF-α induces the recruitment of TBL1 and p65 to the NF-κB target gene promoter. (A) TNF-α-induced p65 binding to the IκBα and c-IAP2 promoters in M4e cells. M4e cells were treated with TNF-α for different times as indicated. ChIP assays were performed with a ChIP assay kit from Upstate Biotechnology with anti-p65 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (B) TNF-α-induced TBL1 binding to the IκBα and c-IAP2 promoters in M4e cells. M4e cells were treated with TNF-α for different time points as indicated. ChIP assays were performed using anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (C) TNF-α induces the recruitment of TBL1 and p65 to the NF-κB target gene promoter in HT1080 cells. TNF-α induced TBL1 and p65 binding to the IκBα and c-IAP2 promoters in HT1080 cells. Cells were treated with TNF-α for 0, 15, and 30 min. ChIP assays were performed with a ChIP assay kit from Upstate Biotechnology with anti-p65 and anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (D) p65 and TBL1 cooccupy the NF-κB target gene promoter, as determined in a re-ChIP assay. M4e cells were treated with TNF-α for 0 and 30 min. The chromatin complexes were first precipitated with anti-p65 antibodies and then subjected to re-ChIP with anti-TBL1 antibodies. Sequentially enriched genomic DNA was amplified by real-time PCR using specific IκB-α and c-IAP2 primer sets. (E) Reverse re-ChIP assays confirming cooccupancy of p65 and TBL1 on the NF-κB target gene promoter. The chromatin complexes were first immunoprecipitated with anti-TBL1 antibodies and then with anti-p65 antibodies.
TBL1 requirement for p65 binding to NF-κB target gene promoters.
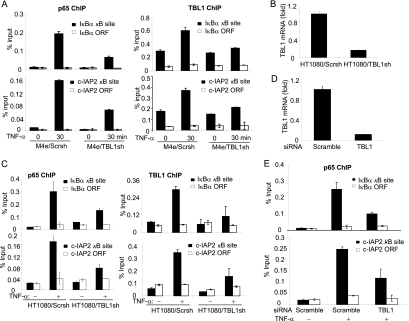
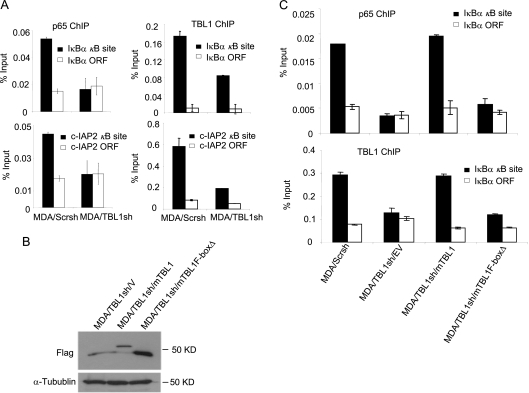
Next, we tested whether TBL1 depletion affected the promoter binding capacity of p65, or vice versa. ChIP assays revealed that the depletion of TBL1 in M4e cells significantly inhibited the recruitment of p65 to the IκBα and c-IAP2 promoters induced by TNF-α (Fig. 4A). Using lentiviruses expressing TBL1 shRNA, we stably knocked down TBL1 in HT1080 cells, as confirmed by RT-PCR (Fig. 4B). The knockdown of TBL1 in HT1080 cells reduced the recruitment of p65 to the IκBα and cIAP-2 promoters induced by TNF-α (Fig. 4C). Also, the knockdown of TBL1 by siRNA that targeted a different TBL1 sequence in HT1080 cells suppressed the recruitment of p65 to the IκBα and c-IAP2 promoters induced by TNF-α, confirming shRNA specificity (Fig. 4D and E). Since NF-κB is constitutively activated in MDA-MB-231 cells, we examined p65 and TBL1 binding activity in both MDA/Scrsh and MDA/TBL1sh cells. We found that the depletion of TBL1 abolished NF-κB binding to the IκBα and c-IAP2 promoters (Fig. 5A). To further confirm our results, we performed rescue experiments in MDA/TBL1sh cells by stably reexpressing the full-length mouse TBL1 (mTBL1) and the F-box-deleted TBL1 mutant (TBL1-F-boxΔ), which are resistant to human TBL1shRNA. Both mTBL1 and TBL1-F-boxΔ expression levels in MDA/TBL1sh cells were confirmed by Western blot analysis (Fig. 5B). As expected, restoration of mTBL1 rescued both TBL1 and p65 recruitment to the IκBα promoter in MDA/TBL1sh cells (Fig. 5C). However, TBL1-F-boxΔ was unable to rescue TBL1 and p65 binding to the IκBα promoter, indicating that the TBL1 F-box domain plays a critical role in the recruitment of NF-κB to chromatin. Taken together, our results suggest that TBL1 is required for the recruitment of p65 to the NF-κB target gene promoters.
FIG. 4.
TBL1 is required for recruitment of p65 to the NF-κB target gene promoter. (A) Knockdown of TBL1 impaired p65 recruitment to the IκBα and c-IAP2 promoters in M4e cells. (Left) ChIP with anti-p65 antibodies; (right) ChIP with anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (B) Knockdown of TBL1 in HT1080 cells. TBL1 knockdown was determined by real-time RT-PCR. (C) Knockdown of TBL1 inhibits p65 binding to the IκBα and c-IAP2 promoters in HT1080 cells. HT1080/Scrsh and HT1080/TBL1sh cells were stimulated with TNF-α for 0 and 30 min. (Left) ChIP with anti-p65 antibodies; (right) ChIP with anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (D) TBL1 was depleted by siRNA in HT1080 cells. HT1080 cells were transfected with scramble and TBL1 siRNA. At 48 h posttransfection, TBL1 expression was assessed by real-time RT-PCR. (E) Depletion of TBL1 in HT1080 cells inhibited p65 binding to the IκBα and c-IAP2 promoters induced by TNF-α. ChIP assays were performed with anti-p65 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment.
FIG. 5.
The TBL1 F-box domain plays a critical role in the recruitment of NF-κB to chromatin. (A) Knockdown of TBL1 inhibits p65 binding to the IκBα and c-IAP2 promoters in MDA-MB-231 cells. ChIP assays were performed with anti-p65 or anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (B) The full-length mTBL1 and mTBL1F-boxΔ were expressed in MDA/TBL1sh cells. MDA/TBL1sh cells were stably transduced with retroviruses expressing Flag-mTBL1, Flag-mTBL1F-boxΔ, or empty vector. TBL1 expression was detected by Western blotting using anti-Flag antibodies. (C) The TBL1 N-terminal F-box domain is required for p65 and TBL1 recruitment to the NF-κB target gene promoter. TBL1 and p65 occupancy levels on the NF-κB target gene promoter in MDA/Scr/V, MDA/TBL1sh/mTBL1, and MDA/TBL1sh/mTBL1F-boxΔ cells were assessed by ChIP assays using anti-p65 and anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment.
To test whether p65 might affect the TBL1 recruitment to the NF-κB target gene promoters, we stably knocked down p65 in M4e cells. As shown in Fig. 6A, over 90% of p65 proteins were depleted by lentiviruses expressing p65 shRNA2, while TBL1 proteins were not affected. We then utilized the M4e cells expressing p65 shRNA2 (M4e/p65sh) for ChIP assays. As predicted, knockdown of p65 reduced p65 binding to the IκBα and c-IAP2 promoters induced by TNF-α in M4e/p65sh cells, compared with M4e cells expressing scramble shRNA (M4e/Scrsh) (Fig. 6B). Importantly, knockdown of p65 in M4e/p65sh cells significantly inhibited the recruitment of TBL1 to the IκBα and c-IAP2 promoters induced by TNF-α (Fig. 6C). In contrast, knockdown of p65 did not modulate TBL1 binding to the promoter of Axin2, a non-NF-κB target gene (data not shown). To rule out any off-target effects, we also stably knocked down p65 in M4e cells by using another shRNA targeting a different p65 sequence (p65shRNA3). Similar to the results noted for p65shRNA2, p65shRNA3 also depleted more than 80% of endogenous p65 proteins in M4e cells, as determined by Western blotting (Fig. 6D). Also, p65shRNA3 inhibited the recruitment of p65 and TBL1 to IκBα and c-IAP2 promoters induced by TNF-α (Fig. 6E and F). However, unlike our findings in M4e cells, we found that the knockdown of p65 in HT1080 cells and MDA-MB-231 cells did not significantly affect the recruitment of TBL1 to the IκBα and c-IAP2 promoters (data not shown).
FIG. 6.
p65 knockdown impairs TBL1 binding to the NF-κB target gene promoters in M4e cells. (A) p65 was knocked down in M4e cells by shRNA. M4e cells were infected with lentiviruses expressing scramble shRNA, p65 shRNA1, or p65 shRNA2. p65 knockdown was determined by Western blot analysis. (B) Knockdown of p65 reduces p65 binding to the IκBα and c-IAP2 promoters in M4e cells. M4e/Scrsh and M4e/p65sh cells were stimulated with TNF-α for 0 and 30 min. ChIP assays were performed with anti-p65 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (C) Knockdown of p65 impaired TBL1 binding to the IκBα and c-IAP2 promoters in M4e cells. M4e/Scrsh and M4e/p65sh cells were stimulated with TNF-α for 0 and 30 min. ChIP assays were performed with anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (D) p65 knocked down in M4e cells by p65 shRNA3. M4e cells were infected with lentiviruses expressing scramble shRNA or p65 shRNA3. p65 knockdown was determined by Western blot analysis. (E) Knockdown of p65 by p65shRNA3 reduces p65 occupancy on the IκBα and c-IAP2 promoters in M4e cells. M4e/Scrsh and M4e/p65sh3 cells were stimulated with TNF-α for 0 and 30 min. ChIP assays were performed with anti-p65 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (F) Knockdown of p65 inhibits TBL1 binding to the IκBα and c-IAP2 promoters in M4e cells. M4e/Scrsh and M4e/p65sh3 cells were stimulated with TNF-α for 0 and 30 min. ChIP assays were performed with anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment.
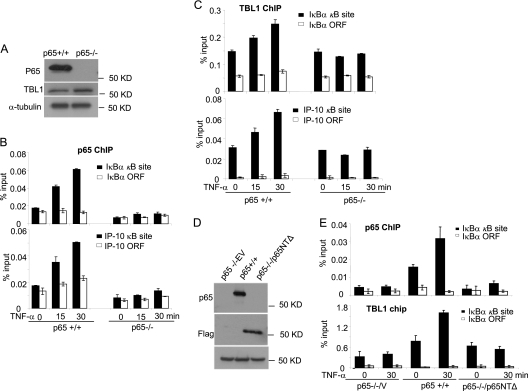
To further examine the role of p65 in TBL1 recruitment, we took advantage of p65+/+ and p65−/− MEFs. Western blot analysis showed that TBL1 was expressed in both p65+/+ MEFs and p65−/− MEFs at similar levels (Fig. 7A). ChIP assays revealed that TNF-α induced the recruitment of p65 and TBL1 to the IκBα and IP-10 gene promoters in p65+/+ MEF cells (Fig. 7B and C; of note, IP-10 is a well-known NF-κB target gene in murine cells). However, TNF-α-induced recruitment of TBL1 to the IκBα and IP-10 promoters was drastically reduced in p65−/− MEFs (Fig. 7C). Since the p65 N-terminal RHD is required for the interaction with TBL1, we also examined whether the p65 N-terminal domain played a role in the recruitment of TBL1 to the NF-κB target gene promoter by stably expressing the p65 N-terminal deletion mutant (p65NTΔ) in p65−/− MEFs (Fig. 7D). Overexpression of p65NTΔ in p65−/− MEFs was unable to rescue the recruitment of TBL1 to the IκBα and IP-10 gene promoters upon TNF-α stimulation (Fig. 7E).
FIG. 7.
p65 is required for TBL1 binding to the NF-κB target gene promoters. (A) p65 and TBL1 expression in p65+/+ MEFs and p65−/− MEFs by Western blot analysis. (B) TNF-α induces p65 binding to the IκBα and IP-10 promoters in p65+/+ MEFS but not in p65−/− MEFs. p65+/+ and p65−/− MEFs were exposed to TNF-α for 0, 15, and 30 min. ChIP assays were performed using anti-p65 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (C) p65 depletion abolishes TBL1 binding to the IκBα and IP-10 promoters. p65+/+ and p65−/− MEFs were exposed to TNF-α for 0, 15, and 30 min. ChIP assays were performed with anti-TBL1 antibodies. Values are means ± standard deviations for triplicate samples from a representative experiment. (D) p65−/− MEFs were stably transduced with retroviruses expressing p65NTΔ (p65 lacking N-terminal amino acids 1 to 190) or control empty vector. p65−/− MEFs expressing p65NTΔ were assessed by Western blotting. (E) The p65 N-terminal domain is required for TBL1 binding to the IκBα promoter. p65+/+, p65−/−-vector, and p65−/−-p65NTΔ MEFs were stimulated with TNF-α for 0 and 30 min. p65 and TBL1 recruitment levels to the IκBα promoter were assessed by ChIP assays. Values are means ± standard deviations for triplicate samples from a representative experiment.
Promoting cell invasion by TBL1-dependepnt NF-κB activation.
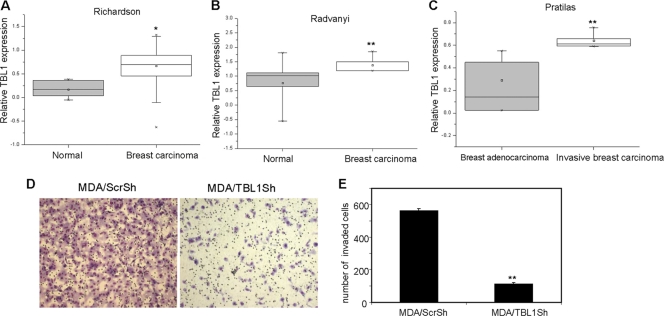
We and others have found that NF-κB plays a critical role in cancer cell invasive growth and metastasis (13, 17, 18, 30, 31, 33, 41). To explore whether TBL1 plays a role in tumor progression and metastasis, we analyzed the level of TBL1 in a variety of human breast cancer tissues by using Oncomine data as described in Materials and Methods. As shown in Fig. 8A and B, analysis of studies by Radvanyl et al. (39) and Richardson et al. (40) revealed that the expression of TBL1 was higher in breast carcinoma than in normal breast tissues. Analysis of studies by Pratilas et al. (38) demonstrated that the expression of TBL1 was significantly higher in invasive breast carcinoma than in breast adenocarcinoma tissues (Fig. 8C), suggesting that TBL1 might play a role in the invasive growth of breast cancer. To further explore whether TBL1 might regulate breast cancer cell invasion through NF-κB, we performed a Matrigel invasion assay using the highly invasive MDA-MB-231 cell line, in which there is constitutive activation of NF-κB (13, 30, 33). Previously, we showed that inhibition of NF-κB abolished MDA-MB-231 cell invasion (33). As depicted in Fig. 8D and E, while MDA/Scrsh cells remained highly invasive, knockdown of TBL1 in MDA/TBL1sh cells resulted in a dramatically reduced invasion of MDA-MB-231 cells through the Matrigel-coated membrane, indicating the importance of TBL1 in the proinvasive functions of NF-κB.
FIG. 8.
Inhibition of TBL1 suppresses breast cancer cell invasion. (A) TBL1 mRNA is higher in breast carcinoma than in normal breast tissues, based on studies reported by Richardson et al. (80). *, P < 0.05. (B) TBL1 mRNA is higher in breast carcinoma than in normal breast tissues, based on studies reported by Radvanyi et al. (39). **, P < 0.01. (C) TBL1 mRNA is highly expressed in invasive breast carcinoma compared with breast adenocarcinoma tissues. **, P < 0.01. (D) Depletion of TBL1 inhibits MDA-MB-231 cell invasion. The experiments were performed in triplicate, and the cells from six random fields were counted and averaged. Values represent means ± standard deviations of triplicate experiments.
DISCUSSION
NF-κB activation plays a critical role in cancer cell survival, transformation, invasion, and metastasis (15, 18, 22, 44). While the systems governing NF-κB activity are complex, the participation of various molecules regulating NF-κB transactivation is becoming better understood. The results presented here identify TBL1 as an important component required for NF-κB-mediated transcriptional activity. Unlike other regulators and interacting molecules related to NF-κB signaling, TBL1 plays a unique role in the recruitment of p65 to the NF-κB target gene promoter. Interestingly, this recruitment appears to be a mutually dependent process in some cells. Therefore, our findings provide important insights into the molecular regulation of NF-κB-dependent gene expression.
It is known that transcriptional activity of NF-κB is mainly regulated by transcription coactivators and corepressors that were originally associated with nuclear receptors (8). The components of the NF-κB corepressor complex include SMRT, NCoR, histone deacetylase 1 (HDAC1), HDAC2, and HDAC3 (4, 7, 19, 20, 23, 24). These proteins are also copurified with several repressor complexes, including TBL1 and TBLR1 complexes (12, 43, 49), indicating the potential role for these proteins in NF-κB-mediated gene regulations. Generally, signaling mechanisms regulate transcription by converging on SMRT/NCoR corepressor complexes to eliminate HDAC activities through a process called derepression, which occurs mostly as a result of the phosphorylation and nuclear export of the corepressor complexes (1, 14, 16, 21, 52). In addition, NCoR is known to be regulated by the proteasome complex that involves chromatin-associated recruitment of the E3 TBL1/TBLR1 and Ubc5 ubiquitin-conjugating enzymes for the exchange of corepressor in order for coactivators to induce transcription (35, 50). Our finding suggests that TBL1 may be an integral component of the NF-κB transcription complex, serving a crucial role in NF-κB promoter recruitment (35).
While Perissi et al. (35) demonstrated that TBL1 is required for the exchange of corepressor/coactivator in liganded nuclear receptors, a significant observation of our study characterizes the connection between the coupling of NF-κB and TBL1 and their subsequent recruitment to the promoter for induction of NF-κB target genes. Since the components of the corepressor complex for NF-κB, including SMRT, NCoR, HDAC1, HDAC2, and HDAC3, are reported to be copurified with TBL1 and TBLR1 (12, 43, 49), there is also a possibility that TBL1 is recruited to the NF-κB target gene promoter along with the NF-κB corepressor complex. However, this interpretation will require further mechanistic studies. Burstein et al. and others (3, 5, 10, 27) reported that COMM domain-containing protein 1 (COMMD1) interacts with NF-κB and negatively regulates its association with chromatin by inducing ubiquitination and degradation. Interestingly, COMMD1 and TBL interact with the same region of p65 (N-terminal amino acids 1 to 180) (3). Hence, it is possible that TBL1 can competitively inhibit COMMD1 binding on p65, thereby protecting chromatin-associated p65 from ubiquitination and degradation. Very recently, TBL1 was found to play a role in protecting β-catenin from Siah-1-mediated ubiquitination and proteasomal degradation (6). Therefore, it is reasonable to speculate that TBL1 also promotes expression of NF-κB target genes by stabilizing p65 on chromatin.
Our findings demonstrate that the mutual dependency between TBL1 and p65 in the recruitment process is restricted to certain cell types. While TBL1 knockdown impaired the recruitment of p65 in all cell lines studied, p65 depletion resulted in reduction of TBL1 promoter binding in M4e and p65−/− MEF cells, but not in HT1080 and MDA-MB-231 cells. As our extensive mapping studies determined the participation of p65 RHD in TBL1 interactions, and all Rel family members are known to have a highly conserved RHD, there is a possibility that other Rel family members play a compensatory role in bringing TBL1 to the NF-κB target gene promoter in p65-depleted HT1080 and MDA-MB-231 cells. In fact, variation in signaling components between cell lines is an expected phenomenon, because cell origin, genetic background, and cellular context determine cell phenotype and behavior. Therefore, the differences seen in such factors among the cell lines used in this study could explain the different outcomes of p65 depletion noted upon TBL1 recruitment.
In conclusion, we report here that a critical component of NF-κB-regulated gene transcription is the coordinated interactions between TBL1 and NF-κB. TBL1 plays a key role in the recruitment of p65 to the NF-κB target gene promoters for the induction of gene transcription. We have also demonstrated that this mechanism is important for the proinvasive functions of NF-κB, as knockdown of TBL1 significantly suppressed the invasiveness of breast cancer cells. Therefore, our findings suggest that targeting the interaction between NF-κB and TBL1 would be an efficient approach for preventing NF-κB signaling and subsequent cancer cell invasion and metastasis.
Acknowledgments
This work was supported by NICDR grants DE015964, DE17684, and DE13848 and NCI grant CA132134 to C.-Y.W.
Footnotes
Published ahead of print on 28 December 2010.
REFERENCES
- 1.Baek, S. H., et al. 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and beta-amyloid precursor protein. Cell 110:55-67. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, A. S. 2001. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J. Clin. Invest. 107:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burstein, E., et al. 2005. COMMD proteins, a novel family of structural and functional homologs of MURR1. J. Biol. Chem. 280:22222-22232. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L. F., W. Fischle, E. Verdin, and W. C. Greene. 2000. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293:1653-1657. [DOI] [PubMed] [Google Scholar]
- 5.de Bie, P., et al. 2006. Characterization of COMMD protein-protein interactions in NF-κB signalling. Biochem. J. 398:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrova, Y. N., et al. 2010. Direct ubiquitination of beta-catenin by Siah-1 and regulation by the exchange factor TBL1. J. Biol. Chem. 285:13507-13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa, L., J. Inglés-Esteve, A. Robert-Moreno, and A. Bigas. 2003. IκBα and p65 regulate the cytoplasmic shuttling of nuclear corepressors: cross-talk between Notch and NFκB pathways. Mol. Biol. Cell 14:491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, Z., et al. 2005. Coactivators and corepressors of NF-κB in IκB alpha gene promoter. J. Biol. Chem. 280:21091-21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garg, A., and B. B. Aggarwal. 2002. Nuclear transcription factor-κB as a target for cancer drug development. Leukemia 16:1053-1068. [DOI] [PubMed] [Google Scholar]
- 10.Geng, H., T. Wittwer, O. Dittrich-Breiholz, M. Kracht, and M. L. Schmitz. 2009. Phosphorylation of NF-κB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 10:381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guenther, M. G., et al. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 13.Helbig, G., et al. 2003. NF-κB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J. Biol. Chem. 278:21631-21638. [DOI] [PubMed] [Google Scholar]
- 14.Hermanson, O., K. Jepsen, and M. G. Rosenfeld. 2002. N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419:934-939. [DOI] [PubMed] [Google Scholar]
- 15.Hoberg, J. E., F. Yeung, and M. W. Mayo. 2004. SMRT derepression by the IκB kinase alpha: a prerequisite to NF-κB transcription and survival. Mol. Cell 16:245-255. [DOI] [PubMed] [Google Scholar]
- 16.Hong, S. H., and M. L. Privalsky. 2000. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol. Cell. Biol. 20:6612-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, M. C., et al. 2004. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117:225-237. [DOI] [PubMed] [Google Scholar]
- 18.Huber, M. A., et al. 2004. NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Invest. 114:569-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, K., P. J. Barnes, and I. M. Adcock. 2000. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell. Biol. 20:6891-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, K., E. Jazrawi, B. Cosio, P. J. Barnes, and I. M. Adcock. 2001. p65-activated histone acetyltransferase activity is repressed by glucocorticoids: mifepristone fails to recruit HDAC2 to the p65-HAT complex. J. Biol. Chem. 276:30208-30215. [DOI] [PubMed] [Google Scholar]
- 21.Jang, M. K., et al. 2001. Ca2+/calmodulin-dependent protein kinase IV stimulates nuclear factor-kappa B transactivation via phosphorylation of the p65 subunit. J. Biol. Chem. 276:20005-20010. [DOI] [PubMed] [Google Scholar]
- 22.Karin, M., Y. Cao, F. R. Greten, and Z. W. Li. 2002. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2:301-310. [DOI] [PubMed] [Google Scholar]
- 23.Kiernan, R., et al. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758-2766. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. K., J. H. Kim, Y. C. Lee, J. Cheong, and J. W. Lee. 2000. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-κB, and serum response factor. J. Biol. Chem. 275:12470-12474. [DOI] [PubMed] [Google Scholar]
- 25.Li, J., and C. Y. Wang. 2008. TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat. Cell Biol. 10:160-169. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., et al. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 9:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maine, G. N., X. Mao, C. M. Komarck, and E. Burstein. 2007. COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J. 26:436-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzawa, S. I., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 29.Mayo, M. W., et al. 2003. Ineffectiveness of histone deacetylase inhibitors to induce apoptosis involves the transcriptional activation of NF-kappa B through the Akt pathway. J. Biol. Chem. 278:18980-18989. [DOI] [PubMed] [Google Scholar]
- 30.Merkhofer, E. C., P. Cogswell, and A. S. Baldwin. 2010. Her2 activates NF-κB and induces invasion through the canonical pathway involving IKKα. Oncogene 29:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakshatri, H., P. Bhat-Nakshatri, D. A. Martin, R. J. Goulet, and G. W. Sledge. 1997. Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol. Cell. Biol. 17:3629-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 33.Park, B. K., et al. 2007. NF-κB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nat. Med. 13:62-69. [DOI] [PubMed] [Google Scholar]
- 34.Pereira, S. G., and L. Oakley. 2008. Nuclear factor-κB1: regulation and function. Int. J. Biochem. Cell Biol. 40:1425-1430. [DOI] [PubMed] [Google Scholar]
- 35.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511-526. [DOI] [PubMed] [Google Scholar]
- 36.Perissi, V., et al. 2008. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol. Cell 29:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins, N. D. 2004. Regulation of NF-κB by atypical activators and tumour suppressors. Biochem. Soc. Trans. 32:936-939. [DOI] [PubMed] [Google Scholar]
- 38.Pratilas, C. A., et al. 2009. BRAF (V600E) is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. U. S. A. 106:4519-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radvanyi, L., et al. 2005. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 102:11005-11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson, A. L., et al. 2006. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9:121-132. [DOI] [PubMed] [Google Scholar]
- 41.Sovak, M. A., et al. 1997. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 100:2952-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita, A., D. R. Buchholz, and Y. B. Shi. 2004. Recruitment of N-CoR/SMRTTBLR1 corepressor complex by unliganded thyroid hormone receptor for gene repression during frog development. Mol. Cell. Biol. 24:3337-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomita, A., D. R. Buchholz, K. Obata, and Y. B. Shi. 2003. Fusion protein of retinoic acid receptor alpha with promyelocytic leukemia protein or promyelocytic leukemia zinc finger protein recruits N-CoR-TBLR1 corepressor complex to repress transcription in vivo. J. Biol. Chem. 278:30788-30795. [DOI] [PubMed] [Google Scholar]
- 44.Vermeulen, K., Z. N. Berneman, and D. R. Van Bockstaele. 2003. Cell cycle and apoptosis. Cell Prolif. 36:165-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, C.-Y., M. Mayo, and A. S. Baldwin. 1996. TNF- and cancer therapy-induced apoptosis potentiation by inhibition of NF-κB. Science 274:784-787. [DOI] [PubMed] [Google Scholar]
- 46.Wang, C.-Y., M. W. Mayo, R. C. Korneluk, D. V. Goeddel, and A. S. Baldwin. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281:1680-1683. [DOI] [PubMed] [Google Scholar]
- 47.Wang, C.-Y., D. C. Guttridge, M. W. Mayo, and A. S. Baldwin. 1999. NF-κB expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol. Cell. Biol. 19:5923-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, C.-Y., J. Cusack, R. Liu, and A. S. Baldwin. 1999. Control of inducible chemoresistance: enhanced anti-tumor therapy via increased apoptosis through inhibition of NF-κB. Nat. Med. 5:412-417. [DOI] [PubMed] [Google Scholar]
- 49.Yoon, H. G., et al. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, J., M. G. Guenther, R. W. Carthew, and M. A. Lazar. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12:1775-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoRHDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, Y., W. Gross, S. H. Hong, and M. L. Privalsky. 2001. The SMRT corepressor is a target of phosphorylation by protein kinase CK2 (casein kinase II). Mol. Cell. Biochem. 220:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]