Abstract
The mitochondrial DNA of Trypanosoma brucei is organized in a complex structure called the kinetoplast. In this study, we define the complete kinetoplast duplication cycle in T. brucei based on three-dimensional reconstructions from serial-section electron micrographs. This structural model was enhanced by analyses of the replication process of DNA maxi- and minicircles. Novel insights were obtained about the earliest and latest stages of kinetoplast duplication. We show that kinetoplast S phase occurs concurrently with the repositioning of the new basal body from the anterior to the posterior side of the old flagellum. This emphasizes the role of basal body segregation in kinetoplast division and suggests a possible mechanism for driving the rotational movement of the kinetoplast during minicircle replication. Fluorescence in situ hybridization with minicircle- and maxicircle-specific probes showed that maxicircle DNA is stretched out between segregated minicircle networks, indicating that maxicircle segregation is a late event in the kinetoplast duplication cycle. This new view of the complexities of kinetoplast duplication emphasizes the dependencies between the dynamic remodelling of the cytoskeleton and the inheritance of the mitochondrial genome.
Faithful inheritance of the genome through successive generations is of fundamental importance to every cell, and sophisticated mechanisms have evolved to perform this function. Trypanosoma brucei is a protozoan parasite of the order Kinetoplastida. During their life cycle, these parasites alternate between the tsetse fly vector and their mammalian host. Differentiation between these forms is accompanied by dramatic changes in metabolism and mitochondrial morphology. Procylic (insect form) T. brucei cells have a single mitochondrion in the form of an extended tubular network. Since there is only one mitochondrion per cell, the duplication cycles of the nucleus and mitochondrion must be coordinated. The mitochondrial genome of Kinetoplastida forms an intricately structured nucleoid, localized within a specialized region of the mitochondrial matrix, a structure termed the kinetoplast. The kinetoplast DNA (kDNA) is composed of two classes of DNA rings. A few dozen maxicircles (23 kb) encode genes for a few mitochondrial proteins (e.g., subunits of respiratory complexes) and rRNA. Several thousand minicircles (∼1 kb) encode a few hundred guide RNAs (usually three per minicircle) that direct the RNA editing (uridylate insertion and deletion) of cryptic maxicircle transcripts; thus, kDNA has many different minicircle species varying in copy number. The most remarkable feature of kDNA is that all of the mini- and maxicircles are topologically linked to form a single planar network. In situ, the T. brucei kDNA network is condensed into a compact disk, approximately 100 nm thick, with a diameter of ∼650 nm, which is anchored to the base of the flagellum through cytoskeletal filaments (36).
Once per cell cycle the kDNA network is replicated and positioned within the cell such that at division, each daughter cell receives one kinetoplast identical to that of the parent. The replication of kDNA has been studied extensively in T. brucei and Crithidia fasciculata, and these data have been integrated into a complex replication model; much of the complexity is required to ensure that each daughter network receives a full minicircle repertoire as well as the appropriate number of maxicircles (for reviews, see references 28 and 43). Replication begins when individual covalently closed minicircles are released from the network into the kinetoflagellar zone (KFZ; the space between the face of the disk and the mitochondrial membrane). It is here that the origin recognition protein (universal minicircle sequence binding protein, or UMSBP [3], and DNA polymerases IB and IC [22]) are located. These and other proteins initiate unidirectional minicircle replication via theta-structure intermediates (9, 11). The minicircle progeny are thought to segregate in the KFZ and then migrate to the antipodal sites, two protein assemblies situated ∼180° apart at opposite poles of the disk (14, 39, 44). These sites contain several enzymes, including structure-specific endonuclease I (10), the helicase TbPIF5 (30), DNA polymerase β (14), and DNA ligase kβ (8), that remove RNA primers and seal most gaps between Okazaki fragments, although at least one gap persists. Of special interest is the topoisomerase II (topo II) named TbTopoIImt, the only mitochondrial topo II encoded in the genome. This enzyme is located predominantly in the antipodal sites (34) and is known to reattach gapped minicircle progeny to the network (48). However, a small amount is situated within the kDNA disk, where it repairs the small holes formed when minicircles are released from the network for replication (27). There also may be TbTopoIImt in the kinetoflagellar zone, where it functions in segregation of minicircle progeny (31). It is not yet known whether this enzyme also releases minicircles from the network prior to their replication.
For each minicircle removed from the network, two are reattached on the network periphery adjacent to the antipodal sites, thus elongating the network and increasing the minicircle copy number. This elongation can be visualized by electron microscopy (EM) or the 4′,6′-diamidino-2-phenylindole (DAPI) staining of isolated networks or of intact cells. When the copy number has doubled, the network becomes bilobed or dumbbell shaped. It then splits in two, and all of the mini- and maxicircle gaps are repaired. The EM of intact cells with segregating kinetoplasts reveals that the sister kinetoplast disks remain linked by a thin filament (a structure termed the nabelschnur) for a short time prior to segregation (16). The two progeny networks finally distribute into daughter cells during cytokinesis. Less is known about maxicircle replication. Unlike minicircles, they remain linked to the network during unidirectional replication as theta structures (6, 18), but like minicircles the parental molecules are covalently closed and progeny contain one or more gaps (29). During the entire replication process, it is remarkable that the higher-order structure of the kinetoplast is maintained, enlarging radially but not changing in thickness. Another interesting feature of kDNA replication is that the pattern of replicated DNA in the network suggests that the kinetoplast disk as a whole undergoes a rotational movement during S phase relative to the antipodal sites, which are thought to remain in a fixed position (32, 37). The mechanism driving this rotation remains unknown.
The replication of kDNA is intimately linked with cell morphogenesis during the trypanosome cell cycle (7, 40, 42, 47, 51). Microtubule cytoskeleton dynamics determine the shape and form of the trypanosome cell throughout its cell and life cycle, and faithful segregation of the nuclear and mitochondrial genome is orchestrated via the mitotic spindle (12) and basal bodies (38), respectively. The basal body in particular has emerged as a master organizer of T. brucei morphogenesis (7). At its distal end, the basal body nucleates the microtubules of the flagellar axoneme. The proximal end of the basal body is linked to the kinetoplast via the filaments of the tripartite attachment complex (TAC) (36). Basal body movement drives the correct segregation and positioning of daughter kinetoplasts in preparation for cytokinesis (38, 40), and the perturbation of the two known TAC proteins p166 (52) and AEP-1 (35) causes failure in kDNA segregation. A cellular electron tomography study of T. brucei recently demonstrated that during the earliest stage of new flagellum growth, the basal body subtending the newly growing flagellum moves around the basal body of the existing mature flagellum in an anticlockwise direction (looking from the basal body toward the tip of the flagellum), resulting in a repositioning from an anterior to a posterior position in the cell (24). Given that basal body duplication and kinetoplast S phase occur almost simultaneously, this raises the question of how the early events of basal body duplication and flagellum formation affect the morphogenesis of two unit-sized kinetoplast disks.
Here, we present a study that explicitly links our understanding of the kDNA minicircle and maxicircle replication processes with the more global cell biological processes of cytoskeletal remodelling and cell morphogenesis. We define a structural model of kinetoplast morphogenesis in situ based on three-dimensional (3D) reconstructions of electron microscopy images and immunofluorescence microscopy. We identified five morphologically distinct stages in the kinetoplast's duplication cycle and discovered that kDNA S phase occurs at the same time as the repositioning of the new basal body from the anterior to the posterior side of the old basal body. This shows that kDNA replication and kinetoplast morphogenesis are intimately tied to basal body dynamics from the earliest stages in their biogenesis. Using fluorescence in situ hybridization (FISH) probes for mini- and maxicircle DNA, we show that in the final stages of kDNA segregation, the progeny disks remain linked by a thread of maxicircle DNA. Cells that overreplicate their maxicircles due to the overexpression of the TbPIF2 helicase (29) have longer and thicker threads than wild-type cells, which persist longer in the cell cycle. We conclude that the nabelschnur structure, defined ultrastructurally, contains maxicircle DNA, and that the unlinking of maxicircles marks the final stage in kinetoplast segregation.
MATERIALS AND METHODS
Cells.
Procyclic forms of Trypanosoma brucei brucei strains LISTER 427, TREU 927, and 29-13 were routinely cultured in SDM-79 medium (5) at 28°C. TREU 927 was used for all wild-type FISH experiments, although similar results were found with 29-13 (data not shown). PIF2-overexpressing cells were made in the 29-13 background (29).
Immunofluorescence studies.
For the analysis of kinetoplast morphology, procyclic trypanosomes were washed in phosphate-buffered saline (PBS), settled on glass slides, and fixed in 2% formaldehyde for 20 min. Following permeabilization with 0.1% NP-40 in PBS, cells were labeled with monoclonal antibody (MAb) YL1/2 (21), followed by secondary antibody tetramethyl rhodamine isocyanate (TRITC) AffiniPure goat anti-rat IgG (Jackson ImmunoResearch Laboratories), as described previously (50). For DNA staining, cells were incubated with 1 μg ml−1 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) before being mounted in glycerol containing 1% (wt/vol) 1,4-diazabicyclo[2.2.2]octane (DABCO) and 10% (vol/vol) 50 mM Na phosphate buffer, pH 8.0. Cells were viewed on a Leitz DM RBE epifluorescence microscope (Leica), and images were captured with a Cool SNAP fx charge-coupled device (CCD) camera (Photometrics). 5-Bromodeoxyuridine (BrdU) labeling was done essentially as described previously (51), except that fixation was in −20°C methanol for 30 min. Cells were labeled with monoclonal antibody BBA4 (50) and rhodamine (TRITC) goat anti-mouse IgM (Millipore) prior to BrdU detection with monoclonal antibody PRB-1 (Molecular Probes) and fluorescein isothiocyanate (FITC) goat anti-mouse IgG (γ-chain specific) (Sigma) and then were stained with DAPI. Images were captured on a DM550 B epifluorescence microscope (Leica) with an Orca CCD camera (Hamamatsu). All image processing was done in Image J (1).
Electron microscopy and generation of kinetoplast models.
Procyclic T. brucei cells were prepared for routine electron microscopy as described previously (25). E-PTA staining was done as described in reference 16. Serial thin sections (light gold interference color, ∼90 nm) were collected on Formvar-coated slot-grids, poststained with lead citrate, and examined in an FEI Tecnai F-12 electron microscope, operating at 80 keV. Digital images were captured on a Gatan Ultrascan 1000 CCD camera with DigitalMicrograph software. Image galleries were assembled using Image J (1), and the brightness and contrast of images were adjusted uniformly. Three-dimensional reconstructions were made with IMOD (http://bio3d.colorado.edu/imod/index.html) (23) using the program Midas for the manual alignment of image stacks and 3dmod for the modeling of the cellular structures.
Fluorescence in situ hybridization and measurements of maxicircle threads.
The minicircle probe was synthesized by a two-step PCR method using isolated kDNA networks from procyclic T. brucei strain 29-13 as the template. First, a 73-bp fragment of the minicircle conserved region was amplified using primers 5′-TATGGGCGTGCAAAAATACA and 5′-CGAAGTACCTCGGACCTCAA. This product was gel purified (Qiagen kit) and used as the template in a second PCR using the same primers and the PCR digoxigenin (DIG) probe synthesis kit (Roche) to incorporate DIG-modified dUTP. The maxicircle probe was labeled by nick translation with biotin-modified dUTP (Roche) using standard protocols (4). The templates for nick translations were plasmids pTKH128, pTKH38, and pTKHR34, a gift from Ken Stuart (Seattle Biomedical Research Institute, Seattle, WA), together representing ∼80% of the maxicircle sequence (46). Reaction conditions were tailored to produce probes smaller than 500 nucleotides to minimize background interference. The three maxicircle probes were pooled for a final concentration of 2.5 ng/μl in the hybridization experiments.
FISH studies were performed as described previously (14, 15, 26, 29), with minor modifications. Briefly, cells were harvested, washed, and resuspended in PBS. Cells then were adhered to poly-l-lysine-coated slides (eight wells, 6 mm diameter) for 10 min. Cells were fixed with 3.5% paraformaldehyde for 10 min, followed by 3.5% paraformaldehyde containing 0.1% Triton X-100 for 10 min. Slides were washed and treated in PBS containing 0.1 M glycine (two times, 5 min each). The slides then were incubated for 30 min in FISH equilibration buffer (50% formamide, 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]). After the removal of this buffer, probe (10 μl per well; 2.5 ng/μl each for both minicircles and maxicircles) was applied in FISH hybridization buffer (10% dextran sulfate, 50% formamide, 2× SSC, 500 μg/ml sheared salmon sperm DNA). Samples were denatured at 70°C for 6 min and then incubated for 18 h at 37°C. Slides then were washed (four times, 5 min each, 40°C) in 50% formamide, 2× SSC, 0.1% Tween 20 and then washed again (three times, 5 min each, 60°C) in 2× SSC, 0.1% Tween 20. Slides were blocked in 5% bovine serum albumin (BSA), 4× SSC, 0.1% Tween 20 (1 h at 37°C) and then incubated with anti-DIG rhodamine (1:50; Roche) and streptavidin-FITC (1:20; Invitrogen) in 1% BSA, 4× SSC, 0.1% Tween 20 (1 h at 37°C). Cells were stained with 2 μg/ml DAPI for 2 min and mounted with Vectashield (Vector Labs, Inc.). To examine total minicircle and maxicircle content by FISH, samples were treated mildly with DNase I prior to hybridization with FISH probes. To do this, cells were adhered to slides, fixed, and permeabilized as described above. A DNase solution was prepared by the dilution of a 2,000-U/ml stock of DNase (NEB) 1:1,000 in DNase buffer (NEB). This solution (10 μl per well) was added to the slides, which were incubated for 10 min at room temperature. The reaction was stopped by washing slides once briefly and then once for 5 min in 1× PBS, 50 mM EDTA, followed by another 5-min wash in PBS. Subsequent equilibration, hybridization, and detections steps were as described above. All images were captured using a Zeiss Axioscop microscope equipped with a 100× phase 3 Plan-Neofluar objective (1.3 numerical aperture; Zeiss) and a Retiga Exi CCD camera (QImaging). Images were acquired and analyzed using IPLab software (Scanalytics) and were adjusted uniformly for brightness and contrast using IPLab software or Adobe Photoshop. The distance between segregating kinetoplasts (from inner edge to inner edge) was measured using Image J (1).
RESULTS
Identification of intermediate stages in the kinetoplast duplication cycle.
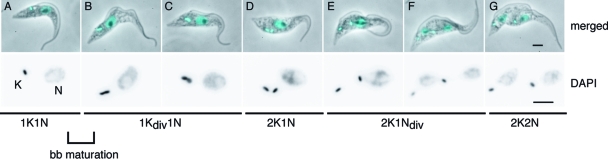
To define the full kinetoplast duplication cycle from a unit-sized disk through S phase, network scission, and the resolution of the nabelschnur structure, we needed to observe kinetoplast morphologies at successive stages in the cell cycle in situ. Since the kinetoplast (K) always divides before the nucleus (N), the position of a trypanosome in the cell cycle can be determined by examining cells stained with DAPI, which allows the visualization of the kinetoplast and nucleus and shows the progression of cells from 1K1N to 2K1N to 2K2N configurations (51). The DAPI staining of T. brucei procyclic cells from exponentially growing asynchronous populations shows clearly that in addition to cells with one- or two-unit-sized kDNA disks, there are several morphologies suggestive of intermediate stages in a duplication process (Fig. 1). To facilitate our subsequent analysis by electron microscopy, we determined the relative frequencies of kinetoplast morphologies in these populations and found that, on average, 41% of the cells in the population had a single kinetoplast (1K), 31% had an elongated or bilobed kinetoplast (1Kdiv), and 28% had two kinetoplasts (2K) (n ≥ 200 cells; three repeats). The high proportion of 1Kdiv cells indicated that kinetoplast replication occurred during an extended period of the cell cycle. We then used the double labeling of cells with DAPI and the monoclonal antibody YL1/2 (49) to correlate kinetoplast morphologies with the early events during growth of the new flagellum (see Fig. S1 in the supplemental material). YL1/2 labels tyrosinated α-tubulin and is a marker for mature basal bodies in trypanosomes (45). The majority (93%) of 1K cells had only one mature basal body, while two mature basal bodies were seen in 99% of Kdiv cells and in all of the 2K cells. This indicates that the maturation of the new basal body occurs during the transition from 1K to 1Kdiv, which is consistent with previous analyses of T. brucei cell cycle timings (42, 51). Importantly for the following analysis, they place the transition from a unit-sized to a bilobed kinetoplast firmly within the time window of basal body maturation and rotation (24).
FIG. 1.
Morphology of the kinetoplast during the cell cycle in T. brucei procyclic forms. The top row of images shows an overlay of the phase-contrast image and DAPI fluorescence (pseudocolored cyan) to show the number and location of kinetoplasts (K) and nuclei (N) in the cell. The second row shows the DAPI signal at twice the magnification to provide a clear view of K shapes. Images are arranged in the order in which the respective K/N configurations occur in the cell cycle. The cell in panel A has a unit-sized kinetoplast (1K), cells in panels B and C have bilobed kinetoplasts (Kdiv), and cells in panels D to G have two kinetoplasts (2K). Ndiv, nucleus with a mitotic spindle. Scale bars, 2 μm. bb, basal body.
Morphogenesis of the kinetoplast during division occurs in five stages.
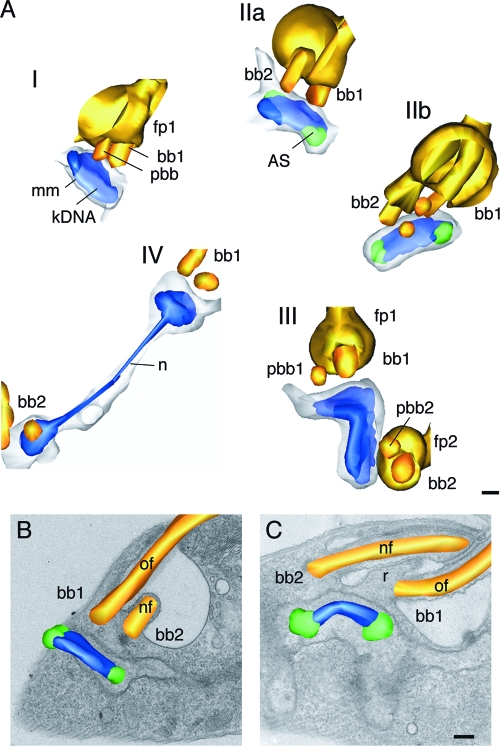
To resolve the precise spatiotemporal relationship between kDNA replication and segregation and basal body duplication, positioning, and flagellum growth, we used an EM-based strategy. We prepared T. brucei procyclic forms from asynchronous cultures for transmission electron microscopy (TEM), collected images of serial thin sections through a total of 30 kinetoplasts at specific stages in their duplication cycle (three examples are shown in Fig. S2 in the supplemental material), and generated 3D reconstructions of kinetoplast morphology. We defined five distinct stages of kinetoplast duplication, I to V (Fig. 2A), based on the shape of the kDNA disk, the presence of antipodal sites, and the replication status of basal bodies and flagellar pocket. In stage I, the kinetoplast disk is of unit size, has no visible antipodal sites, and is associated with one basal body/probasal body pair (Fig. 2A). In stage II, the kinetoplast disk is associated with two basal bodies and has prominent antipodal sites (indicative of kinetoplast S phase). The antipodal sites on opposite poles of the disk are oriented such that one is always nearer the posterior end of the cell than the other. This geometry is consistent with previous measurements of antipodal site position in T. brucei (32). In stage IIa, the newly forming flagellum still is contained entirely within the existing flagellar pocket, positioned anterior to the mature flagellum (Fig. 2A and B); in stage IIb, the kDNA disk has a domed shape (but is not yet a bilobe). The kinetoplast is associated with two basal body/probasal body pairs, with the growing new flagellum now positioned posterior to the mature flagellum (Fig. 2A and C). A ridge has formed between the old flagellar pocket and the bud of the new flagellar pocket, but the tip of the new flagellum still is contained within the preexisting pocket (Fig. 2A and C). In stage III, the kinetoplast consists of two joined disks, forming a bilobed shape (Fig. 2A); the planes of the disks are at an angle (giving the appearance of a V shape when viewed from the side or a shape resembling a twisted figure eight when viewed from the top). Antipodal sites occasionally are observed on one or both of the disks. The kinetoplast is associated with two basal body/probasal body pairs; two separate flagellar pockets are present. In stage IV, the kinetoplasts appear as two unit-sized disks that have moved apart, but they still are linked by the nabelschnur (Fig. 2A). In stage V, the two kinetoplasts are fully separated; their morphology is the same as that in stage I.
FIG. 2.
Model of the kinetoplast duplication cycle. T. brucei procyclic forms were prepared for TEM. Images were captured of serial thin sections through kinetoplasts at different stages in their duplication cycle, and 3D reconstructions were generated from these images. (A) The duplication cycle can be divided into five distinct stages, I to V, based on the morphology of the kDNA disk and the replication status of the basal body (bb)/probasal body (pbb) and adjacent flagellar pocket (fp); mm, mitochondrial membrane. bb1 denotes the basal body subtending the mature flagellum, and bb2 is the basal body of the new flagellum. In stage I, the kDNA forms a unit-sized disk; in stage II, the kDNA disk has prominent antipodal sites (AS); stage III kinetoplasts are bilobed and flagellar pocket duplication is complete; stage IV kinetoplasts are connected by a nabelschnur (n); and stage V kinetoplasts (not shown) are morphologically the same as stage I kinetoplasts. (B) Model of a stage IIa kinetoplast superimposed onto a single image from the series (oriented such that the anterior end of the cell is facing right). In this cell, bb2 is in a position anterior to bb1. (C) Model of a stage IIb kinetoplast superimposed onto a single image from the series (oriented as in panel B). In this cell, bb2 is now in a position posterior to bb1. nf, new flagellum; of, old flagellum; r, ridge forming between the old and new flagellar pocket. Scale bars, 200 nm.
Kinetoplast S phase coincides with the anticlockwise rotation of the new basal body around the old.
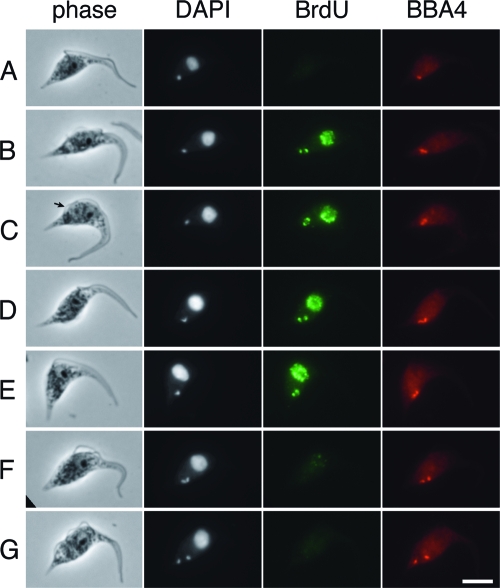
Progression from stage IIa to stage IIb, described above (Fig. 2A to C), coincides precisely with the repositioning of the new basal body to a posterior position, indicating that kinetoplast S phase occurs while the rotation of the new basal body around the old basal body takes place. To obtain further evidence for this, we incubated cells with the thymidine analogue BrdU for 15 min and used immunofluorescence microscopy to visualize the sites of the newly incorporated DNA with anti-BrdU antibodies and the basal bodies with monoclonal antibody BBA4 (50) (Fig. 3). We detected BrdU in 32% of kinetoplasts (n = 152 cells), and all BrdU-labeled kinetoplasts showed the characteristic pattern of BrdU incorporation at the two antipodal sites. The kinetoplasts that had incorporated BrdU were a subset of cells with 1K1N (Fig. 3B to E); none of the 2K1N or 2K2N cells had labeled kinetoplasts (Fig. 3F and G and data not shown). 1K1N cells with a unit-sized kinetoplast, which had not yet formed new probasal bodies (Fig. 3B and C), represented the earliest stage in the cell cycle in which BrdU was detected at kinetoplast poles. Cells with a bilobed kinetoplast, two basal body/probasal body pairs, and a short new flagellum (Fig. 3D and E) represented the latest stage in the cell cycle with BrdU-labeled kinetoplast poles. This pattern of labeling is consistent with our earlier observations by TEM, where antipodal sites were seen in stage IIa and IIb and a subset of stage III kinetoplasts (Fig. 2). Taken together, these data demonstrate that S phase and the repositioning of the basal body occur simultaneously. This is interesting, because it has been shown that the T. brucei kinetoplast undergoes rotational movement during the replication of the kDNA network (32), yet it is not known what drives this movement. The precise temporal correlation between basal body positioning and kDNA rotation shown here suggests that the rotational movements of the two structures are causally linked.
FIG. 3.
BrdU labeling of antipodal sites. T. brucei procyclic cells in asynchronous culture were incubated with BrdU for 15 min before processing for the immunofluorescence detection of incorporated BrdU (MAb PRB-1) and basal bodies (MAb BBA4). DNA was stained with DAPI. (A to G) Cells in successive stages of the cell cycle. BrdU incorporation was detected at the poles of kinetoplasts in a subset of 1K1N cells (B and C) and 1Kdiv1N cells (D and E). The arrow in panel C indicates the short new flagellum. Cells with BrdU-positive kinetoplasts also had numerous BrdU foci throughout the nucleus. Scale bar, 5 μm.
Segregation of maxicircle DNA marks the final stage in kinetoplast division.
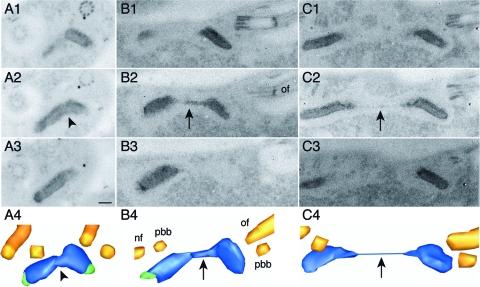
Following the positioning of the new basal body to a posterior position, a lateral movement of the new basal body toward the posterior pole of the cell segregates the two daughter kinetoplasts, reaching a final interbasal body distance of ∼4.5 μm (40). Segregating kinetoplasts remain connected by a nabelschnur, which can reach at least 1 μm (16). Ethanolic phosphotungstic acid (E-PTA) staining provides views of the nabelschnur with higher contrast compared to that in standard TEM preparations, and for this reason E-PTA-stained cells were used to generate serial section images and 3D reconstructions of kinetoplasts with a nabelschnur (Fig. 4). The resulting images showed that the nabelschnur emanates from a zone of diffuse electron density at the inner edges of the disks, and as the distance between the disks increases the nabelschnur becomes longer and thinner (compare Fig. 4B and C). Taken together, the TEM data suggest that the nabelschnur is formed from material present in the midzone of bilobed (stage III) kinetoplasts, which is stretched out as the kinetoplasts are segregated.
FIG. 4.
3D reconstructions of the nabelschnur in segregating kinetoplasts. T. brucei procyclic forms were prepared for TEM, stained with E-PTA, and used to generate 3D reconstructions from serial sections, as described for Fig. 2. (A) Bilobed kinetoplast (the arrowhead marks the midzone between the lobes); (B and C) kinetoplasts with a short and a long nabelschnur, respectively (arrows). A1 to A3, B1 to B3, and C1 to C3 show adjacent serial sections; A4, B4, and C4 show the full 3D reconstruction of the corresponding kinetoplasts. of, old flagellum; nf, new flagellum; pbb, probasal body. Scale bar, 100 nm.
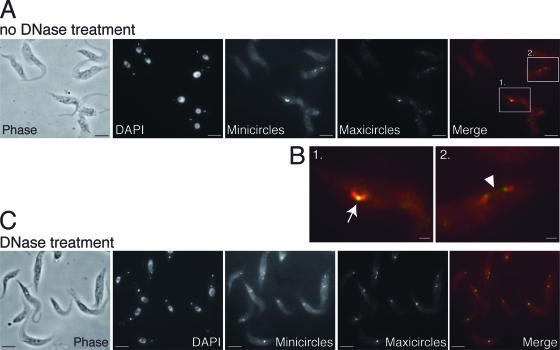
By the careful examination of the 2K1N cells stained with DAPI, we found that a subset of these cells had a thin thread of DNA connecting the two kinetoplasts, but the intensity of the DAPI signal from the DNA thread was very low (Fig. 1D). Our next objective was to determine which class of kDNA was present in these threads. We therefore probed wild-type cells with FISH probes specific for mini- or maxicircle DNA. In cells not treated with DNase I, only nicked or gapped circles are denatured and accessible to the hybridization probe, thus labeling only postreplication minicircles and maxicircles where gap repair has not been completed. Figure 5A shows two examples. The first cell (Fig. 5B, boxed) is 1Kdiv1N, with a bilobed kinetoplast. The gapped minicircle DNA is detected throughout the disk, while gapped maxicircle DNA appeared to be concentrated in the midzone. The second cell (enlarged view in Fig. 5B) is 2K1N, and in this cell the gapped minicircle DNA again is detected throughout the disks, while gapped maxicircle DNA is concentrated at the disk inner edges, with a thin thread of maxicircle DNA stretched between the kinetoplasts. The treatment of trypanosomes with the nicking enzyme DNase I before FISH enables the hybridization of all DNA irrespective of gap repair status (Fig. 5C). In 1K1N cells, the signal from the mini- and maxicircle DNA was overlapping in the kDNA disk. In 2K1N cells the distribution of minicircles and maxicircles did not differ appreciably from that found in cells without DNase treatment (Fig. 5C). The kinetoplasts in 2K2N cells as well as late-stage 2K1N cells, which were undetectable by FISH in the absence of DNase, were labeled in DNase-treated samples, and their FISH patterns resembled those of kinetoplasts in 1K1N cells. The localization pattern of maxicircle DNA thus fits precisely the localization and behavior of the nabelschnur seen in electron micrographs (Fig. 4), and we conclude that the nabelschnur is the ultrastructural correlate of maxicircle threads with associated proteins.
FIG. 5.
Visualization of replicating and nonreplicating kDNA networks by FISH. (A) A representative field showing cells prepared using the standard FISH protocol. Only minicircles and maxicircles that are nicked or gapped due to replication are accessible to FISH probes. In the merged images minicircles are shown in red and maxicircles in green. Scale bar, 5 μm. Areas outlined by the white boxes in the merged image are enlarged in panel B. (B) The numbers refer to the order in which they are discussed in the text. Arrow indicates concentration of maxicircles in the center of a dumbbell-shaped kDNA network. The arrowhead indicates a thin thread of maxicircles that connects two networks that have already separated. Scale bar, 1 μm. (C) A representative field showing cells that were nicked by DNase treatment prior to the FISH procedure. Here, all kDNA networks regardless of cell cycle stage are labeled by the minicircle and maxicircle FISH probes. Scale bar, 5 μm.
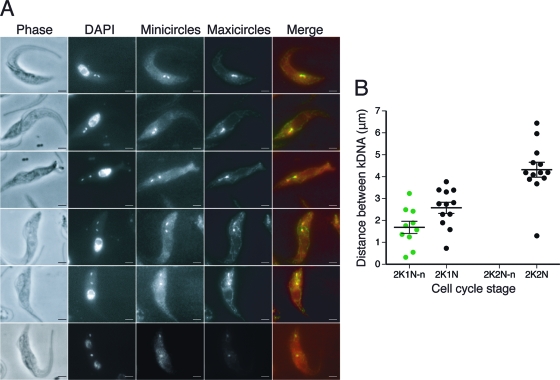
We then focused on the population of 2K1N and 2K2N cells to determine the frequency of maxicircle threads and measured the distance between the disks with and without threads. This analysis showed that maxicircle DNA threads occurred in a subpopulation of the 2K1N cells, and no threads were observed in 2K2N cells (Fig. 6A, B). In 2K1N cells, disks with a thread were closer together than disks without a thread (average distance with thread, 1.68 μm [standard deviations, 0.89]; average distance without thread, 2.58 μm [standard deviations, 0.88]; n = 22). The circumference of a T. brucei maxicircle is ∼6.3 μm (13). The length of maxicircle threads in the wild type never exceeded the length of a stretched out maxicircle (i.e., no thread was longer than half the maxicircle circumference). The average distance between the disks increased further, to 4.32 μm (standard deviations, 1.22; n = 13), in 2K2N cells. These data show that the segregation of the maxicircle DNA occurs during the 2K1N stage, later than the segregation of the minicircle networks but before the partitioning of the nuclear DNA.
FIG. 6.
Quantitation of frequency and length of maxicircle threads in wild-type (927) cells. (A) The first five rows show representative FISH images of cells containing threads of various lengths. Cells were treated with the nicking enzyme DNase I prior to FISH. The last row shows a 2K2N cell lacking a thread. Scale bar, 2 μm. (B) Quantitation of frequency and length of threads in various stages of the cell cycle. In all cells containing two kinetoplasts (K), the distance between them was measured in microns. Cells then were scored according to whether or not they contained a thread. Green dots, cells containing a thread. Black dots, cells lacking a thread. A thread was never observed in 2K2N cells. Horizontal bars indicate the means. Error bars show standard errors of the means. N, nucleus; K, kinetoplast; n, nabelschnur/maxicircle thread.
Increase in maxicircle copy number delays maxicircle segregation.
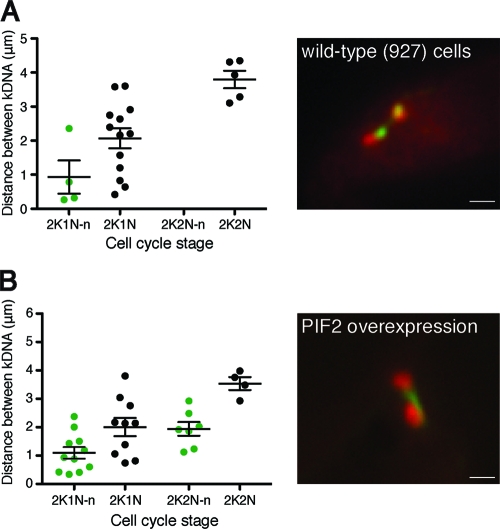
The overexpression of the mitochondrial helicase TbPIF2 leads to the spectacular overreplication of maxicircles, characterized by prominent maxicircle DNA threads and kinetoplast segregation defects (29). Further analysis of these cells by FISH showed that the frequency of maxicircle threads in the mutants was increased compared to that of the wild type (Fig. 7A, B). Importantly, in cells overexpressing TbPIF2, but not in wild-type cells, threads also were observed in a subpopulation of 2K2N cells. This suggests that the increased number of maxicircles either delays or blocks their segregation.
FIG. 7.
Frequency and length of threads increases during overexpression of the mitochondrial helicase PIF2. (A) Quantitation of length and frequency of maxicircle threads in wild-type (927) cells (n = 22). FISH was performed in the absence of DNase treatment, meaning that threads were observed less frequently, most likely because some gaps in the replicated maxicircles are filled prior to maxicircle division, rendering them inaccessible to our FISH probes. Analysis was performed as described for Fig. 6. Green dots, cells containing a thread. Black dots, cells lacking a thread. Horizontal bars indicate the means. Error bars show standard errors of the means. K, kinetoplast; N, nucleus; n, nabelschnur/maxicircle thread. The image shows a 2K1N cell with a very fine maxicircle thread (minicircles, red; maxicircles, green). Scale bar, 1 μm. (B) A similar experiment was performed on cells that had overexpressed TbPIF2 for 48 h. TbPIF2 is a mitochondrial helicase involved in maxicircle replication. The overexpression of this protein leads to an increase in the number of maxicircles. In these cells (n = 32), the frequency and length of the maxicircle thread increases and can now be observed in 2K2N cells. The nabelschnur also appears thicker. The image shows a 2K1N cell (minicircles, red; maxicircles, green). Scale bar, 1 μm.
Taken together, these data provide a structural model of a full kinetoplast duplication cycle in situ and relate this to events in basal body duplication and positioning. We show that at the beginning of the cycle, kinetoplast S phase coincides with the rotation of the basal body subtending the new flagellum around the old flagellum. This correlation suggests a role for the basal body in organizing kinetoplast morphogenesis in S phase that precedes its well-described function in kinetoplast segregation. At the end of the duplication cycle it is the segregation of maxicircle DNA that marks the final stage in the duplication process.
DISCUSSION
Many previous studies have focused on individual events in the replication and segregation of the kDNA network, including the replication mechanisms of mini- and maxicircles and the individual contributions of many kDNA replication enzymes (28, 33, 43). In this study, we have examined how the kinetoplast structure in situ changes during the cell cycle and how these individual events of kDNA replication are coordinated with other events in cellular morphogenesis. Our data indicate that cytoskeleton-mediated cell morphogenesis orchestrates the whole kinetoplast duplication cycle, from S phase to the segregation of the mini- and maxicircle networks.
Our structural model of the kinetoplast duplication cycle revealed that the anticlockwise rotation of the new basal body around the old occurs while kinetoplast S phase is in progress. The evidence that these two events happen simultaneously is strong. The serial-section EM reconstructions enabled us to order kinetoplasts according to the progression of the basal body movement by comparing the length of the new axoneme, the formation of new probasal bodies, and the budding of the new flagellar pocket from the old. The elongation of the new axoneme starts when the new basal body is still in an anterior position. We found that kinetoplasts at this stage already had antipodal sites (stage IIa kinetoplasts) (Fig. 2A, B), which indicates that minicircle replication is under way. The growth of the new axoneme then continues while new probasal bodies are formed and the basal body subtending the new flagellum migrates around the old basal body to a posterior position in the cell (24, 42). We observed antipodal sites in kinetoplasts where the new flagellum had reached a position halfway through the flagellar pocket and kinetoplasts where the rotation had been completed (stage IIb kinetoplasts) (Fig. 2A, C). In an alternative approach, we visualized antipodal sites and nearby regions of the network by BrdU labeling, which is one of the most direct indicators of S phase applicable to whole cells. The data shown in Fig. 3 are fully consistent with the ultrastructural analysis, indicating that kinetoplast S phase coincides with the duplication of the basal bodies and continues for some period after the emergence of the new flagellum from the cell. Importantly, this means that the basal body and kinetoplast, two structures that are tethered to one another, undergo a rotational movement at the same time. While we have not yet identified the motor that drives the rotational movement of the basal body or the kinetoplast, this suggests that these movements are causally linked.
A temporal link between kinetoplast rotational dynamics and basal body movement during S phase.
Previous studies, mainly involving the fluorescence microscopy of isolated networks labeled metabolically with BrdU, provided the first indication of network rotation in T. brucei (32). With short pulses (3 min) of BrdU, labeling occurred at two points on the network periphery, 180° apart. With a longer pulse (15 min), each point expanded into a larger fraction of the periphery. This arc of labeling, covering a limited region of the periphery, then would expand in thickness as additional rows of minicircles were added. These studies led to the conclusion that the attachment of the additional rows in this limited region of the periphery occurred by the oscillation of the kinetoplast disk between fixed antipodal sites. The rotation of the kinetoplast then would allow minicircle attachment to the adjacent region of the periphery. These data also suggested that the kinetoplast rotation is not continuous and unidirectional and that sometimes rotation involved a jump to a nonadjacent region of the network periphery. How this occurs is not yet clear.
In principle, the tethering of the basal bodies to the kDNA through the TAC could suffice to translate the anticlockwise rotation of the new basal body around the old directly into a rotational movement of the disk. The inhibition of basal body duplication by the RNA interference (RNAi)-mediated ablation of POLO-like kinase (19) and the inhibition of basal body segregation by treatment with the antimicrotubule drug ansamitocin (38) prevented kinetoplast segregation. Kinetoplasts in the affected cells remained associated with the old flagellum, and DAPI intensity increased over time, indicating that kDNA synthesis continued in the unsegregated kinetoplasts. Similarly, the RNAi-mediated ablation of TAC component p166 caused the accumulation of cells with kinetoplasts up to 10-fold larger than normal, which remained associated with the old basal body (52). These experiments clearly demonstrate that kDNA synthesis itself is independent of a functional link to basal bodies. Interestingly, however, in the p166 mutants the attachment of newly replicated minicircles to the network was severely disorganized (52). Moreover, in POLO-like kinase mutants the replication of kDNA was delayed compared to that of control cells (19). This is consistent with a model in which the TAC-basal body connection has a role in directing the ordered replication of the network and perhaps in facilitating the delivery of replicated free daughter minicircles to the appropriate network pole for reattachment. If this were correct, we would predict that mutant cells that cannot build a new basal body would exhibit an aberrant pattern of minicircle incorporation similar to that of the p166 mutants. Future studies of TAC components no doubt will provide interesting new insights into the specific mechanisms of kDNA replication and the proximal functions of basal bodies linking cytoskeletal dynamics to genome inheritance.
Segregation of maxicircle DNA is the final step in the scission of the daughter networks.
Much less is known about the replication mechanism and in situ organization of maxicircles than about minicircles. In nonreplicating kinetoplasts, T. brucei maxicircles are distributed throughout the disk (15). Furthermore, all of the maxicircles are interlocked with each other, forming a so-called network within a network (41). In contrast to minicircles, maxicircles remain attached to the network during replication (6, 18). During replication, the maxicircles gradually accumulate in the middle of the network (15) and in bilobed kinetoplasts in situ, and in isolated dumbbell-shaped networks they are clustered in the midzone (15, 17, 20). Consistently with these previous reports, we saw the clustering of maxicircles in the midzone of bilobed kinetoplasts (Fig. 5A). In addition, we detected thin maxicircle DNA threads, devoid of detectable quantities of minicircle DNA (although they could contain bound protein), stretched between segregating disks (Fig. 5 and 6). While we never detected minicircles in these thread structures, the minicircle and maxicircle probes were labeled using different techniques (see Materials and Methods); therefore, we cannot exclude the possibility that the maxicircle probe is more sensitive. However, the findings from numerous other studies that maxicircles cluster at the center of dividing T. brucei networks (15, 17, 20) are consistent with our results and our model. We conclude that the persistence of this maxicircle thread explains the phenomenon of the nabelschnur (16), a thread of electron-dense material within the midzone of the bilobe, which was captured in several of the 3D reconstructions (Fig. 4); these are stage IV kinetoplasts.
Increased maxicircle content delays resolution of the nabelschnur structure.
Networks from trypanosomes overexpressing the TbPIF2 helicase have a 3- to 6-fold increase in maxicircles with little change in minicircle level. We previously showed by DAPI staining and FISH that maxicircle threads were prominent in these cells (29). Our current FISH analyses confirmed that the thick DNA threads in these mutants contained maxicircles. Nevertheless, these threads differed from those in wild-type cells by their increased length (although never exceeding the length of a single stretched-out maxicircle) and persistence in 2K2N cells. We speculate that the increased number of maxicircles saturates the enzymatic activity responsible for resolving the nabelschnur. While we do not know the identity of this enzyme, a likely candidate is the sole mitochondrial topoisomerase II, TbTopoIImt. In fact, the first observation of threads by DAPI staining was in trypanosomes treated with teniposide, a topo II inhibitor (38). This possibility is difficult to address experimentally, as TbTopoIImt has several well-defined roles at multiple and earlier stages in the kDNA duplication cycle (27, 31, 48). Because of this, the TbTopoIImt RNAi phenotype is complex, and the kinetoplast structure becomes so dramatically altered that kinetoplasts in RNAi cells never reach stage IV, in which two clearly defined kinetoplasts are connected by a thin thread of maxicircles.
A thread-like structure also has been described in cells in which the TbHslVU protease is depleted by RNAi (26). This protease controls minicircle and maxicircle copy number by degrading master positive regulators of their synthesis; TbPIF2 is the maxicircle regulator (29), and the minicircle regulator is not yet known. These cells have giant kDNA networks with minicircle copy numbers increased 15- to 20-fold and those of maxicircles increased 2.8-fold. As kDNA replication proceeds in an uncontrolled manner, kinetoplast organization is lost and the networks are unable to segregate. This results in the large kinetoplasts becoming stretched out between the two basal bodies as they begin to move apart. In some cases, these elongated kinetoplasts can resemble threads; however, the threads in the HslVU phenotype differ fundamentally from the nabelschnur found in wild-type cells, in that they can contain both minicircles and maxicircles.
The kinetoplast duplication cycle in the context of cytoskeleton-mediated cell morphogenesis.
Studies of cytoskeleton dynamics in T. brucei procyclics have identified a number of key morphogenetic markers for successive stages in the cell cycle, and we are now able to place the kinetoplast duplication cycle in this context. Measurements of interbasal body distances have shown that in T. brucei procyclics, basal body segregation is biphasic (2, 7, 40). During the first phase the basal bodies move up to ∼2 μm apart, while the new flagellum extends alongside the old. Kinetoplast replication stages II to IV fall within this period. During the second phase, the flagellum connector that joins the tip of the new flagellum to the side of the old flagellum remains arrested (at the critical point, 0.5 along the old flagellum length [7]); during the latter part of flagellum growth, the increase in interbasal body distance is more rapid until the basal bodies are positioned ∼4.5 μm apart, assuming their final precytokinesis position. The transition from slow to accelerated basal body movement has been termed the cryptic transition point (40). Our measurements showing that the resolution of the maxicircle thread occurred at interkinetoplast distances of ∼2 μm (Fig. 6) provide strong evidence that this cryptic transition point is the point at which the final unlinking of the maxicircle DNA happens (transition from stage IV to V kinetoplasts). The segregation of the kDNA may simply facilitate a faster movement of the basal bodies. Alternatively, accelerated basal body movement may provide a necessary signal for the completion of network segregation. In the TbPIF2 overexpressers, some 2N2K cells still had a maxicircle thread, and in these cells the basal bodies remained closer together than in 2K2N cells where the thread had been resolved (Fig. 7). This evidence suggests that the persistence of a maxicircle thread had no influence on the first phase of basal body segregation but was sufficient to restrict basal body movement during the second phase.
The discrete distribution of different groups of proteins around the kinetoplast is a defining feature of kDNA replication, and it seems likely that cytoskeletal elements such as the basal bodies and components of TAC play a role in the structural organization of this process. Based on these new data and their integration with earlier studies, our overview of network replication is as follows. Prior to the start of kDNA replication, maxicircles and different species of minicircles are randomly and uniformly distributed throughout the unit-sized network. After covalently closed minicircles are individually released from the network into the kinetoflagellar zone for replication, their gapped progeny are reattached to the network periphery adjacent to the antipodal sites. Thus, the minicircle copy number increases and the network elongates. Since minicircles are removed from the central region of the network and their progeny attached at or near the network poles, the maxicircle network gradually and passively concentrates in the central region of the elongating network. Furthermore, when the last covalently closed minicircles have been released from the network and their progeny reattached, the minicircles in each lobe of the double-sized network are linked to each other but not to those in the other lobe. Thus, a distinct mechanism is not needed for the segregation of the minicircle networks, as the removal of the final unreplicated minicircle from the network for replication effectively segregates the two daughter minicircle networks. At this point the two lobes are held together only by a centrally located maxicircle network, which may be double sized due to replication; since FISH on networks untreated with DNase I reveals gapped maxicircles in the midzone (Fig. 5B, image 1), at least some of them must have undergone replication by this time. The cleavage of this thread by TbTopoIImt or an unknown enzyme is thus the final step in the scission of the daughter networks. When the networks have completely segregated, the sister networks have maxicircles clustered on one side of the network as visualized by EM (20). Finally, by further topo II action, the network then must undergo remodelling, randomizing the location of maxicircles and presumably all the different minicircle species.
Supplementary Material
Acknowledgments
We thank Mike Shaw, University of Oxford, for assistance with electron microscopy and Beiyu Liu for helpful comments on the manuscript.
Work in the laboratory of K.G. was supported by a Wellcome Trust program grant and the E. P. Abraham Trust. The laboratory of P.T.E. was supported by NIH grant AI058613.
Footnotes
Published ahead of print on 20 December 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Absalon, S., et al. 2007. Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS One 2:e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu-Elneel, K., D. R. Robinson, M. E. Drew, P. T. Englund, and J. Shlomai. 2001. Intramitochondrial localization of universal minicircle sequence-binding protein, a trypanosomatid protein that binds kinetoplast minicircle replication origins. J. Cell Biol. 153:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, A. L., S. G. Ballard, and D. C. Ward. 1990. Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. U. S. A. 87:7757-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brun, R., and Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 6.Carpenter, L. R., and P. T. Englund. 1995. Kinetoplast maxicircle DNA replication in Crithidia fasciculata and Trypanosoma brucei. Mol. Cell. Biol. 15:6794-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidge, J. A., et al. 2006. Trypanosome IFT mutants provide insight into the motor location for mobility of the flagella connector and flagellar membrane formation. J. Cell Sci. 119:3935-3943. [DOI] [PubMed] [Google Scholar]
- 8.Downey, N., J. C. Hines, K. M. Sinha, and D. S. Ray. 2005. Mitochondrial DNA ligases of Trypanosoma brucei. Eukaryot. Cell 4:765-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drew, M. E., and P. T. Englund. 2001. Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J. Cell Biol. 153:735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel, M. L., and D. S. Ray. 1999. The kinetoplast structure-specific endonuclease I is related to the 5′ exo/endonuclease domain of bacterial DNA polymerase I and colocalizes with the kinetoplast topoisomerase II and DNA polymerase beta during replication. Proc. Natl. Acad. Sci. U. S. A. 96:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Englund, P. T. 1979. Free minicircles of kinetoplast DNA in Crithidia fasciculata. J. Biol. Chem. 254:4895-4900. [PubMed] [Google Scholar]
- 12.Ersfeld, K., and K. Gull. 1997. Partitioning of large and minichromosomes in Trypanosoma brucei. Science 276:611-614. [DOI] [PubMed] [Google Scholar]
- 13.Fairlamb, A. H., P. O. Weislogel, J. H. Hoeijmakers, and P. Borst. 1978. Isolation and characterization of kinetoplast DNA from bloodstream form of Trypanosoma brucei. J. Cell Biol. 76:293-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, M., A. F. Torri, D. C. Ward, and P. T. Englund. 1992. In situ hybridization to the Crithidia fasciculata kinetoplast reveals two antipodal sites involved in kinetoplast DNA replication. Cell 70:621-629. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, M. L., A. F. Torri, D. Perez-Morga, D. C. Ward, and P. T. Englund. 1994. Kinetoplast DNA replication: mechanistic differences between Trypanosoma brucei and Crithidia fasciculata. J. Cell Biol. 126:631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gluenz, E., M. K. Shaw, and K. Gull. 2007. Structural asymmetry and discrete nucleic acid subdomains in the Trypanosoma brucei kinetoplast. Mol. Microbiol. 64:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guilbride, D. L., and P. T. Englund. 1998. The replication mechanism of kinetoplast DNA networks in several trypanosomatid species. J. Cell Sci. 111:675-679. [DOI] [PubMed] [Google Scholar]
- 18.Hajduk, S. L., V. A. Klein, and P. T. Englund. 1984. Replication of kinetoplast DNA maxicircles. Cell 36:483-492. [DOI] [PubMed] [Google Scholar]
- 19.Hammarton, T. C., S. Kramer, L. Tetley, M. Boshart, and J. C. Mottram. 2007. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol. Microbiol. 65:1229-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeijmakers, J. H., and P. J. Weijers. 1980. The segregation of kinetoplast DNA networks in Trypanosoma brucei. Plasmid 4:97-116. [DOI] [PubMed] [Google Scholar]
- 21.Kilmartin, J. V., B. Wright, and C. Milstein. 1982. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93:576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klingbeil, M. M., S. A. Motyka, and P. T. Englund. 2002. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol. Cell 10:175-186. [DOI] [PubMed] [Google Scholar]
- 23.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 24.Lacomble, S., et al. 2010. Basal body movements orchestrate membrane organelle division and cell morphogenesis in Trypanosoma brucei. J. Cell Sci. 123:2884-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacomble, S., et al. 2009. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 122:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Z., M. E. Lindsay, S. A. Motyka, P. T. Englund, and C. C. Wang. 2008. Identification of a bacterial-like HslVU protease in the mitochondria of Trypanosoma brucei and its role in mitochondrial DNA replication. PLoS Pathog. 4:e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsay, M. E., E. Gluenz, K. Gull, and P. T. Englund. 2008. A new function of Trypanosoma brucei mitochondrial topoisomerase II is to maintain kinetoplast DNA network topology. Mol. Microbiol. 70:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, B., Y. Liu, S. A. Motyka, E. E. Agbo, and P. T. Englund. 2005. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 21:363-369. [DOI] [PubMed] [Google Scholar]
- 29.Liu, B., et al. 2009. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol. Cell 35:490-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, B., J. Wang, G. Yildirir, and P. T. Englund. 2009. TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase involved in processing of minicircle Okazaki fragments. PLoS Pathog. 5:e1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, B., et al. 2010. TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication. J. Biol. Chem. 285:7056-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Y., and P. T. Englund. 2007. The rotational dynamics of kinetoplast DNA replication. Mol. Microbiol. 64:676-690. [DOI] [PubMed] [Google Scholar]
- 33.Lukes, J., et al. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melendy, T., C. Sheline, and D. S. Ray. 1988. Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell 55:1083-1088. [DOI] [PubMed] [Google Scholar]
- 35.Ochsenreiter, T., S. Anderson, Z. A. Wood, and S. L. Hajduk. 2008. Alternative RNA editing produces a novel protein involved in mitochondrial DNA maintenance in trypanosomes. Mol. Cell. Biol. 28:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogbadoyi, E. O., D. R. Robinson, and K. Gull. 2003. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 14:1769-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez-Morga, D. L., and P. T. Englund. 1993. The attachment of minicircles to kinetoplast DNA networks during replication. Cell 74:703-711. [DOI] [PubMed] [Google Scholar]
- 38.Robinson, D. R., and K. Gull. 1991. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731-733. [DOI] [PubMed] [Google Scholar]
- 39.Robinson, D. R., and K. Gull. 1994. The configuration of DNA replication sites within the Trypanosoma brucei kinetoplast. J. Cell Biol. 126:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, D. R., T. Sherwin, A. Ploubidou, E. H. Byard, and K. Gull. 1995. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 128:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro, T. A. 1993. Kinetoplast DNA maxicircles: networks within networks. Proc. Natl. Acad. Sci. U. S. A. 90:7809-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherwin, T., and K. Gull. 1989. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos Trans. R. Soc. Lond. B Biol. Sci. 323:573-588. [DOI] [PubMed] [Google Scholar]
- 43.Shlomai, J. 2004. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 4:623-647. [DOI] [PubMed] [Google Scholar]
- 44.Simpson, A. M., and L. Simpson. 1976. Pulse-labeling of kinetoplast DNA: localization of 2 sites of synthesis within the networks and kinetics of labeling of closed minicircles. J. Protozool. 23:583-587. [DOI] [PubMed] [Google Scholar]
- 45.Stephan, A., S. Vaughan, M. K. Shaw, K. Gull, and P. G. McKean. 2007. An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic 8:1323-1330. [DOI] [PubMed] [Google Scholar]
- 46.Stuart, K. D., and S. B. Gelvin. 1982. Localization of kinetoplast DNA maxicircle transcripts in bloodstream and procyclic form Trypanosoma brucei. Mol. Cell. Biol. 2:845-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaughan, S. 2010. Assembly of the flagellum and its role in cell morphogenesis in Trypanosoma brucei. Curr. Opin. Microbiol. 13:453-458. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Z., and P. T. Englund. 2001. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 20:4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wehland, J., H. C. Schroder, and K. Weber. 1984. Amino acid sequence requirements in the epitope recognized by the alpha-tubulin-specific rat monoclonal antibody YL 1/2. EMBO J. 3:1295-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woods, A., et al. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491-500. [DOI] [PubMed] [Google Scholar]
- 51.Woodward, R., and K. Gull. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 95:49-57. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, Z., M. E. Lindsay, A. Roy Chowdhury, D. R. Robinson, and P. T. Englund. 2008. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 27:143-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.