Abstract
The histone H3 variant Cse4 specifies centromere identity in Saccharomyces cerevisiae by its incorporation into a special nucleosome positioned at CEN DNA and promotes the assembly of the kinetochore complex, which is required for faithful chromosome segregation. Our previous work showed that Cse4 is also associated with the partitioning locus STB of the 2μm circle—a multicopy plasmid that resides in the yeast nucleus and propagates itself stably. Cse4 is essential for the functional assembly of the plasmid partitioning complex, including the recruitment of the yeast cohesin complex at STB. We have located Cse4 association strictly at the origin-proximal subregion of STB. Three of the five directly repeated tandem copies of a 62-bp consensus sequence element constituting this region are necessary and sufficient for the recruitment of Cse4. The association of Cse4 with STB is dependent on Scm3, the loading factor responsible for the incorporation of Cse4 into the CEN nucleosome. A chromosomally integrated copy of STB confers on the integration site the capacity for Cse4 association as well as cohesin assembly. The localization of Cse4 in chromatin digested by micrococcal nuclease is consistent with the potential assembly of one Cse4-containing nucleosome, but not more than two, at STB. The remarkable ability of STB to acquire a very specialized, and strictly regulated, chromosome segregation factor suggests its plausible evolutionary kinship with CEN.
The 2μm plasmid of Saccharomyces cerevisiae is an example of a highly optimized, circular, multicopy extrachromosomal selfish DNA element (25, 27, 49). The plasmid does not seem to contribute to the host's fitness under standard laboratory growth conditions. However, at its steady-state copy number of 40 to 60 molecules per cell, any growth disadvantage imposed by the plasmid is rather small (16, 30). The most remarkable attribute of this selectively almost neutral entity is its ability to propagate with nearly chromosome-like stability at its steady-state copy number. The entire genetic makeup of the 2μm circle is devoted to three functions: efficient replication by the host machinery, equal segregation, and maintenance of the copy number. The plasmid accomplishes these goals with minimal metabolic encumbrance to its host.
Direct visualization of fluorescence-tagged reporter plasmids in live cells suggests that 2μm circle molecules are organized in the nucleus as 3 to 5 dynamic foci that form a close-knit cluster (47). The plasmid also segregates as a clustered entity; sister clusters part from each other and move away at the anaphase stage of the cell cycle. Population analysis and time lapse assays have revealed close similarities between the 2μm circle and the yeast chromosomes or a centromere plasmid (minichromosome) in their dynamics and kinetics of segregation (17, 47). The relevant inference from a variety of experiments is that plasmid segregation is tightly coupled to chromosome segregation, perhaps by attachment of duplicated plasmid clusters to a pair of sister chromatids (17, 31, 47).
Because of the large reduction in the effective copy number caused by clustering, efficient plasmid segregation is dependent on an active partitioning system comprising two plasmid proteins, Rep1 and Rep2, and the cis-acting locus STB. The Rep-STB system overcomes the mother bias to which ARS plasmids, replication competent but lacking partitioning machinery, are subjected (34). Available evidence suggests that negation of mother bias and coupling of plasmid segregation to chromosome segregation are manifestations of the same underlying mechanism. A number of mutations that affect the fidelity of chromosome segregation cause the 2μm circle to missegregate in tandem with the chromosomes (31, 47). This chromosome-coupled behavior is abrogated by inactivation of either or both of the Rep proteins or by deletion of the STB locus.
The 2μm plasmid also harbors an amplification system, consisting of the Flp site-specific recombinase and its target sites (FRTs) (31), which can correct a drop in the copy number resulting from rare missegregation events. A Flp-mediated DNA inversion event during bidirectional replication, changing the direction of one fork with respect to the other and thus preventing termination, is believed to be the basis for DNA amplification (15, 39, 48). The amplified DNA may be resolved into individual plasmid copies by Flp-mediated or homologous recombination. A fourth plasmid-encoded protein, Raf1, a positive regulator of FLP expression, ensures that the amplification response, when required, is rapidly triggered (36, 42). A combination of negative and positive regulatory circuits involving Rep1, Rep2, and Raf1 serves to minimize deviations of the plasmid copy number from the steady-state value. Recent evidence suggests that modification of Flp by the host SUMO attachment system is important in preventing aberrant amplification of the 2μm circle (6, 51).
A number of observations suggest that the 2μm plasmid partitioning system channels chromosome segregation factors into the plasmid segregation pathway (17, 18, 20, 31, 32, 52). This partitioning system assists the plasmid in utilizing the mitotic spindle and the spindle-associated Kip1 motor to localize to its partitioning center in the nucleus (10). The STB chromatin is associated with the centromere-specific histone H3 variant Cse4p (CenH3), and the functional state of STB is established by the RSC2 chromatin-remodeling complex (20, 23, 50). The yeast cohesin complex is assembled at STB in a Rep1- and Rep2-dependent manner and mediates the bridging of sister plasmid clusters harboring duplicated plasmid copies (17, 18, 31). The disassembly of cohesin during anaphase is essential for the segregation of sister clusters into daughter cells (31). Assays using single-copy derivatives of STB reporter plasmids suggest that plasmid pairing by cohesin occurs in a sister-to-sister fashion and that sisters are dispatched in opposite directions upon the dissolution of the cohesin bridge (17, 18). Thus, despite the multicopy nature of the plasmid, there must be a high level of organization within the plasmid cluster to coordinate DNA replication with sister plasmid pairing.
The association of Cse4, presumed to be an exclusive component of centromeric nucleosomes, with the partitioning locus of an extrachromosomal and nonessential entity such as the 2μm plasmid is quite intriguing. Our present analyses demonstrate the authenticity, specificity, and CEN independence of Cse4-STB association and locate it in the origin-proximal region of STB (STB-proximal) containing five tandem direct copies of an AT-rich 62-bp consensus element. The loading factor for Cse4 at CEN, Scm3, interacts with STB in a metastable fashion. Depletion of Scm3 causes abrogation of the Cse4-STB association. A chromosomally integrated copy of STB mimics a plasmid-borne STB locus in its association with Cse4. The site of integration, which does not recruit cohesin in its native state, becomes competent for cohesin assembly. Our results, in toto, are consistent with a common evolutionary ancestry for the point centromere of budding yeast and the 2μm plasmid partitioning locus (29).
MATERIALS AND METHODS
Plasmids.
The plasmids employed as reporters or for integration into a chromosomal site are described below.
(i) Plasmid pSG5.
The construction of the multicopy STB reporter plasmid pSG5, used in several of the pulldown assays, has been described previously (18). This plasmid, containing the 2μm replication origin and STB along with the TRP1 marker, was composed solely of yeast sequences.
(ii) Plasmid pCH5.
In the multicopy STB reporter plasmid pCH5, the origin-proximal AvaI-HpaI STB segment was flanked by four BamHI sites adjacent to the AvaI site and four BglII sites adjacent to the HpaI site. The template for the construction of pCH5 was pSG5-1 (18), which was also the immediate precursor for pSG5 (see the preceding section). First, the entire pSG5-1 plasmid was amplified as a linear fragment using two oppositely oriented primers spanning the HpaI site, one of which contained four tandem copies of the BglII recognition sequence. The amplified DNA was cut with HpaI and was self-ligated to generate plasmid pCH5-1, retaining the BglII sites. Next, pCH5-1 was amplified in its full-length linear form by using two inverse primers spanning the AvaI site, one of which contained four iterations of the BamHI recognition sequence. Both primers contained, in addition, a copy of the SmaI recognition sequence, such that SmaI digestion and self-ligation generated pCH5-2, harboring four BamHI sites next to AvaI. Digestion of pCH5-2 by SalI and self-ligation generated pCH5, from which all nonyeast sequences were eliminated. After the transformation of yeast with the ligation mixture, Trp+ transformants were screened by PCR and Southern blot analysis of total DNA to identify those that had acquired pCH5.
(iii) The pSTB2-5 series of plasmids.
The series of four plasmids comprising pSTB2 to pSTB5 (pSTB2-5 plasmids) contained, in addition to the 2μm circle origin, two, three, four, and all five copies, respectively, of the 62-bp repeat element that is the building block for STB-proximal. The repeat elements were present in these plasmids in their native order, starting with the repeat element closest to the origin. The steps in their construction were as follows. The STB-proximal plus ORI fragments were amplified using a fixed primer adjacent to the origin and distinct individual primers, within or at the far border of STB-proximal, marked by an HpaI site. The primer pairs were designed to contain BamHI recognition sequences adjacent to their annealing regions. Because of the repeated nature of the STB region that served as the template for PCR, the amplified DNA contained, in addition to the authentic product, extra products due to primer annealing at secondary sites. Amplification reaction products were fractionated by agarose gel electrophoresis, and DNA bands of the expected sizes were isolated and purified by chloroform-phenol extraction and ethanol precipitation. The TRP1 marker without its associated ARS1 sequence was amplified from a template plasmid using a pair of primers both of which contained a BamHI recognition sequence at their nonannealing termini. Each of the STB fragments was digested with BamHI and was ligated to a similarly digested TRP1 fragment. The pSTB2-5 plasmids were recovered from the corresponding ligation mixtures by transformation in yeast, followed by Southern blotting and PCR analysis of total DNA prepared from the transformants.
(iv) pSH-STB, pSH-STB-Prox, and pSH-STB-Dist.
Plasmids pSH-STB, pSH-STB-Prox, and pSH-STB-Dist were used to integrate the full STB locus and the origin-proximal or origin-distal region of STB into the HIS3 locus on chromosome XV. The STB, STB-proximal, and STB-distal regions, bordered by PstI-AvaI, HpaI-AvaI, and PstI-HpaI restriction enzyme sites in the 2μm circle genome, were amplified by PCR using DNA prepared from a [cir+] yeast strain. The forward and reverse primer pairs employed in the amplification reaction were designed to contain an EcoRI and a BamHI recognition sequence, respectively, within their nonannealing terminal regions. After digestion of the amplified DNA with EcoRI and BamHI, the fragments were cloned into the EcoRI-BamHI backbone fragment from the yeast integrative plasmid pRS403. The resulting plasmids were linearized by cutting within the HIS3 marker using NdeI and were used to transform a [cir0] his3 recipient strain harboring a galactose-inducible REP1-REP2 cassette. The correct integrants among the His+ transformants were identified by PCR screening of total DNA prepared from them.
(v) p5015.
p5015 is a CEN reporter plasmid, provided by D. Ivanov and K. Nasmyth (24), harboring CEN4 as well as TRP1. It was employed as a control in a number of assays.
Plasmid pulldown using an anti-Myc antibody.
Pulldown assays were carried out in [cir+], [cir0], or [cir0]-PGAL-REP1-REP2 host strains expressing Myc12-tagged Cse4. The modified CSE4 gene was present at the normal chromosomal location and was driven by the native promoter. In a standard assay, spheroplasts obtained from 250 ml of a log-phase culture (optical density at 600 nm [OD600], ∼0.8) by lyticase treatment were lysed in 2.5 ml of lysis buffer according to published protocols (24). The crude lysate was spun at 12,000 × g and 4°C to produce the cleared lysate. Five hundred microliters of the cleared lysate was saved as the input fraction, and the remainder was incubated with an anti-Myc antibody overnight at 4°C. Protein A-Dynabeads (500 μl) were added, and incubation was continued for 5 h at 4°C. The beads were harvested, and 500 μl of the supernatant was saved as the “unbound” fraction. The beads were washed three times with lysis buffer containing 200 mM NaCl. Finally, DNA was eluted from the beads using 1% sodium dodecyl sulfate (SDS) elution buffer (24) at 65°C. All the fractions were adjusted to a final concentration of 1% SDS, extracted with phenol-chloroform, and precipitated with ethanol. DNA from the input, unbound, and bound fractions was dissolved in 50 μl 1× Tris-EDTA (TE). Aliquots from these fractions were analyzed by electrophoresis in agarose gels followed by Southern blot analysis. The input and unbound fractions corresponded to one-eighth of the 1× bound fractions.
Restriction enzyme digestion of plasmids following pulldown.
After the pulldown of a reporter plasmid, the protein A-Dynabeads were resuspended in 1 ml of lysis buffer (see above) and were incubated with or without appropriate restriction enzymes for 4 h at 4°C. An excess of enzymes was employed to ensure nearly complete digestion of plasmid DNA. Beads were harvested, and the supernatant was collected. The different fractions were processed further, and DNA was recovered from them by ethanol precipitation as described above for the plasmid pulldown assays. Samples were subjected to agarose gel electrophoresis and Southern hybridization using specific DNA probes.
Micrococcal nuclease digestion of chromatin and analysis of nucleosomes for Cse4 association.
Nuclei were isolated from 500-ml (large-scale) or 200-ml (small-scale) log-phase cultures (OD600, ∼0.8 to 1.0) by following published protocols (13). The nuclear pellet from the small-scale culture was resuspended in 1 ml SPC buffer [1 M sorbitol, 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.3), 5 mM CaCl2] and was then digested with 200 U of micrococcal nuclease (Worthington Biochemicals) at 37°C for 30 min. The digestion buffer included 1 mM phenylmethylsulfonyl fluoride (PMSF) as well as 1× LPC (10 μg per ml each of leupeptin, pepstatin A, and chymostatin). These conditions were standardized to yield predominantly mono- and dinucleosomes. After the addition of 10 mM EDTA, the reaction mixture was spun down at 13,200 rpm for 10 min in an Eppendorf centrifuge (model 5415R) at 4°C. The supernatant was incubated with an anti-Myc antibody; the mixture was adsorbed on protein A-Dynabeads; and the bound DNA was isolated as described under “Plasmid pulldown using an anti-Myc antibody” above. The input DNA and bound DNA were analyzed by PCR using a defined set of primer pairs. For each primer pair, signal intensities were estimated from reactions employing a set of dilutions of the immunoenriched and input DNAs as templates. Corrections were made for differences in the amplification efficiencies of primer pairs by normalizing the signals from the immunoprecipitated samples to those from the corresponding input DNAs.
Depletion of Scm3.
The strain expressing SCM3 from the GAL promoter was grown in galactose to an OD600 of ∼0.3 and was then shifted to glucose medium for 3 h. Under these conditions, the vast majority of cells were arrested in the G2/M phase.
Cell cycle arrest using α-factor, nocodazole, or the ndc10-1 mutation.
For G1 arrest, cells grown to mid-log phase were treated with 15 μg per ml of α-factor. The efficiency of arrest was nearly 90 to 95%. For spindle depolymerization and G2/M arrest, nocodazole was added as a solution in dimethyl sulfoxide (DMSO) to mid-log-phase cells at 20 μg/ml. More than 85% of the cells were arrested in the large budded state with a single nucleus at the mother-bud neck. The corresponding control cells were treated with the same amount of DMSO, but without nocodazole. A fresh culture of the ndc10-1 strain, seeded by an overnight inoculum, was grown to early-log phase at 26°C and was shifted to 37°C for 2.5 h to inactivate kinetochore function.
ChIP.
Chromatin immunoprecipitation (ChIP) analyses were performed as described previously (20). In assays using multiple primer pairs, PCR signals yielded by immunoprecipitated DNA were normalized to the corresponding signals given by the input DNA, as described under “Micrococcal nuclease digestion of chromatin and analysis of nucleosomes for Cse4 association” above.
Miscellaneous protocols.
Routine experimental protocols, such as bacterial and yeast transformations, total yeast DNA and plasmid DNA preparations, and curing of native 2μm circles from [cir+] yeast strains, are described on the web page of the Jayaram laboratory (http://www.sbs.utexas.edu/jayaram/jayaramlab.htm).
Yeast strains and oligonucleotides.
The yeast strains used in this study are listed in Table S1 in the supplemental material. The oligonucleotides employed as primers in PCRs pertaining to key experiments are listed in Tables S2 and S3 in the supplemental material. Sequences of additional oligonucleotides, not listed in these tables, are available on request.
RESULTS
An STB reporter plasmid can be pulled down by a Cse4-directed antibody in the absence of kinetochore function: concordance between ChIP and pulldown assays.
As already pointed out, the presence at STB of Cse4, which is the hallmark of the centromeric nucleosome, is quite surprising. A previous demonstration of Cse4-STB association was based on ChIP analyses employing formaldehyde-mediated DNA-protein cross-linking (20). Since the 2μm plasmid cluster is localized close to the spindle pole body (10, 32), often overlapping clustered centromeres, cross-linking of Cse4 present at CEN with STB DNA due to their proximity could be a potential pitfall. To rule out such an artifact, we carried out pulldown of STB and CEN reporter plasmids using a Cse4-directed antibody in the absence of cross-linking and under conditions that keep these loci active or inactive.
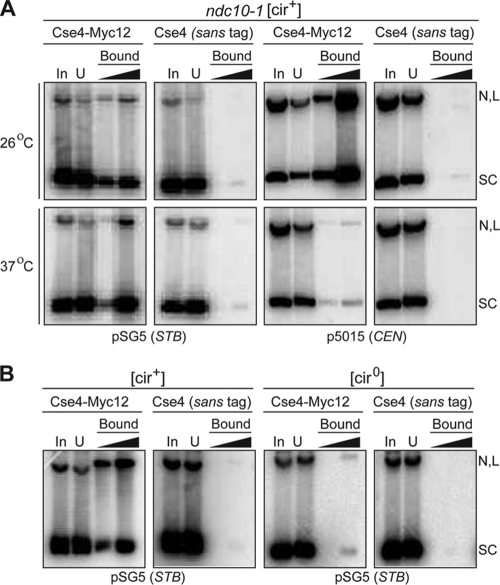
In a [cir+] ndc10-1 host strain expressing Cse4-Myc12 and supplying Rep1 and Rep2 from endogenous 2μm circle molecules, both the STB and CEN reporter plasmids were pulled down by the anti-Myc antibody at the permissive temperature, 26°C (Fig. 1A). At the nonpermissive temperature, 37°C, the STB plasmid, but not the CEN plasmid, was pulled down. Consistent with the requirement of Rep1 and Rep2 proteins for Cse4-STB association (20), the STB reporter plasmid could not be pulled down in a [cir0] strain, lacking native 2μm circles and thus lacking the Rep proteins (Fig. 1B).
FIG. 1.
A Cse4-specific antibody brings down an STB reporter plasmid in a Rep1- and Rep2-dependent and Ndc10 independent manner. Plasmid pulldowns in this set of assays and those for which results are shown in subsequent figures were performed as described in Materials and Methods. The antibody employed was directed to the Myc12 epitope fused to Cse4. Plasmid DNA was detected by Southern blot analysis using a radioactively labeled hybridization probe. The DNA load in the second lane of the bound fraction was five times that in the first. In, input; U, unbound; SC, supercoiled plasmid; N, L, nicked and linear plasmids, respectively.
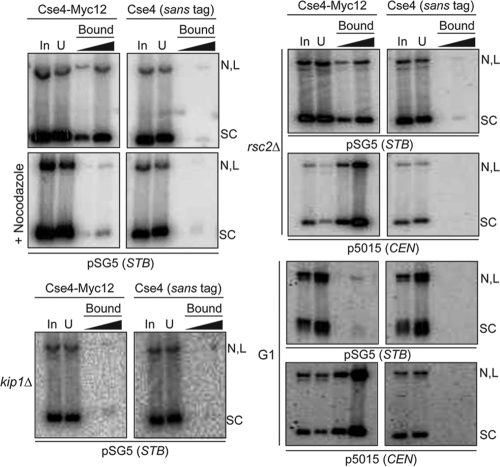
Localization of the 2μm plasmid to its specific nuclear address, assisted by the mitotic spindle and the Kip1 motor protein, appears to be an important spatial regulatory step in the plasmid partitioning pathway (10). Consistent with such a mechanism, Cse4 could not be detected at STB by ChIP in a kip1Δ strain or in nocodazole-treated cells (20). Furthermore, ChIP assays suggested the dissociation of Cse4 from STB during late telophase of the cell cycle, in contrast to the persistence of Cse4 at CEN through the ensuing G1 phase (20). Finally, according to ChIP data, the establishment of the functional state of the STB chromatin by the RSC2 chromatin-remodeling complex (23, 50) occurs after the recruitment of Cse4 at STB (20). In agreement with the ChIP conclusions, the Cse4-directed antibody failed to bring down the STB reporter plasmid in G1-arrested or nocodazole-treated cells, as well as in cells lacking Kip1 function (Fig. 2). In contrast, the rsc2Δ mutation had no effect on plasmid pulldown. Significantly, G1 arrest did not impede the pulldown of the CEN reporter plasmid; the rsc2Δ mutation was in this regard also innocuous.
FIG. 2.
Spindle depolymerization, G1 arrest, and the kip1Δ mutation, but not the rsc2Δ mutation, block Cse4-STB association. Pulldown of the STB or CEN reporter plasmid and DNA analysis were performed as described for Fig. 1.
The results described above attest to the authenticity of Cse4-STB association, which is not dependent on chemical cross-linking and is not affected by inactivation of centromere function. They argue in favor of the hypothesis that Cse4 is a genuine component of the 2μm plasmid partitioning complex.
We have noted that the average pulldown efficiency of the STB reporter plasmid was less than 10% that of the CEN reporter plasmid. This discrepancy can be readily seen by comparing the lanes for the “input” and “unbound” fractions for the two plasmids in the top panels (26°C) of Fig. 1. Perhaps Cse4-STB association is not mediated through a nucleosome, or this association is substoichiometric. These alternative possibilities are being investigated.
Only the STB-containing DNA fragment of a reporter plasmid stays associated with the Cse4-directed antibody after restriction enzyme digestion.
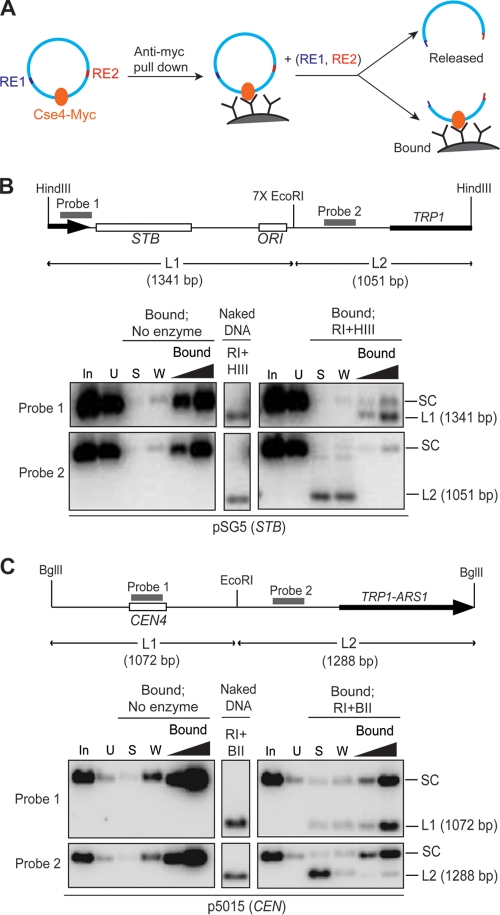
Previous results on the salt extractability of Cse4 from STB, as well as differential restriction enzyme sensitivities of STB in the presence and absence of functional Cse4, were interpreted as being consistent with the presence of Cse4 as a nucleosome component at STB (20). However, since STB is the site of multiple protein associations, DNA accessibility could be restricted as a result of cooperative DNA-protein and/or protein-protein interactions. As a result, a nonnucleosomal mode of Cse4 residence at STB cannot be ruled out. Regardless of the precise nature of Cse4-STB association, the pulldown assays for which results are shown in Fig. 1 and 2 demonstrate that a reporter plasmid can be baited by the Cse4 antibody only when the plasmid-borne STB locus is functional. A reasonable expectation, then, is that fragmentation of the reporter plasmid should free any DNA piece lacking STB from its association with the Cse4-directed antibody (Fig. 3A).
FIG. 3.
Cse4 association is specific to STB-containing DNA. (A) Schematic diagram of the rationale of the analysis. If Cse4 association is specific to a plasmid locale, DNA digestion with the restriction enzymes RE1 and RE2 should release the fragment lacking Cse4 from its association with the Cse4-directed antibody. (B and C) The STB (B) or CEN (C) reporter plasmid employed in the assays is schematically diagrammed at the top. Restriction enzyme digestions were performed on protein A-Dynabeads, and DNAs in the released or retained fractions were analyzed by gel electrophoresis and Southern hybridization using the indicated probes. For reference, enzyme digestions performed on total DNA (“naked” DNA) prepared from the experimental strains were analyzed similarly. Hybridization profiles from the relevant portions of the gels are displayed. RI, HIII, and BII stand for EcoRI, HindIII, and BglII, respectively. In, input; U, unbound; S, supernatant; W, wash; SC, supercoiled plasmid.
We therefore digested an STB or a CEN reporter plasmid following pulldown so as to split the STB- or CEN-containing fragment from the plasmid backbone fragment. We then probed the retention of the resulting DNA fragments by the Cse4-directed antibody, or their release from that antibody (Fig. 3B and C). In the case of the STB reporter plasmid, the STB-containing HindIII-EcoRI fragment (L1; 1,341 bp) remained preferentially associated with the Cse4-directed antibody on protein A beads, whereas the fragment lacking STB (L2; 1,051 bp) was released into the supernatant (Fig. 3B). In the control assay with a CEN reporter plasmid, the CEN-containing EcoRI-BglII fragment (L1; 1,072 bp) stayed bound to the beads, and the fragment lacking CEN (L2; 1,288 bp) dissociated from them (Fig. 3C).
Thus, Cse4 association is restricted to DNA regions harboring STB or CEN in the respective reporter plasmids and is not affected by digestion of the DNA with restriction enzymes. Such a localized and stable DNA-protein interaction would be consistent with the occupancy of STB by a Cse4-containing nucleosome(s).
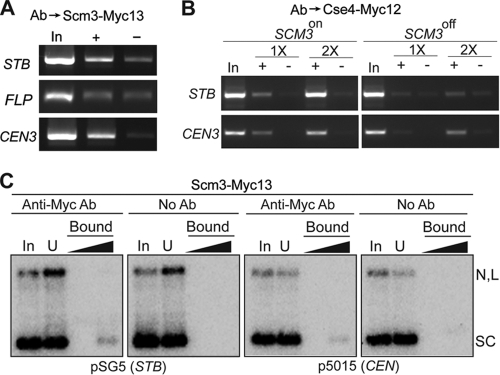
Scm3 associates with STB and is required for Cse4 recruitment at STB.
Scm3 is an essential protein in S. cerevisiae that is associated with CEN, interacts with Cse4, and is required for the localization of Cse4 at CEN (4, 33, 43). There is controversy as to whether Scm3 is a kinetochore protein that functions as a loading factor for Cse4 or an authentic constituent of the centromeric nucleosome (5, 33). The presence of Cse4 at STB raises the question of whether Cse4 recruitment by the 2μm plasmid is also dependent on Scm3.
Scm3 was detected at STB, as at CEN3, by ChIP (Fig. 4A). Scm3-STB association was specific, as suggested by the absence of Scm3 at the FLP locus of the 2μm plasmid. There was a marked diminution of Cse4 association at both STB and CEN3 when cells were depleted of Scm3 (Fig. 4B). However, neither a CEN nor an STB reporter plasmid could be pulled down using an antibody against Scm3 (Fig. 4C).
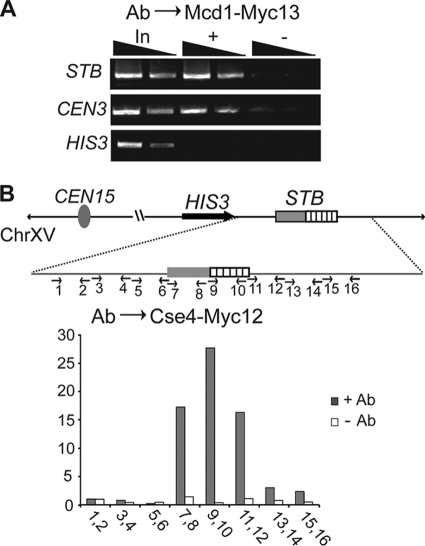
FIG. 4.
Scm3 interacts with STB and promotes Cse4-STB association. (A and B) ChIP assays were performed in [cir+] host strains using antibodies (Ab) against Scm3-Myc13 (A) and Cse4-Myc12 (B). The immunoprecipitated DNA was probed using primers specific to the STB and FLP loci of the 2μm plasmid and CEN3, as indicated. “In,” “+,” and “−” refer to input, immunoprecipitated, and mock-immunoprecipitated (without antibody) DNA samples, respectively. For panel B, the expression of SCM3, under the control of the GAL promoter, was turned on or off in the presence of galactose or glucose, respectively, as the carbon source. 1× and 2× refer to the relative amounts of template DNA used in the PCRs. (C) The pulldown analyses of the STB and CEN reporter plasmids were similar to those described in the legend to Fig. 1.
Considered together, the ChIP and pulldown results suggest that while Scm3 promotes Cse4-STB association, the interaction between Scm3 and STB is weaker than that between Cse4 and STB. Furthermore, the sharply contrasting behaviors of a CEN reporter plasmid during pulldown by antibodies targeting Cse4 or Scm3 argue against Scm3 being an integral component of the nucleosome core at CEN.
A chromosomally integrated copy of STB is competent for Cse4 recruitment in its ectopic context.
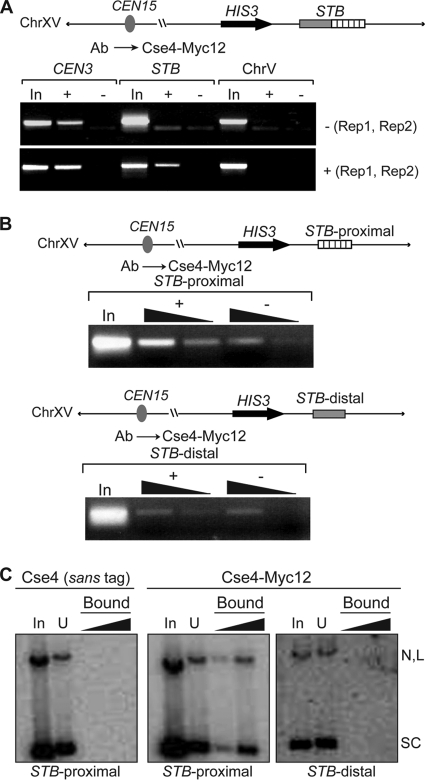
As noted before, the precise nuclear address of the 2μm plasmid appears to be a critical determinant in its equal segregation. We wondered whether the partitioning function of STB is context specified, that is, whether it is active only in its native extrachromosomal state. We integrated a copy of STB at the HIS3 locus on the right arm of chromosome XV and assayed its ability to recruit Cse4. The host strain was [cir0] and harbored a galactose-inducible REP1-REP2 expression cassette.
The association of Cse4 with the integrated STB copy was detected by ChIP, but only under conditions of REP1 and REP2 induction (Fig. 5A). When the expression of REP1 and REP2 was repressed by glucose, Cse4 was absent at STB.
FIG. 5.
Cse4 associates with STB or STB-proximal, but not with STB-distal, integrated at the HIS3 locus on chromosome XV. (A and B) The sequence contexts of chromosomally integrated STB, STB-proximal, and STB-distal are schematically diagrammed. ChIP assays were performed in a [cir0] host strain harboring an integrated REP1-REP2 cassette controlled by the GAL promoter. “−(Rep1, Rep2)” and “+(Rep1, Rep2)” denote the glucose-repressed and galactose-induced states of the REP1-REP2 loci, respectively. “ChrV” refers to a location on the chromosome V arm where cohesin is assembled but Cse4 is not present. (C) The STB-proximal and STB-distal sequences harbored by the pulled down reporter plasmids were the same as those present in the corresponding integrated versions of these loci.
Thus, the competence of STB to acquire Cse4 is not lost by displacing it from its normal location within the 2μm plasmid. The chromosomally located STB, like its plasmid counterpart, is dependent on Rep1 and Rep2 for Cse4 acquisition. The integrated STB fragment does not include the 2μm circle replication origin, which is approximately 400 bp away from the STB border proximal to it. The location of STB with respect to the origin or the intervening 2μm circle DNA between them does not appear to have a strong influence on Cse4-STB association.
Cse4-STB association is specific to the ORI-proximal region of STB.
The STB locus has a bipartite organization, a proximal and a distal region with respect to the origin of replication of the 2μm plasmid. As noted previously, STB-proximal is made up of a tandem array of five directly repeated units of a 62-bp AT-rich consensus sequence element (35). STB-distal contains a transcription termination signal that blocks two 2μm circle transcripts (1,650 nucleotides [nt] and 600 nt) spanning the RAF1 locus from entering STB-proximal (35, 45). Furthermore, STB has been shown to mediate transcriptional silencing with the assistance of the Sir2 to Sir4 proteins (37). Consistent with this ability, STB-distal contains an element that can downregulate transcription from upstream promoters (35). We wanted to know whether STB-proximal and STB-distal are functionally distinct with regard to Cse4 association.
ChIP analyses performed on STB-proximal and STB-distal integrated at the chromosomal HIS3 locus revealed that the occupancy of Cse4 was limited to STB-proximal (Fig. 5B). The signal from STB-distal was nearly the same as the background. The ChIP results were concordant with the results of pulldown assays performed on reporter plasmids containing STB-proximal or STB-distal (Fig. 5C).
The specific association of Cse4 with STB-proximal is consistent with the observation that this region is sufficient to confer stability on a reporter plasmid in a Rep1-Rep2-dependent manner (26, 35). However, the efficacy of this minimal STB region can differ depending on the sequence context in which it is placed. The transcriptional insulation provided by STB-distal may be important for realizing the full potential of STB in plasmid stability.
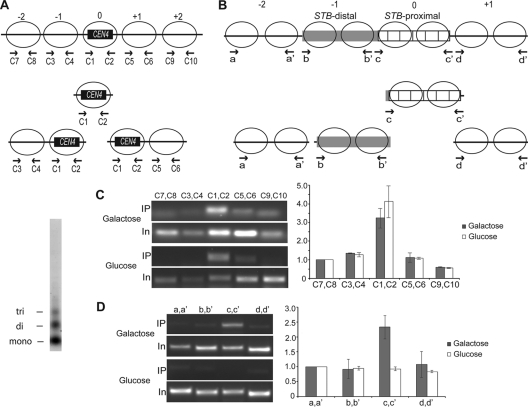
Localization of Cse4 in nucleosome fractions produced by micrococcal nuclease digestion.
The experimental evidence from Fig. 5 localizes Cse4 to STB-proximal but does not rule out the possibility that STB-distal may acquire competence for Cse4 association when it is adjacent to STB-proximal. We also do not know whether such competence, if valid, might spread from STB-proximal to any DNA sequence placed next to it. Since Cse4 is incorporated into a nucleosome in its only known, and well-characterized, association with CEN DNA, we wanted to delimit the region spanning STB-proximal and adjacent DNA occupied by Cse4 with respect to nucleosome markers.
Previous analyses employing DNase I and micrococcal nuclease digestion suggested that the organization of STB chromatin, which appears to have a relatively low occupancy of nucleosomes, is modulated by the Rep1 protein and the RSC2 chromatin-remodeling complex (12, 46, 50). The size of STB-proximal or STB-distal, ∼300 bp, can accommodate one or two standard H3-containing or Cse4-containing nucleosomes, which wrap ∼150 bp and ∼125 bp of DNA, respectively, around them. In the experiments described below, we enriched Cse4-associated nucleosome fractions from micrococcal-nuclease-digested chromatin and investigated the presence of STB-proximal and adjacent DNA sequences by a PCR-based assay. The rationale was adapted from analogous assays employed to demonstrate the exclusive localization of Cse4 to a single nucleosome present at a centromere (13).
Nuclei were digested with micrococcal nuclease by using the [cir0] strain containing the integrated copy of STB (see Fig. 5), and inducible REP1-REP2, to yield a nucleosome population consisting predominantly of mono- and dinucleosomes (≥85%), with a significantly smaller fraction of trinucleosomes (≤15%). DNA isolated from the immunoenriched subpopulation of Cse4-associated nucleosomes was analyzed by PCR using a series of primer pairs directed to CEN4 (as a control), STB, and DNA regions immediately flanking these loci on either side. Since the position of the CEN nucleosome is clearly defined, resolution at the single-nucleosome level was possible for CEN analysis (Fig. 6A). In contrast, STB analysis could not be as precise. Information on exact nucleosome placements at and around STB is lacking. Furthermore, because STB-proximal is composed of repeated DNA elements, primer pairs that would specifically amplify selected subregions within this region were difficult to design. Given these caveats, better resolution than that at the dinucleosome level was not possible. As indicated in the schematic in Fig. 6B, the primer pairs for STB analysis were spaced two nucleosome lengths of DNA (∼300 bp) apart. Although the depiction of nucleosomes within and outside STB is rather arbitrary, it is consistent with published micrococcal nuclease digestion results (50). Two nuclease-sensitive sites have been mapped to the borders of STB-proximal, with a weaker one at the center, suggesting the presence of internucleosome DNA linkers at these locales. Therefore, two nucleosomes are likely positioned at STB-proximal. Assuming no discontinuity in nucleosome spacing or phasing, STB-distal, which is nearly identical in size to STB-proximal, would also contain two nucleosomes.
FIG. 6.
Association of Cse4 with STB probed in chromatin digested with micrococcal nuclease to yield predominantly mono- and dinucleosomes. The assays were conducted in the STB-integrated strain, with REP1 and REP2 expression either repressed by glucose or induced by galactose. As a control, the CEN region of chromosome IV was analyzed in parallel. (A) Expected pattern of mono- and dinucleosomes associated with Cse4 in chromatin spanning CEN4. Since Cse4 is present exclusively in the nucleosome positioned at CEN(0), only one mononucleosome (0) and two types of dinucleosomes (0, +1 and 0, −1) will be pulled down by the Cse4-directed antibody. (B) Based on their sizes, STB-proximal and STB distal can each potentially accommodate two adjacent nucleosomes. The placement of two nucleosomes at STB-proximal was prompted by the presence of a clear micrococcal-nuclease-sensitive site at each of its borders, plus a weaker one at its center (50). If Cse4 residence is confined to STB-proximal, only the c-c′ primer pair is expected to yield a positive PCR signal. (C and D) (Left) Amplification outputs from one assay for a given amount of the immunoprecipitated DNA (IP) and a fixed dilution of the input DNA (In) using the indicated primer pairs that probe CEN4 (C) or STB (D) as well as the DNA regions on either side of them. (Right) The cumulative data are displayed as bar graphs. In this representation, the signal given by each primer pair from the immunoprecipitated DNA was normalized to that from the input DNA, and a value of 1 was assigned for the leftmost primer pairs: C7, C8 in CEN4 analysis and a, a′ in STB analysis. (Far left) Nucleosome profile of the chromatin digest used for the analyses.
The signal intensities of the PCR products were plotted after they were normalized to the corresponding values from reactions with the input DNA as the template (Fig. 6C and D). As expected, a sharp peak signifying Cse4 occupancy marked the CEN4 DNA (Fig. 6C). A similar peak was positioned at STB-proximal in the presence of Rep1 and Rep2, with a drop-off in the signal at STB-distal as well as flanking DNAs to the left and right of STB (Fig. 6D). The authenticity of this peak was certified by its absence when the expression of REP1 and REP2 by the host strain was repressed by glucose.
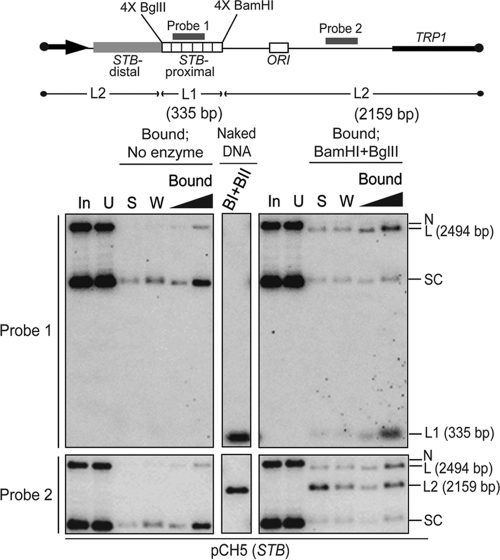
As a final verification of the Cse4 residence zone, we performed pulldown assays, followed by restriction enzyme digestion, of a reporter plasmid in which STB-proximal was bounded by four tandem copies each of BglII and BamHI recognition sequences at its left and right ends, respectively. After digestion with BamHI plus BglII, the STB-proximal DNA fragment (L1; 335 bp) remained quantitatively associated with the Cse4-directed antibody in the “bound” fraction (Fig. 7, top). More than 70% of the rest of the plasmid DNA, including STB-distal (L2; 2,159 bp), was disjoined from the antibody and appeared in the “supernatant”/“wash” fractions (Fig. 7, bottom).
FIG. 7.
Only the STB-proximal segment can retain association with the Cse4 directed antibody following pulldown and restriction enzyme digestion of a reporter plasmid. The four tandem copies of BamHI and BglII recognition sites that cordon off STB-proximal in the reporter plasmid are diagrammed schematically. Plasmid pulldown and Southern blot analysis of the BamHI- plus BglII-digested DNA were conducted as described in the legend to Fig. 3. BI + BII (above the lane showing results for the digestion of naked DNA) refer to BamHI and BglII, respectively.
In summary, the association of Cse4 with STB is mediated almost exclusively through the STB-proximal region. By analogy to centromeres, this association could involve a Cse4-containing nucleosome, or perhaps two such nucleosomes. However, the possibility that the Cse4-STB interaction does not conform to the conventional Cse4-CEN interaction cannot be ruled out entirely.
A subset of the STB-proximal consensus elements is necessary and sufficient for Cse4 recruitment.
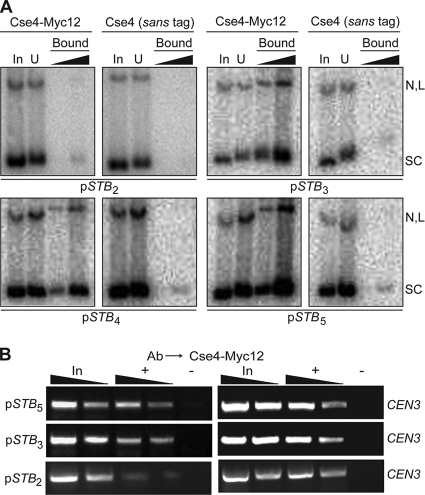
In order to determine whether Cse4-STB association is dependent on the full complement of STB-proximal, or whether a subset of the 62-bp repeat units would suffice, we tested reporter plasmids containing 2 to 5 tandem copies of the repeat unit in pulldown as well as ChIP assays. The deletions were introduced at the far end of STB-proximal from the 2μm origin of replication. All of the constructs retained the same native plasmid sequence from their intact STB borders to the origin.
An array of at least three repeat units was necessary and sufficient to elicit optimal Cse4-STB association (Fig. 8). There was a sharp drop in the pulldown efficiency (Fig. 8A), as well as in the ChIP signal (Fig. 8B), for a reporter plasmid containing only a two-repeat-unit array.
FIG. 8.
A subset of three of the 62-bp repeat elements within STB-proximal is sufficient for Cse4-STB association. Cse4-STB association was assayed by pulldown (A) and ChIP (B) assays. The subscript assigned to STB denotes the number of intact repeat elements, starting with the origin-proximal repeat element, present in a given reporter plasmid.
The requirement of three repeat elements (∼180 bp) for Cse4-STB association suggests that the minimal DNA region that interacts with Cse4 is sufficiently long to be wrapped around at least one Cse4-containing nucleosome (∼125 bp).
The chromosomally integrated copy of STB promotes cohesin assembly.
A critical, and perhaps the final, step in the pathway for the assembly of the plasmid partitioning complex is the recruitment of the cohesin complex at STB (17, 18, 31). It is not clear how cohesion between sister molecules, observed with the help of a fluorescence-tagged single-copy reporter plasmid (17, 18), translates into cohesion between two sister clusters containing multiple molecules of the 2μm plasmid. In one model, every pair of sister plasmids coheres, and each sister within such a pair is distributed one-to-one between sister clusters. It is as if the original plasmid cluster acts as a template for the formation of its sister. A plausible alternative model, in which sister clusters are held together by cohesin bridges formed by a subset of plasmid sisters, cannot be ruled out. Nevertheless, every condition so far tested that blocks cohesin-STB association leads to plasmid missegregation, suggesting that cohesin is an authentic component of the plasmid partitioning complex (10, 17, 20, 31, 32). Although Cse4 association satisfies one important criterion for the functionality of the chromosomally integrated STB, we wanted to know whether this STB is also competent for cohesin assembly. A positive result would imply that the entire pathway for organizing the plasmid partitioning complex is unaffected by the transplantation of STB from plasmid to chromosome.
As revealed by ChIP, the integrated STB copy was capable of cohesin assembly, whereas the proximal HIS3 locus was not (Fig. 9A). Consistent with this result, a ChIP walk performed along a portion of the integrated region revealed a Cse4 peak positioned over STB (Fig. 9B).
FIG. 9.
STB integrated at the HIS3 locus promotes cohesin recruitment at this chromosome locale. ChIP assays were carried out in two derivatives of the STB-integrated strain, one expressing MCD1-MYC13 and the other expressing CSE4-MYC12 from their normal chromosomal locations. Rep1 and Rep2 were supplied by turning on, in the presence of galactose, the GAL promoter-controlled REP1-REP2 cassette harbored by these strains. (A) The immunoprecipitated DNA was probed using primer pairs specific to STB and to the chromosomal CEN3 and HIS3 loci, respectively. (B) The indicated primer pairs (each pair encompassing ∼300 bp) were used to march along the indicated DNA region. The PCR signals were normalized to those from the input DNA and were plotted on an arbitrary scale, with the signal from the “1, 2” primer pair set as equal to 1.
The ability of STB to license a chromosomal site that normally does not associate with either Cse4 or the cohesin complex to acquire these factors required for 2μm circle segregation suggests that the functionality of the Rep-STB system in assembling the partitioning complex is preserved in this ectopic environment.
DISCUSSION
In this study, we have characterized the functional association between Cse4, a histone H3 variant regarded as unique to specialized nucleosomes assembled at centromeres (7, 29), and the partitioning locus of the 2μm plasmid. Cse4 is recruited to STB not only in its native plasmid context but also in an ectopic chromosomal context. The localization of Cse4 strictly to STB-proximal, containing five iterations of a 62-bp repeat element, and the DNA length requirement for this localization are consistent with the potential occupancy of STB chromatin by a Cse4-containing nucleosome, or perhaps two such nucleosomes. Furthermore, the retention of function by STB placed within a chromosome makes it possible to analyze the plasmid partitioning system without the complexities of multiple copies and the clustered organization of STB in its extrachromosomal state. We consider below the broader implications of the unanticipated presence of the same histone variant at the partitioning loci of chromosomes and a nonessential extrachromosomal DNA element.
Appropriation of a critical chromosome segregation factor by the multicopy 2μm plasmid: a functional paradox?
Replacement of the canonical histone H3 by members of the CenH3 family, including Cse4, exclusively at centromeres permits unequivocal distinction between centromeric and noncentromeric DNA and provides the spatial signal for the assembly of the kinetochore complex (21, 29, 44). Whether that distinction is imparted by altered histone stoichiometry of the CEN nucleosome core or through distinct structural motifs conferred by CenH3 on a histone octamer, or is due to the contrary positive writhing of CEN DNA around the CenH3-containing nucleosome, is controversial (5, 14, 33, 40).
Consistent with its highly specialized role in chromosome segregation, Cse4 is present only within the single nucleosome that resides at the short ∼125-bp centromere of each S. cerevisiae chromosome (13). The cell guards against the adverse consequences of Cse4 association with noncentromeric DNA (2, 9, 41) and potential assembly of ectopic kinetochores by actively turning over such mislocalized Cse4 via the ubiquitn-proteasome pathway (8, 22, 38). Excess Cse4 not bound to chromatin may also be subject to degradation. It is not clear whether the spread of Cse4 nucleosomes beyond CEN DNA is prevented by the establishment of chromatin barriers. Nevertheless, the strict regulation of Cse4 would limit its overall availability. Under this restriction, and since three to four times as many 2μm plasmids as chromosomes are present in the nucleus, the association of Cse4 with STB was unexpected. Yet the specificity of this interaction and its abrogation under conditions that inactivate STB, as revealed in this study, support a functional role for Cse4 in plasmid partitioning. The mechanism by which a high-copy-number selfish DNA element copes with the tight cellular economy of a host factor on which it is dependent is currently being investigated.
Cse4 signals distinct partitioning mechanisms at CEN and STB.
Despite the association of Cse4 with STB, we have not detected the presence of any of the kinetochore components at STB (31; also unpublished data). Conversely, neither Rep1 nor Rep2 interacts with CEN (31). There is also no evidence for direct spindle-mediated segregation of the 2μm plasmid. The presence of two copies of STB on a reporter plasmid does not induce instabilities analogous to those observed in dicentric minichromosomes (unpublished observations). Currently available evidence would be consistent with a model in which the plasmid cluster segregates in a chromosome-tethered fashion. The pairing of duplicated clusters by the cohesin complex would enhance the probability of their tethering to sister chromatids. Upon the disassembly of cohesin, sister clusters would hitchhike on sister chromatids to daughter cells. Within such a scheme, Cse4-STB association would fit into the general strategy by which the 2μm circle partitioning system appropriates chromosome segregation factors in order to achieve chromosome-coupled plasmid segregation.
Cse4 associations with CEN and STB are functionally analogous in that they promote assemblies of partitioning complexes at both these loci. At the same time, they are functionally distinct in that one of these (at CEN) is devoted to chromosome segregation and the other (at STB) to plasmid segregation. The two separate programs of high-order protein assembly, each temporally ordered and functionally hierarchical on its own, may be set into motion by specific, yet distinct, DNA-protein interactions initiated at these loci. The binding of the CBF3 complex to the CDEIII sequence within CEN is thought to provide the trigger for kinetochore assembly (11). Similarly, interactions of the Rep proteins, and perhaps host factors, with STB likely nucleate the assembly of the plasmid partitioning complex (19, 31). Critical dependence on specific Cse4-DNA interactions is a shared attribute of both pathways. At CEN, this interaction is mediated through a specialized nucleosome. The situation is not as clear-cut at STB, although current evidence is consistent with STB harboring one or two Cse4-containing nucleosomes. As far as we know, STB is the only noncentromeric site, chromosomal or episomal, in a eukaryotic cell where CenH3 recruitment is sequence specific, regulated, and functionally relevant.
The presence of two functional centromeres in a minichromosome in yeast gives rise to frequent nondisjunction or DNA breakage events, presumably due to opposing forces generated by their attachment to spindles from opposite poles. These instabilities can be effectively prevented by decreasing the distance between the competing centromeres (28). Since STB-mediated plasmid segregation does not appear to follow the CEN-kinetochore mechanism of spindle attachment (see above), the potential assembly of two Cse4 nucleosomes at STB does not pose the threat of instabilities. Furthermore, by analogy to CEN, the immediate proximity of the nucleosomes would have prevented such instabilities, even if the plasmid had relied on direct spindle-mediated segregation at some point in its evolutionary history. The mechanistically distinct contributions of the mitotic spindle to the segregation modes of CEN and STB plasmids may also account for the large differences in their respective copy numbers.
Is the point centromere of budding yeast derived from the partitioning locus of an ancestral 2μm plasmid?
The simple, genetically defined point centromere of S. cerevisiae and a related group of fungi belonging to the Saccharomycetaceae lineage represents a major evolutionary transition from the larger, more complex and epigenetically specified “regional” centromeres found in most other eukaryotes, including the vast majority of fungi (29). Yet both classes of centromeres carry the common epigenetic mark of CenH3-containing nucleosomes. The switch to point centromeres appears to have occurred contemporaneously with the loss of most or all of the machinery required for heterochromatin assembly and RNA interference (1). Quite remarkably, the presence of stably propagating 2μm circle-related nuclear plasmids appears to be unique to the Saccharomycetaceae lineage as well (3). It is reasonable, therefore, that the partitioning locus of an ancestral plasmid, following chromosomal integration, could have served as the progenitor of the point centromere (29). It is also noteworthy that the repeat elements in STB-proximal can be aligned as two tandem units of 124 bp with 97% homology between them (35) and that each unit is almost identical in size to the 125-bp S. cerevisiae centromere.
By the reasoning presented above, the ancestral Rep1 and Rep2 plasmid partitioning proteins are the plausible precursors for the scaffold proteins that support the assembly of the kinetochore complex (29). It is significant in this regard that components of the CBF3 complex, such as Ndc10 and Ctf13, have no identifiable homologues outside the Saccharomycetaceae among fungi and other eukaryotes. Similarly, Rep1 and Rep2 homologues are limited to the family of 2μm circle-related plasmids. The Rep2 proteins are barely recognizable as homologues by primary sequence alignment, perhaps suggesting their coevolution with their respective hosts.
Despite a common origin, the rapid divergence between the chromosomal and plasmid partitioning systems would make it virtually impossible at present to recognize their evolutionary kinship at the DNA and protein levels. The finite, though small, fitness penalty to be paid for bearing the plasmid burden would have provided the drive for the chromosome segregation machinery to evolve away from the plasmid partitioning system. Yet the conservation of Cse4 association and cohesin assembly at CEN and STB may not only point to the common ancestry of these loci but may also signify the plasmid's counterstrategy to ensure its efficient propagation by exploiting the chromosome segregation pathway.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health award GM-064363. Partial support was provided by grant F-1274 from the Robert F. Welch Foundation.
We thank Jennifer Gerton, Kim Nasmyth, Munira Basrai, and Sue Biggins for the gifts of yeast strains and plasmids. We acknowledge with thanks the help provided by Hong Cui with one set of ChIP assays. We are grateful for the comments from referees, which provided the inducement to clarify subtle but important features of 2μm plasmid partitioning.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Aravind, L., H. Watanabe, D. J. Lipman, and E. V. Koonin. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 97:11319-11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au, W. C., M. J. Crisp, S. Z. DeLuca, O. J. Rando, and M. A. Basrai. 2008. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179:263-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaisonneau, J., F. Sor, G. Cheret, D. Yarrow, and H. Fukuhara. 1997. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid 38:202-209. [DOI] [PubMed] [Google Scholar]
- 4.Camahort, R., et al. 2007. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26:853-865. [DOI] [PubMed] [Google Scholar]
- 5.Camahort, R., et al. 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35:794-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X. L., A. Reindle, and E. S. Johnson. 2005. Misregulation of 2μm circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 25:4311-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo, K. H. 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1:165-177. [DOI] [PubMed] [Google Scholar]
- 8.Collins, K. A., S. Furuyama, and S. Biggins. 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14:1968-1972. [DOI] [PubMed] [Google Scholar]
- 9.Crotti, L. B., and M. A. Basrai. 2004. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. EMBO J. 23:1804-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, H., S. K. Ghosh, and M. Jayaram. 2009. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J. Cell Biol. 185:251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Wulf, P., A. D. McAinsh, and P. K. Sorger. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagrelius, T. J., A. D. Strand, and D. M. Livingston. 1987. Changes in the DNase I sensitivity of DNA sequences within the yeast 2μm plasmid nucleoprotein complex effected by plasmid-encoded products. J. Mol. Biol. 197:415-423. [DOI] [PubMed] [Google Scholar]
- 13.Furuyama, S., and S. Biggins. 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 104:14706-14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuyama, T., and S. Henikoff. 2009. Centromeric nucleosomes induce positive DNA supercoils. Cell 138:104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futcher, A. B. 1986. Copy number amplification of the 2μm circle plasmid of Saccharomyces cerevisiae. J. Theor. Biol. 119:197-204. [DOI] [PubMed] [Google Scholar]
- 16.Futcher, A. B., and B. S. Cox. 1983. Maintenance of the 2μm circle plasmid in populations of Saccharomyces cerevisiae. J. Bacteriol. 154:612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh, S. K., S. Hajra, and M. Jayaram. 2007. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc. Natl. Acad. Sci. U. S. A. 104:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh, S. K., C. C. Huang, S. Hajra, and M. Jayaram. 2010. Yeast cohesin complex embraces 2μm plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res. 38:570-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadfield, C., R. C. Mount, and A. M. Cashmore. 1995. Protein binding interactions at the STB locus of the yeast 2μm plasmid. Nucleic Acids Res. 23:995-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajra, S., S. K. Ghosh, and M. Jayaram. 2006. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-μm circle partitioning locus and promotes equal plasmid segregation. J. Cell Biol. 174:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henikoff, S., and Y. Dalal. 2005. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15:177-184. [DOI] [PubMed] [Google Scholar]
- 22.Hewawasam, G., et al. 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40:444-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, J., J. M. Hsu, and B. C. Laurent. 2004. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell 13:739-750. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov, D., and K. Nasmyth. 2005. A topological interaction between cohesin rings and a circular minichromosome. Cell 122:849-860. [DOI] [PubMed] [Google Scholar]
- 25.Jayaram, M., S. Mehta, D. Uzri, Y. Voziyanov, and S. Velmurugan. 2004. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-μm yeast plasmid. Prog. Nucleic Acid Res. Mol. Biol. 77:127-172. [DOI] [PubMed] [Google Scholar]
- 26.Jayaram, M., A. Sutton, and J. R. Broach. 1985. Properties of REP3: a cis-acting locus required for stable propagation of the Saccharomyces cerevisiae plasmid 2μm circle. Mol. Cell. Biol. 5:2466-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayaram, M., X. M. Yang, S. Mehta, Y. Voziyanov, and S. Velmurugan. 2004. The 2μm plasmid of Saccharomyces cerevisiae, p. 303-324. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 28.Koshland, D., L. Rutledge, M. Fitzgerald-Hayes, and L. H. Hartwell. 1987. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell 48:801-812. [DOI] [PubMed] [Google Scholar]
- 29.Malik, H. S., and S. Henikoff. 2009. Major evolutionary transitions in centromere complexity. Cell 138:1067-1082. [DOI] [PubMed] [Google Scholar]
- 30.Mead, D. J., D. C. Gardner, and S. G. Oliver. 1986. The yeast 2μm plasmid: strategies for the survival of a selfish DNA. Mol. Gen. Genet. 205:417-421. [DOI] [PubMed] [Google Scholar]
- 31.Mehta, S., et al. 2002. The 2μm plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 158:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta, S., X. M. Yang, M. Jayaram, and S. Velmurugan. 2005. A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2μm plasmid-cohesin association. Mol. Cell. Biol. 25:4283-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuguchi, G., H. Xiao, J. Wisniewski, M. M. Smith, and C. Wu. 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129:1153-1164. [DOI] [PubMed] [Google Scholar]
- 34.Murray, A. W., and J. W. Szostak. 1983. Pedigree analysis of plasmid segregation in yeast. Cell 34:961-970. [DOI] [PubMed] [Google Scholar]
- 35.Murray, J. A., and G. Cesareni. 1986. Functional analysis of the yeast plasmid partition locus STB. EMBO J. 5:3391-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray, J. A., M. Scarpa, N. Rossi, and G. Cesareni. 1987. Antagonistic controls regulate copy number of the yeast 2μm plasmid. EMBO J. 6:4205-4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papacs, L. A., Y. Sun, E. L. Anderson, J. Sun, and S. G. Holmes. 2004. REP3-mediated silencing in Saccharomyces cerevisiae. Genetics 166:79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranjitkar, P., et al. 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40:455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds, A. E., A. W. Murray, and J. W. Szostak. 1987. Roles of the 2μm gene products in stable maintenance of the 2μm plasmid of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:3566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekulic, N., E. A. Bassett, D. J. Rogers, and B. E. Black. 2010. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Som, T., K. A. F. Armstrong, C. Volkert, and J. R. Broach. 1988. Autoregulation of 2μm circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell 52:27-37. [DOI] [PubMed] [Google Scholar]
- 43.Stoler, S., et al. 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. U. S. A. 104:10571-10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan, B. A., M. D. Blower, and G. H. Karpen. 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2:584-596. [DOI] [PubMed] [Google Scholar]
- 45.Sutton, A., and J. R. Broach. 1985. Signals for transcription initiation and termination in the Saccharomyces cerevisiae plasmid 2μm circle. Mol. Cell. Biol. 5:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veit, B. E., and W. L. Fangman. 1985. Chromatin organization of the Saccharomyces cerevisiae 2μm plasmid depends on plasmid-encoded products. Mol. Cell. Biol. 5:2190-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velmurugan, S., X. M. Yang, C. S. Chan, M. Dobson, and M. Jayaram. 2000. Partitioning of the 2-μm circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell Biol. 149:553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volkert, F. C., and J. R. Broach. 1986. Site-specific recombination promotes plasmid amplification in yeast. Cell 46:541-550. [DOI] [PubMed] [Google Scholar]
- 49.Volkert, F. C., D. W. Wilson, and J. R. Broach. 1989. Deoxyribonucleic acid plasmids in yeasts. Microbiol. Rev. 53:299-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, M. C., S. R. Scott-Drew, M. J. Hayes, P. J. Howard, and J. A. Murray. 2002. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2μm plasmid maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:4218-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong, L., X. L. Chen, H. R. Silver, N. T. Ahmed, and E. S. Johnson. 2009. Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2μm circle plasmid. Mol. Biol. Cell 20:1241-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, X. M., S. Mehta, D. Uzri, M. Jayaram, and S. Velmurugan. 2004. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol. Cell. Biol. 24:5290-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.