Abstract
Activation of the small GTPase RhoA following angiotensin II stimulation is known to result in actin reorganization and stress fiber formation. Full activation of RhoA, by angiotensin II, depends on the scaffolding protein β-arrestin 1, although the mechanism behind its involvement remains elusive. Here we uncover a novel partner and function for β-arrestin 1, namely, in binding to ARHGAP21 (also known as ARHGAP10), a known effector of RhoA activity, whose GTPase-activating protein (GAP) function it inhibits. Using yeast two-hybrid screening, a peptide array, in vitro binding studies, truncation analyses, and coimmunoprecipitation techniques, we show that β-arrestin 1 binds directly to ARHGAP21 in a region that transects the RhoA effector GAP domain. Moreover, we show that the level of a complex containing β-arrestin 1 and ARHGAP21 is dynamically increased following angiotensin stimulation and that the kinetics of this interaction modulates the temporal activation of RhoA. Using information gleaned from a peptide array, we developed a cell-permeant peptide that serves to inhibit the interaction of these proteins. Using this peptide, we demonstrate that disruption of the β-arrestin 1/ARHGAP21 complex results in a more active ARHGAP21, leading to less-efficient signaling via the angiotensin II type 1A receptor and, thereby, attenuation of stimulated stress fiber formation.
The Rho-GTPases are a subfamily of the Ras superfamily of GTP-hydrolyzing enzymes that have a unique role in the spatial and temporal control of many vital cellular processes. They act as binary switches to toggle between the activated GTP-bound state of the protein and the inactive GDP conformation, depending on the application of a variety of extracellular stimuli (19) Active Rho proteins interact with a number of effectors to transduce specific signals, which modulate a number of important biological functions, such as reorganization of the actin cytoskeleton, membrane trafficking, and cell cycle regulation (19, 37). Critical to the Rho signal transduction mechanism is the ability to switch between the active and inactive states. The correct balance is achieved by three classes of regulatory proteins: Rho guanine nucleotide exchange factors (GEFs), Rho guanine nucleotide dissociation inhibitors (GDIs), and Rho GTPase-activating proteins (GAPs) (19). Rho GAPs share a homologous region of sequence (Rho GAP domain) that allows binding of GTP-bound Rho proteins to accelerate their GTPase activity and to promote hydrolysis of the bound GTP to GDP. Rho GAP domains are unique to Rho subfamily members and consequently cannot turn off other classes of GTPases (19).
One group of clinically important Rho GAPs, which consists of over 30 members, is the ARHGAP family (22). Genetic mutation of the genes that encode these proteins invariably results in carcinogenesis via aberrant signaling through Rho-GTPases such as Rho, Rac, and Cdc42 (9, 21). One member of the ARHGAP family that has received much attention of late is ARHGAP21 (also known as ARHGAP10) (5). ARHGAP21 contains an N-terminal PDZ domain, a central pleckstrin homology (PH) domain, and a C-terminally located Rho GAP domain. ARHGAP21 is known to locate to the Golgi apparatus via direct association with ADP-ribosylation factor 1 (ARF1), where it can act as a GAP for Cdc42, RhoA, and Rac1 (13, 29). ARF1 can also recruit ARHGAP21 to the plasma membrane, where it can modulate cell surface Cdc42 dynamics to regulate endocytosis (24). Most recently, ARHGAP21 has been shown to act as a dynamic molecular scaffold for focal adhesion kinase (FAK) and protein kinase C (PKC) during cardiac hypertrophy (7). Translocation of ARHGAP21 to the Z-lines of cardiac myocytes during hypertrophic stress appears to regulate focal adhesion turnover and cytoskeleton assembly as part of the cellular adaptive response.
β-Arrestins are cytosolic proteins that can be recruited to interact with various activated G-protein-coupled receptors at the plasma membrane, enabling receptor desensitization and downstream signaling (12). However, an increasing body of evidence suggests that β-arrestins can also act as platforms to orchestrate proteins involved in cytoskeletal reorganization (reviewed in reference 11) and other processes involving small GTPases such as ARF6 (10). Indeed, β-arrestin 1 has been shown to be crucial in the activation of RhoA following ligation of the angiotensin II type 1A receptor (4). Robust RhoA activation coordinated by β-arrestin 1 and Gαq was sufficient to trigger downstream stimulation of Rho-associated kinase (ROCK) and to induce stress fiber formation. Although the activation of Rho signaling via the angiotensin II type 1A receptor has been investigated extensively (17, 18, 32, 34), the mechanism behind the involvement of β-arrestin 1 has not been elucidated. Here we show for the first time that β-arrestin 1 interacts directly with ARHGAP21 by binding within its Rho GAP domain. We show that a β-arrestin 1/ARHGAP21 complex is formed in cells and that, in response to stimulation of the angiotensin II type 1A receptor, this complex facilitates RhoA activation and stress fiber formation.
MATERIALS AND METHODS
Materials.
The Bradford reagent was from Bio-Rad. Alexa Fluor 488 phalloidin, the Lipofectamine 2000 transfection reagent, sodium pyruvate-free Dulbecco's modified Eagle medium (DMEM), and Opti-MEM I reduced-serum medium were from Invitrogen. Antibodies against β-arrestin 1 and β-arrestin 2 (sc-53780 and sc-13140) and against RhoA (sc-418) were purchased from Santa Cruz. The antibody against ARHGAP6 (WH0000395M1) was from Sigma-Aldrich; that against ArfGAP3 (ab85971) was from Abcam; those against P44/42 mitogen-activated protein kinase (MAPK) (extracellular signal-related kinases 1 and 2 [ERK 1/2]) and total ERK 1/2 (9106 and 9126) were from Cell Signaling. The antibodies against PDE4D have been published previously (6). Small interfering RNA (siRNA) against ARHGAP21 and control siRNA have been described previously (14). All other biochemicals were sourced from Sigma. A stock solution of angiotensin II was prepared in sterile deionized water. The β-arrestin-1/Arhgap21 disruptor peptide and the control peptide were prepared in dimethyl sulfoxide (DMSO). In order to detect and immunopurify ARHGAP21, we developed our own antibody against a peptide derived from the ARHGAP21 sequence from residues 521 to 540 as previously described (14).
Cell culture.
HEK 293 cells stably expressing the myc-tagged angiotensin II type 1A receptor (At1AR HEK 293 cells) were cultured in sodium pyruvate-free DMEM supplemented with 10% fetal bovine serum and the antibiotics hygromicin B and blasticidin S. Cells were plated out either in 6-well plates at ∼60% confluence for xCELLigence work or on round coverslips in 6-well plates at ∼40% confluence for immunofluorescence work. Cells were pretreated with 2 μg/ml doxycycline for 48 h to induce AT1AR expression prior to further experimental procedures.
Cdc42 activation assays.
The activation of Cdc42 was determined using the G-LISA Cdc42 Activation Assay Biochem kit (colorimetric format) from Cytoskeleton (Denver, CO) according to the manufacturer's instructions.
GST-Rhotekin pulldown assay.
Glutathione S-transferase (GST)-Rhotekin beads were prepared according to the manufacturer's instructions (Upstate Cell Signaling, United Kingdom). Following stimulation, cells were lysed in 1× ice-cold Mg2+ lysis buffer (125 mM HEPES [pH 7.5], 750 mM NaCl, 5% Igepal CA-630, 50 mM MgCl2, 5 mM EDTA, 10% glycerol, 10 μg/ml aprotonin, and 10 μg/ml leupeptin) (Upstate Cell Signaling, United Kingdom). Lysed cells were then incubated on ice for ∼10 min, after which they were scraped into prechilled 1.5-ml microcentrifuge tubes. Lysates were centrifuged at 15,800 × g for 5 min. A volume of lysate containing ∼65 to 80 μg of protein was then pipetted into a fresh, prechilled 1.5-ml microcentrifuge tube and was mixed with 30 μg of GST-Rhotekin beads in a final volume of ∼300 μl. Lysates with beads were allowed to rotate for 1 h at 4°C before the beads were washed 3 times with 0.5 ml of 1× Mg2+ lysis buffer. Following the last wash, the majority of the supernatant was removed by pipette, and the beads were aspirated to dryness with a flat gel-loading tip (Marsh Biomedical, Rochester, NY).
GST pulldown assays.
Appropriate amounts of GST fusion proteins were mixed with a 100-μl slurry of 50% (vol/vol) phosphate-buffered saline (PBS)-washed glutathione resin (Amersham Biosciences) for 2 h at 4°C. The beads were pelleted by centrifugation and were washed twice with PBS containing 1% Triton X-100 before the addition of 20 μg of the His-tagged β-arrestin 1 fusion protein in a 1-ml solution of PBS and Triton X-100 containing 5 mM dithiothreitol. After a 2-h incubation at 4°C, the beads were collected, washed three times with a PBS-Triton X-100 solution, and eluted in 100 μl of 2× Laemmli buffer. Eluates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the GST fusion proteins and His-tagged β-arrestin 1 were detected by immunoblotting.
xCELLigence RTCA.
For xCELLigence real-time cell assays (RTCA), the E-Plate 96 (Roche) was used (38). This comprises a 96-well plate with integral sensor electrode arrays to measure cell impedance. The xCELLigence system was used according to the instructions of the supplier (Roche Applied Science). The core of the xCELLigence system is the E-plate 96: this is a single-use, disposable device used for performing cell-based assays on the RTCA SP instrument. The E-plate 96 is a 96-well microtiter plate with incorporated gold cell sensor arrays in the bottom, which allow the impedance inside each well to be monitored and assayed in real time. The changes in impedance correlate with cell shape changes (short term) or the growth (long term) of the cells in each well (2, 23, 31, 43). The E-plate 96 has a low-evaporation lid design. The diameter of each well is 5.0 mm ± 0.05 mm, with a total volume of 243 ± 5 μl, and approximately 80% of the bottom area of each well is covered by circle-on-line electrodes, which are designed to be used in an environment of +15 to +40°C, with a maximum relative humidity of 98%, without condensation (38). The voltage applied to the electrodes during RTCA measurement is about 20 mV. The impedance measured between electrodes in an individual well depends on electrode geometry, the ion concentration in the well, and whether or not cells are attached to the electrodes (38). In the absence of cells, electrode impedance is determined mainly by the ion environment both at the electrode-solution interface and in the bulk solution. In the presence of cells, cells attached to the electrode sensor surfaces will act as insulators and thereby alter the local ion environment at the electrode-solution interface, leading to an increase in impedance (43). Thus, the more cells are growing on the electrodes, the larger the electrode impedance value. Small changes in impedance can also be recorded when cells temporally change shape in response to agonists (43). The RTCA-associated software allows users to obtain parameters such as the average value, maximum and minimum values, standard deviation (SD), half-maximum effective concentration (EC50), half-maximum inhibitory concentration (IC50), and cell index (CI) (38). Fifty microliters of pyruvate-free DMEM supplemented with 2 μg/ml doxycycline was added to each well for calibration of the plate. For the β-arrestin 1/ARHGAP21 disruptor peptide experiments, the wells were seeded with 2,500 AT1AR HEK 293 cells/well, and the cell index was measured for 48 h to ensure linear growth of the cells, while the cells were also pretreated with doxycycline to ensure AT1AR expression. Following the 48 h of incubation, cells in appropriate wells either were left untreated (negative control) or were treated with either 10 μM β-arrestin 1/ARHGAP21 disruptor peptide or 10 μM control peptide for 2 h. This was followed by treatment with 40 nM angiotensin II. These cells were then counted and added to the E-Plate 96, and the cell index was measured for 48 h before the cells were treated with 40 nM angiotensin II. Cell index measurements were normalized to that at the start of angiotensin II treatment, and data were analyzed at the 30-min time point following angiotensin II treatment.
Microscopy.
AT1AR HEK 293 cells grown on glass coverslips in 6-well plates treated with disruptor peptides or siRNA knockdown of ARHGAP21 were incubated with 40 nM angiotensin II for 30 min. Cells were washed 3 times with PBS and were fixed in 4% paraformaldehyde for 10 min before being washed again with PBS. Cells were permeabilized in 0.2% Triton X-100 in Tris-buffered saline (TBS) (10 mM Tris [pH 7.5], 150 mM NaCl) for 15 min. Cells were washed as before and were incubated in Alex Fluor 488 phalloidin for F-actin staining for 20 min according to the manufacturer's instructions. After a wash, cells were mounted on glass microscope slides with Prolong Gold antifade reagent and 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Cells were visualized using the Zeiss Pascal laser-scanning confocal microscope. For visual quantification of stress fiber formation, slides were examined using a Zeiss Axiovision fluorescent imaging microscope at a magnification of ×100. Images of 100 random fields were taken from each category of cell treatments and were assigned by persons other than the person who performed the experiment to one of two categories: cells either showing stress fiber formation or not following angiotensin II treatment.
Statistical analysis.
Statistical analysis of quantified data was performed using GraphPad Prism (version 4.0) software. One-way analysis of variance (ANOVA) with Tukey's multiple-comparison posttest analysis was used, and a P value of <0.05 was taken as indicating a statistically significant difference.
Yeast 2-hybrid screen.
The yeast 2-hybrid screen was performed by Hybrigenics (Paris, France) using full-length β-arrestin 1 (residues 1 to 419) fused to LexA at the C terminus as bait to screen the human brain RP1 library. Two-hybrid screens were performed using a cell-to-cell mating protocol (25). 5′ and 3′ sequences were determined for all positive clones, and the latest release of GenBank was searched for these sequences using BLASTN (1).
Peptide arrays and alanine scans.
Peptide libraries covering the entire sequences of both β-arrestin 1 and ARHGAP21 were produced by automatic SPOT synthesis and were probed as described previously by us (3, 6, 26, 27, 30). They were synthesized on continuous cellulose membrane supports on Whatman 50 cellulose membranes using Fmoc (9-fluorenylmethyloxycarbonyl) chemistry with the AutoSpot-Robot ASS 222 peptide synthesizer (Intavis Bioanalytical Instruments AG, Cologne, Germany). Alanine-scanning peptide libraries were constructed by taking the residues in positive spots and sequentially changing each residue to alanine (or, if alanine was the natural amino acid at that position, to aspartate). The interaction of spotted peptides with purified recombinant GST and with GST-β-arrestin 1 and GST-ARHGAP21 fusion proteins was determined by overlaying the cellulose membranes with 10 μg/ml recombinant protein. Bound recombinant proteins were then detected following wash steps with rabbit anti-GST, and detection was performed with a horseradish peroxidase-coupled anti-rabbit secondary antibody.
Expression of GST fusions in Escherichia coli.
Cultures of E. coli JM109 containing pGEX-β-arrestin 1, pGEX, and PGEX-ARHGAP21 truncation constructs were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (Roche Molecular Biochemicals) for 4 h at 30°C. Bacteria were harvested by centrifugation at 6,000 × g for 15 min at 4°C, and the bacterial pellet was frozen at −80°C overnight. The bacterial pellets were resuspended in 10 ml of ice-cold resuspension buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and Complete protease inhibitor mixture) and were sonicated with 4 30-s bursts at the maximal setting. Triton X-100 was added to a final concentration of 0.02%, and cell debris was then removed by centrifugation at 15,000 × g for 10 min at 4°C. The cleared supernatant was incubated with 1/10 volume of preequilibrated glutathione-Sepharose beads on an orbital shaker for 30 min at 4°C. The beads were collected by centrifugation at 13,000 × g for 1 min and were washed three times with ice-cold resuspension buffer. The fusion proteins were eluted by the addition of 5 mM glutathione-50 mM Tris-HCl (pH 8.0) and were dialyzed three times against 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 5% glycerol. The purified fusion proteins were stored at −80°C until required.
RESULTS
ARHGA21 is a binding partner for β-arrestin 1.
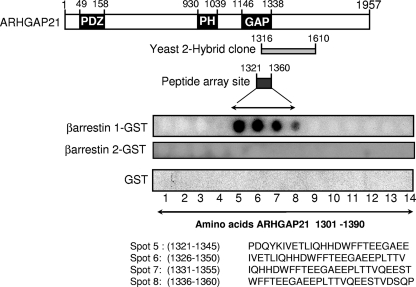
Using a yeast 2-hybrid approach with human β-arrestin 1 to probe a human brain RP1 library, ARHGAP21 was identified as a partner of β-arrestin 1 (Hybrigenics). Reliable interactions were filtered based on a predicted biological score, which is a computed technical parameter that gives the level of confidence in the interaction detected. All of the ARHGAP21 clones identified as interactors (25 in total) achieved the manufacturer's highest confidence rating, “A” (Hybrigenics). ARHGAP21 is a very large protein of 1,957 amino acid residues, containing an N-terminal PDZ domain, a central pleckstrin homology (PH) domain, and a C-terminal RhoGAP domain, which has a role in the regulation of actin assembly (14, 29) Because ARHGAP21 is such an extremely large protein, it is unlikely that full-length species will be found in libraries. Indeed, all the positive clones isolated encoded fragments that contained regions of ARHGAP21 sequence spanning amino acids 1316 to 1610 with 100% homology (Fig. 1).
FIG. 1.
Association of ARHGAP21 and β-arrestin 1 by yeast 2-hybrid and peptide array analyses. (Top) Schematic diagram showing the functionally relevant areas of ARHGAP21. Overlapping sequences within ARHGAP21 that were shown to interact with β-arrestin 1 by yeast 2-hybrid analysis (shaded rectangle) and peptide array (filled rectangle) are indicated. (Bottom) Immobilized peptide spots of overlapping 25-mer peptides, each shifted along by 5 amino acids in the entire sequence of ARHGAP21, probed for interaction with either β-arrestin 1-GST, β-arrestin 2-GST, or GST alone. Positive interactions were detected in amino acid sequences spanning positions 1321 to 1360 of ARHGAP21.
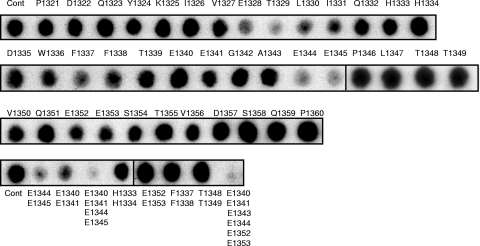
As an independent means of verifying the direct interaction of β-arrestin 1 within this region of ARHGAP21, we used peptide array technology. This can provide a novel and powerful method for investigating the molecular nature of protein-protein interactions in a cell-free system. Indeed, we have used this technology to considerable advantage to exhaustively map sites of interaction between each of the scaffold proteins, β-arrestin and RACK1, and the cyclic AMP (cAMP) hydrolyzing phosphodiesterase, PDE4D5 (3, 6). In the study we report here, we generated a library of overlapping peptides (25-mers), each shifted by 5 amino acids, which spans the entire 1,957-amino-acid sequence of ARHGAP21. These were spot-synthesized on cellulose membranes to generate an immobilized peptide library that was then probed with a purified recombinant GST fusion protein of β-arrestin 1 or β-arrestin 2. The binding of β-arrestin 1 to individual peptide spots was assessed immunologically, with positive interactions identified as dark spots (Fig. 1). In agreement with the yeast 2-hybrid analysis, a cluster of peptides (spots 5 to 8) covering ARHGAP21 amino acids 1321 to 1360 was observed for GST-β-arrestin 1 binding but not with GST alone or β-arrestin 2 when used as a probe. This sequence was subjected to further analysis by screening these parent peptides with single substitutions (mutations to alanine) of successive amino acids to generate a family of “mutant” peptide progeny from the parent species (Fig. 2). From such experiments, we observed that β-arrestin 1 binding was attenuated upon alanine substitution within two regions of ARHGAP21. One is a region containing E1328, T1329, L1330, and I1331, and the other region contains E1344 and E1345, where the substitution of alanine for each of these residues compromised β-arrestin 1 binding. Additionally, the double substitution of alanine for E1344 and E1345 also attenuated the interaction of the 25-mer peptide, which encompassed these residues, with β-arrestin 1 (Fig. 2). These regions are contained either within or adjacent to the sequence identified as the Rho-GAP domain of ARHGAP21 (amino acids 1146 to 1338) (29). This suggests that the direct interaction with β-arrestin 1 but not with β-arrestin 2 may alter the GAP function of ARHGAP21.
FIG. 2.
Alanine scanning arrays of the β-arrestin 1 binding site on ARHGAP21. The alanine substitution array for the P1321-to-P1360 peptide (Cont) (top leftmost spot) from ARHGAP21was probed with β-arrestin 1-GST. A1343 was replaced with an aspartate residue. Multiple substitutions were also undertaken as indicated (bottom).
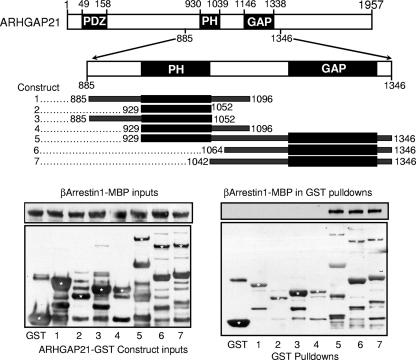
The 2-hybrid and peptide array analyses provide strong evidence for a direct interaction between β-arrestin 1 and ARHGAP21. Although we can generate purified recombinant β-arrestin 1, the size of ARHGAP21 militates against the cloning and expression of a full-length recombinant species, as discussed by others previously (14). Others, however, have generated a library of truncated ARHGAP21 proteins fused to GST (14), of which we have taken advantage to probe for direct interaction with the purified β-arrestin 1-maltose-binding protein (MBP) fusion protein (Fig. 3). Pulled down GST fusion proteins were then probed for coimmunoprecipitating β-arrestin 1-MBP by Western blotting (Fig. 3). β-Arrestin 1-MBP was shown to associate with all GST-ARHGAP21 constructs containing the Rho-GAP domain but not with GST alone or with ARHGAP21 constructs devoid of the Rho-GAP domain. In agreement with the data from both the yeast 2-hybrid and peptide array analyses, it would appear that β-arrestin 1 binds directly to the Rho-GAP domain of ARHGAP21.
FIG. 3.
β-Arrestin 1 binds to the GAP domain of ARHGAP21. (Top) Purified GST fusions with the indicated regions of the sequence of ARHGAP21 (amino acids 885 to 1346) and GST alone were mixed with purified MBP fusions of β-arrestin 1. (Bottom) Inputs of purified β-arrestin 1-MBP and ARHGAP21-GST constructs (left) and pulled down GST fusion proteins (right) were blotted for the presence of β-arrestin 1-MBP. Asterisks indicate bands that correspond either to an ARHGAP21-GST construct (constructs 1 to 7) or to GST alone (first lane).
Dynamic interaction between β-arrestin 1 and ARHGAP21 in At1AR HEK 293 cells.
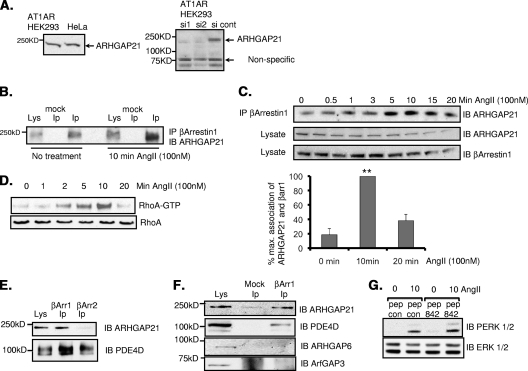
There is growing evidence for a role of β-arrestins in facilitating small-GTPase-mediated events (11). Indeed, it has been shown that β-arrestin 1 is required to activate the small GTPase RhoA, leading to the reorganization of stress fibers following the activation of the angiotensin II type 1A receptor in human embryonic kidney 293 (HEK 293) cells stably expressing the AT1AR (AT1AR HEK 293 cells) (4). Using these cells, as well as lysates from HeLa cells, we detected the expression of endogenous ARHGAP21 as a single 250-kDa species (Fig. 4A, left), consistent with the size of endogenous ARHGAP21 detected previously by others in HeLa, MCF-7 (29), and Caco2 (13) cells. Our antibody was shown to interact specifically with ARHGAP21, since siRNA constructs that had previously been shown to be effective in silencing ARHGAP21 (13) drastically depleted immunoreactivity (Fig. 4A, right).
FIG. 4.
Dynamic interaction between β-arrestin 1 and ARHGAP21 in AT1AR HEK 293 cells. (A) (Left) Cell lysates from AT1AR HEK 293 cells and HeLa cells were blotted for ARHGAP21. (Right) The specificity of the antibody was verified using siRNA silencing of ARHGAP21. cont, control. (B) Immunoprecipitates (IP) of β-arrestin 1 from AT1AR HEK 293 cells were blotted (IB) for ARHGAP21 before or after treatment with angiotensin II (AngII) (100 nM). (C) (Top) Immunoprecipitates of β-arrestin 1 from AT1AR HEK 293 cells were blotted for ARHGAP21 following a time course of treatment with angiotensin II (100 nM). (Center panels) Equal amounts of ARHGAP21 and β-arrestin 1 in inputs. (Bottom) Bar chart showing quantifications of the upper trace. max., maximum; βarr1, β-arrestin 1. (D) RhoA activation in AT1AR HEK 293 cells as measured by a GST-Rhotekin pulldown assay following a time course of angiotensin II treatment. (E) Immunoprecipitates of β-arrestin 1 or β-arrestin 2 from AT1AR HEK 293 cells were blotted for ARHGAP21 and PDE4D. (F) Immunoprecipitates of β-arrestin 1 from AT1AR HEK 293 cells were blotted for ARHGAP21, PDE4D, ARHGAP6, and ArfGAP3. (G) Determination of total and phospho-ERK 1 and 2 in lysates of AT1AR HEK 293 cells following angiotensin II treatment after pretreatment with peptide 842 (pep 842) or a control peptide (pep con). PERK, phospho-ERK.
The association of endogenously expressed ARHGAP21 and β-arrestin 1 was demonstrated in AT1AR HEK 293 cells by immunopurifying β-arrestin 1 and then blotting for ARHGAP21 (Fig. 4B). In doing this, we noted that more ARHGAP21 was immunopurified with β-arrestin 1 following treatment of these cells with angiotensin (100 nM) (Fig. 4B). This suggests that AT1AR stimulation can dynamically enhance the association between β-arrestin 1 and ARHGAP21. To further test this notion, we immunopurified β-arrestin 1 over an angiotensin treatment time course and probed the samples for ARHGAP21 (Fig. 4C). Indeed, by doing this, we see that the association of β-arrestin 1 with ARHGAP21 could be transiently increased in an agonist-dependent fashion. Moreover, the time course of increased association of the two proteins mirrored that of angiotensin II-mediated RhoA activation as measured by a GST-Rhotekin pulldown assay (Fig. 4D). These data suggest that increased complex formation between β-arrestin 1 and ARHGAP21 may facilitate the activation of RhoA in these cells consequent to angiotensin challenge. In this regard, we propose that the sequestration of β-arrestin 1 to the GAP domain of ARHGAP21 likely serves to inhibit its GAP function and thereby to facilitate RhoA activation. In agreement with our peptide array data, no association between ARHGAP21 and β-arrestin 2 could be detected (Fig. 4E). Since β-arrestin 1 interacts with the highly conserved GAP domain of ARHGAP21, we were interested in determining whether β-arrestin 1 could also bind to other proteins with Rho-type GAP domains or indeed non-Rho-type GAPs via their GAP domains. We were unable to detect any association of β-arrestin 1 with either ARHGAP6 or ArfGAP3, suggesting that there is some selectivity for ARHGAP21 (Fig. 4F). Additionally, ARHGAP21 was the only GAP detected in our yeast 2-hybrid screen using β-arrestin 1 as bait.
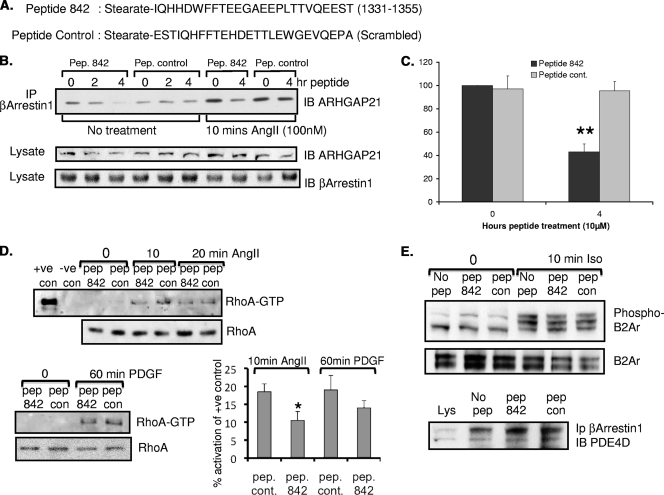
Disruption of the β-arrestin 1-ARHGAP21 complex promotes stress fiber formation and changes in cell shape following angiotensin challenge.
Our scanning peptide array approach coupled with 2-hybrid analyses identified the putative docking site for β-arrestin 1 as being located on the GAP domain of ARHGAP21 in a region spanning amino acids 1321 to 1360 (Fig. 1). Alanine-scanning analyses highlighted several key residues in this domain (E1328, T1329, L1330, I1331, E1344, and E1345) as being of potential importance in allowing for β-arrestin 1 association. Previous work from our laboratory has shown that cell-permeant analogues of 25-mer peptides, identified in this manner, can be used to disrupt signaling complexes within cells so as to affect specific functional outputs such as the phosphorylation of the β2-adrenergic receptor by PKA (36) and the phosphorylation of β-arrestin 1 by ERK (30). Here we utilized a sequence from ARHGAP21, encompassing residues 1331 to 1355 (Fig. 1, spot 7), implicated in β-arrestin 1 docking to generate a stearoylated, cell-permeant peptide (peptide 842) to be assessed as a potential disruptor of β-arrestin 1/ARHGAP21 complexes (Fig. 5A). We also synthesized a scrambled version of stearoylated peptide 842 with the same net weight and charge to provide a control (Fig. 5A). Both these peptides contained an N-terminal stearate group to confer passage into cells (30). Using these reagents, we showed that the test peptide, peptide 842, but not the control peptide attenuated the association between ARHGAP21 and β-arrestin 1 as determined by immunoprecipitation experiments from cell lysates (Fig. 5B). Maximum disruption of the complex, significantly (P < 0.01) reducing the ability of angiotensin challenge to promote the association of ARHGAP21 with β-arrestin 1, was observed after treatment of cells with peptide 842 for 4 h (Fig. 5B, 4 rightmost lanes, and C). In decreasing the association between ARHGAP21 and β-arrestin 1, peptide 842 also significantly attenuated the activation of RhoA following angiotensin treatment (Fig. 5D, top and bottom right) and decreased the activation of RhoA following treatment with platelet-derived growth factor (PDGF) (Fig. 5D, bottom left) but had no effect on angiotensin-triggered Cdc42 activation (data not shown) or ERK activation (Fig. 4G). Peptide 842 did not affect the association of β-arrestin 1 with another known partner, PDE4D5 (Fig. 5E, bottom), nor did it alter the ability of β-arrestin 1 to desensitize the β2-adrenergic receptor, as assayed using antibodies to PKA phosphosites on the receptor (Fig. 5E, top).
FIG. 5.
Peptide disruption of the β-arrestin 1/ARHGAP21 complex in HEK AT1AR cells. (A) Sequences of the cell-permeant ARHGAP21/β-arrestin 1 disruptor peptide (peptide 842) based on the sequence of ARHGAP21 (amino acids 1331 to 1355) and a scrambled control peptide. (B) (Top) Immunoprecipitates (IP) of β-arrestin 1 from AT1AR HEK 293 cells pretreated with peptide (Pep.) 842 or a peptide control for the indicated times were blotted for ARHGAP21 before or after treatment with angiotensin II (AngII) (100 nM). (Center and bottom) Equal amounts of ARHGAP21 and β-arrestin 1 in inputs. (C) Densitometric analysis of the data shown in the four rightmost lanes of panel B. Results of 3 experiments are shown. **, P < 0.01. (D) (Top) RhoA activation in AT1AR HEK 293 cells as measured by a GST-Rhotekin pulldown assay following a time course of angiotensin II (100 nM) treatment with and without pretreatment (2 h) with peptide 842 or a control (con) peptide. +ve, positive; −ve, negative. (Bottom left) RhoA activation in AT1AR HEK 293 cells as measured by a GST-Rhotekin pulldown assay following PDGF (20 ng/ml) treatment with or without pretreatment (2 h) with peptide 842 or a control peptide. (Bottom right) Quantification by densitometry of the data shown on the left. The results of 3 experiments are shown. *, P < 0.05. (E) (Top and center) Western blot analyses of HEKB2 293 cell lysates following isoprenaline (10 μM) treatment with or without pretreatment (2 h) with peptide 842 or a control peptide. Samples were blotted for the β2-adrenergic receptor (B2Ar) and the phospho-β2-adrenergic receptor (Ser345/346). (Bottom) Immunoprecipitates of β-arrestin 1 from AT1AR HEK 293 cells pretreated with peptide 842 or a peptide control were blotted for PDE4D.
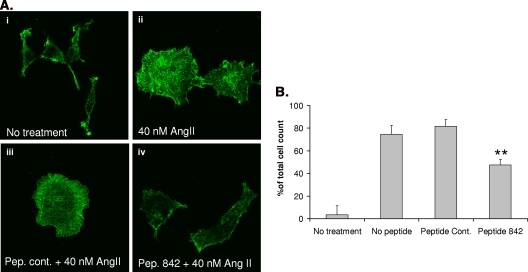
Having demonstrated that we can disrupt the ARHGAP/β-arrestin 1 complex and attenuate RhoA activation by treating cells with peptide 842 (Fig. 5), we set out to determine if this disrupting peptide might be able to attenuate downstream RhoA signaling induced by angiotensin stimulation of AT1AR HEK cells. We utilized two methods to address this: first, a traditional approach in which we visually assessed the formation of stress fibers in fixed cells and second, a novel approach employing xCELLigence technology, which measures impedance in living cells on the surfaces of cell culture plates. The latter technique provides a dynamic, noninvasive, label-free analysis in living cells of parameters such as cell shape changes (15) and cell growth (38, 41).
As previously observed (4), treatment of AT1AR HEK 293 cells with angiotensin II provoked robust F-actin reorganization and stress fiber formation in approximately 75% of cells (Fig. 6). Pretreatment of cells with the control peptide had no significant effect on stress fiber formation, with 81% ± 5% of cells showing actin reorganization upon angiotensin II stimulation. However, following treatment with peptide 842, significantly fewer cells (a total of 47.5% ± 5%; P < 0.01) produced stress fibers (Fig. 6), suggesting that the disruption of the ARHGAP21-β-arrestin 1 complex resulted in a more active ARHGAP21 and, consequently, a less active RhoA. Neither peptide 842 nor the control peptide induced stress fiber formation on its own (data not shown).
FIG. 6.
Disruption of the β-arrestin 1/ARHGAP21 complex affects angiotensin II (AngII)-stimulated stress fiber formation. (A) Visualization of angiotensin II-mediated stress fiber formation in AT1AR HEK 293 cells following pretreatment with peptide (Pep.) 842 or a peptide control (cont.). (B) Analysis of the percentage of total cells containing angiotensin II-mediated stress fibers following pretreatment with peptide 842 or a peptide control. Asterisks indicate a significant difference (P < 0.01) from the peptide control.
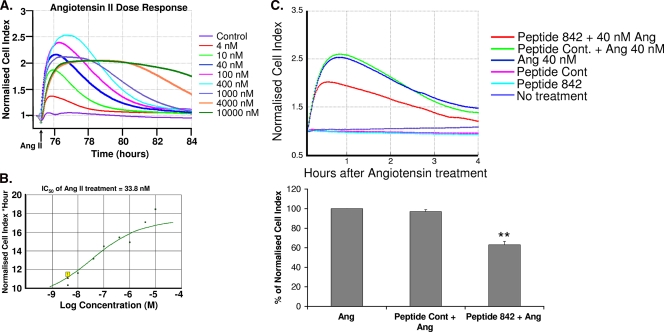
To confirm the role of the ARHGAP21-β-arrestin 1 complex in angiotensin-induced RhoA signaling, we utilized xCELLigence technology to monitor cell shape changes in real time. The xCELLigence system provides sensitive and functional monitoring of receptor-mediated signaling by recording short-term changes in impedance (31, 43). Since stress fibers are expected to have transient effects on cell shape, we set out to verify that xCELLigence technology could detect changes induced by angiotensin II signaling. We first carried out real-time dose-response experiments using a range of angiotensin concentrations (4 nM to 10 μM). The peak response time (Fig. 7A) following a 1-h angiotensin stimulation matched that seen for stress fiber formation (data not shown), and the IC50 (33 nM) (Fig. 7B) was similar to that previously recorded (10 nM) for the activation of RhoA by angiotensin II in these cells (4). In addition, the concentration of angiotensin that resulted in the maximal activation of RhoA in these cells (100 nM [4]) also closely matched what we observed here for angiotensin in causing the maximum change in short-term impedance as measured by increases in the cell index (Fig. 7A).
FIG. 7.
Disruption of the β-arrestin 1/ARHGAP21 complex affects AT1AR signaling. (A) Real-time detection of dose-response curves from AT1AR HEK 293 cells treated with angiotensin II as detected using xCELLigence technology. The arrow indicates the time of addition of angiotensin II (Ang II). (B) Determination of IC50s for the angiotensin II response in AT1AR HEK 293 cells using data from panel A. (C) Evaluation of the effects of pretreatment with peptide 842 or a peptide control (Cont.) on the response to angiotensin II treatment (1 h) in AT1AR HEK 293 cells by using xCELLigence technology. Asterisks indicate a significant difference (P < 0.01) from the value for the peptide control.
Since the real-time cell electronic sensing system could effectively monitor RhoA-mediated responses to angiotensin II, we utilized it to determine if disruption of the ARHGAP21-β-arrestin 1 complex would affect RhoA signaling in real time (Fig. 7C). Wells that were not treated with angiotensin failed to produce any response (in terms of an increase in the cell index), whereas control wells (no peptide) and those pretreated with the peptide control produced identical responses to angiotensin with respect to timing and amplitude (Fig. 7C). In contrast, the cell index for wells pretreated with peptide 842 (Fig. 7C, red line) peaked earlier, with significantly lower (P < 0.01) increases. These data suggest that disruption of the ARHGAP21/β-arrestin 1 complex attenuates angiotensin-induced RhoA signaling by releasing the inhibition conferred by β-arrestin 1 on the ARHGAP21 RhoGAP domain.
DISCUSSION
β-Arrestins are multifunctional scaffolds that were originally characterized for their role in desensitizing seven-transmembrane receptors (7TMRs) but are now known to be versatile adaptors that transduce signals via multiple effector pathways (11, 35). Signals directed by distinct 7TMRs can have different functional outputs depending on whether the signal is relayed by the G protein associated with the receptor or by β-arrestin proteins themselves (40). Such a system allows for “biased” signaling whereby novel agonists of certain 7TMRs can promote signaling via only the G protein or β-arrestins (33). An early example of biased ligand signaling was demonstrated for the angiotensin II type 1A receptor, where SII angiotensin failed to activate Gαq but could still drive ERK MAP kinase responses in a β-arrestin 2-dependent manner (42). Subsequent investigation of the angiotensin II type 1A receptor demonstrated that activation of RhoA, as well as subsequent actin reorganization, required β-arrestin 1 but not β-arrestin 2 expression following angiotensin II treatment (4). More-recent work also suggests that both β-arrestins 1 and 2 are required for membrane blebbing resulting from the coupling of angiotensin to RhoA (16). Clearly, different isoforms of β-arrestins can initiate the transduction of discrete signals via distinct pathways following the activation of a single receptor type. However, until now, the mechanism by which β-arrestin 1 activates RhoA following ligation of the angiotensin II type 1A receptor has remained unknown. RhoA activation can result from the activation of a RhoA exchange factor (GEF) and/or the inhibition of a Rho GTPase-activating protein (GAP). Here we demonstrate, for the first time, that the molecular mechanism underpinning such RhoA activation is likely to reflect the direct association of β-arrestin 1 with a recently discovered and as yet poorly appreciated GAP for RhoA, namely, ARHGAP21 (13, 29).
We first uncovered the possibility that ARHGAP21 might provide a partner for β-arrestin 1 by using a 2-hybrid approach where all ARHGAP21 sequences in the 2-hybrid clones that encoded species positively interacting with β-arrestin 1 were shown to include the GAP domain of ARHGAP21 (Fig. 1). This suggested to us that the β-arrestin 1 interaction site might well lie within the GAP domain of ARHGAP21 (Fig. 1). A direct association between ARHGAP21 and β-arrestin 1 was verified by peptide array mapping and pulldown experiments utilizing fragments of recombinant ARHGAP21 and full-length recombinant β-arrestin 1 that were expressed in E. coli (Fig. 2, 3, and 4). The interaction seems to be β-arrestin 1 specific, since we could find no evidence to suggest that β-arrestin 2 could bind in the same manner (Fig. 1 and 4). We presume that the interaction of β-arrestin 1 with ARHGAP21 inhibits its GAP activity, permitting a more robust activation of RhoA (Fig. 5D). In many instances, the activation of RhoA, in response to extracellular stimuli, is regulated by the activation of Rho GEFs (19). Here, however, we highlight a novel mechanism of Rho activation whereby the intrinsic GTPase activity of Rho is increased by the inhibition of a Rho GAP. Previous studies have shown that even in the absence of receptor stimulation, pharmacological inhibition of RhoGAP activity can be sufficient to promote Rho-driven biological responses (39). A recent report investigating angiotensin II responses in vascular smooth muscle cells demonstrated that another RhoGAP, p190A, was dephosphorylated and inactivated following stimulation of AT1AR. The transient p190A phosphorylation was responsible for early RhoA activation, suggesting that posttranslational modification of Rho GAPs may play a role in the modulation of RhoA activity. In contrast, our study shows that angiotensin II, by increasing the levels of a RhoGAP and β-arrestin 1 complex (Fig. 4), temporally controls RhoA activation and represents the first instance of modulation of a RhoGAP in such a way. The mechanistic basis of this increase, however, remains to be determined, since β-arrestin 1 provides an extremely complex and challenging system to dissect. Thus, β-arrestin 1 can potentially bind a very large number of partner proteins, some of which can be expected to sterically interdict the binding of others. This indicates that there will be subpopulations of β-arrestin 1 with different cohorts of partner proteins. Thus, an angiotensin-mediated increase in the cohort binding ARHGAP21 might result from conformational changes occurring upon the recruitment of β-arrestin 1 to the AT1AR that lead either to enhanced affinity for ARHGAP21 or to the loss of other partner proteins. However, the increase in the binding of β-arrestin 1 to ARHGAP21 could result from any angiotensin-triggered modification of either β-arrestin 1 or ARHGAP21, by phosphorylation or ubiquitination, for example.
Signaling through RhoA has been increasingly investigated as a possible route for the development of new therapeutic agents directed at a number of diseases, such as neurological disorders and cancer (28). Inhibitor compounds have been described for Rho kinase (ROCK) (8), an important downstream effector of RhoA, and peptide-derived ligands that target the p75 neurotrophin receptor upstream of RhoA have also been developed to modulate RhoA/ROCK activity (20). To date, however, there has been a lack of compounds directed at either RhoGEFs or RhoGAPs (28). We describe here, for the first time, a novel peptide agent that allows for the modulation of a RhoA-driven cellular response (Fig. 5D). Using information from our peptide array mapping approach, we have developed a cell-permeant peptide, based on the presumed β-arrestin 1 docking region of ARHGAP21 (Fig. 5), that effectively disrupts the ARHGAP-β-arrestin 1 complex to rescue RhoGAP activity, which controls RhoA activity resulting from either angiotensin or PDGF treatment (Fig. 5D). Using traditional methods to visualize stress fiber formation (Fig. 6) and novel xCELLigence technology to monitor the response to angiotensin II (Fig. 7), we show that our peptide disruptor acts as a RhoGAP activator by concomitantly dampening angiotensin II signaling via the AT1AR and attenuating stress fiber formation. Intriguingly, our peptide acts against a defined fraction of β-arrestin 1 that exists in complex with ARHGAP21, so that the disruptor provides a highly specific tool with which to dissect the role of β-arrestin 1 in the signaling involved following angiotensin II stimulation of the AT1AR. Our study uncovers a new molecular function for β-arrestin 1 and suggests that inhibitors of protein-protein interactions may represent a novel strategy for the development of specific pharmaceutical modulators of Rho GTPase signaling for therapeutic intervention in cancer and neurological disorders.
Acknowledgments
This work was supported by grants from the Medical Research Council (United Kingdom; G0600765) and Fondation Leducq (06CVD02).
We thank Philippe Chavrier, Institut Curie, CNRS, Paris, France, for the GST-ARHGAP21 constructs.
Footnotes
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atienza, J. M., et al. 2006. Dynamic and label-free cell-based assays using the real-time cell electronic sensing system. Assay Drug Dev. Technol. 4:597-607. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, G. S., et al. 2007. Mapping binding sites for the PDE4D5 cAMP-specific phosphodiesterase to the N- and C-domains of β-arrestin using spot-immobilized peptide arrays. Biochem. J. 404:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, W. G., et al. 2005. β-Arrestin 1 and Gαq/11 coordinately activate RhoA and stress fiber formation following receptor stimulation. J. Biol. Chem. 280:8041-8050. [DOI] [PubMed] [Google Scholar]
- 5.Bassères, D. S., E. V. Tizzei, A. A. Duarte, F. F. Costa, and S. T. Saad. 2002. ARHGAP10, a novel human gene coding for a potentially cytoskeletal Rho-GTPase activating protein. Biochem. Biophys. Res. Commun. 294:579-585. [DOI] [PubMed] [Google Scholar]
- 6.Bolger, G. B., et al. 2006. Scanning peptide array analyses identify overlapping binding sites for the signalling scaffold proteins, β-arrestin and RACK1, in cAMP-specific phosphodiesterase PDE4D5. Biochem. J. 398:23-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borges, L., et al. 2008. ARHGAP21 associates with FAK and PKCζ and is redistributed after cardiac pressure overload. Biochem. Biophys. Res. Commun. 374:641-646. [DOI] [PubMed] [Google Scholar]
- 8.Burthem, J., et al. 2007. The rho-kinase inhibitors Y-27632 and fasudil act synergistically with imatinib to inhibit the expansion of ex vivo CD34+ CML progenitor cells. Leukemia 21:1708-1714. [DOI] [PubMed] [Google Scholar]
- 9.Carles, A., et al. 2006. Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display. Oncogene 25:1821-1831. [DOI] [PubMed] [Google Scholar]
- 10.Claing, A., et al. 2001. β-Arrestin-mediated ADP-ribosylation factor 6 activation and β2-adrenergic receptor endocytosis. J. Biol. Chem. 276:42509-42513. [DOI] [PubMed] [Google Scholar]
- 11.DeFea, K. A. 2007. Stop that cell! β-Arrestin-dependent chemotaxis: a tale of localized actin assembly and receptor desensitization. Annu. Rev. Physiol. 69:535-560. [DOI] [PubMed] [Google Scholar]
- 12.DeWire, S. M., S. Ahn, R. J. Lefkowitz, and S. K. Shenoy. 2007. β-Arrestins and cell signaling. Annu. Rev. Physiol. 69:483-510. [DOI] [PubMed] [Google Scholar]
- 13.Dubois, T., and P. Chavrier. 2005. ARHGAP10, a novel RhoGAP at the cross-road between ARF1 and Cdc42 pathways, regulates Arp2/3 complex and actin dynamics on Golgi membranes. Med. Sci. (Paris) 21:692-694. (In French.) [DOI] [PubMed] [Google Scholar]
- 14.Dubois, T., et al. 2005. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat. Cell Biol. 7:353-364. [DOI] [PubMed] [Google Scholar]
- 15.Ehlers, A., S. Stempin, R. Al-Hamwi, and A. Lampen. 2010. Embryotoxic effects of the marine biotoxin okadaic acid on murine embryonic stem cells. Toxicon 55:855-863. [DOI] [PubMed] [Google Scholar]
- 16.Godin, C. M., and S. S. Ferguson. 2010. The angiotensin II type 1 receptor induces membrane blebbing by coupling to Rho A, Rho kinase, and myosin light chain kinase. Mol. Pharmacol. 77:903-911. [DOI] [PubMed] [Google Scholar]
- 17.Higashi, M., et al. 2003. Long-term inhibition of Rho-kinase suppresses angiotensin II-induced cardiovascular hypertrophy in rats in vivo: effect on endothelial NAD(P)H oxidase system. Circ. Res. 93:767-775. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, H. H., et al. 2008. Mechanisms of angiotensin II signaling on cytoskeleton of podocytes. J. Mol. Med. 86:1379-1394. [DOI] [PubMed] [Google Scholar]
- 19.Jaffe, A. B., and A. Hall. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247-269. [DOI] [PubMed] [Google Scholar]
- 20.James, S. E., et al. 2008. Anti-cancer drug induced neurotoxicity and identification of Rho pathway signaling modulators as potential neuroprotectants. Neurotoxicology 29:605-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh, M., and M. Katoh. 2004. Characterization of human ARHGAP10 gene in silico. Int. J. Oncol. 25:1201-1206. [PubMed] [Google Scholar]
- 22.Katoh, M., and M. Katoh. 2003. FNBP2 gene on human chromosome 1q32.1 encodes ARHGAP family protein with FCH, FBH, RhoGAP and SH3 domains. Int. J. Mol. Med. 11:791-797. [PubMed] [Google Scholar]
- 23.Kirstein, S. L., et al. 2006. Live cell quality control and utility of real-time cell electronic sensing for assay development. Assay Drug Dev. Technol. 4:545-553. [DOI] [PubMed] [Google Scholar]
- 24.Kumari, S., and S. Mayor. 2008. ARF1 is directly involved in dynamin-independent endocytosis. Nat. Cell Biol. 10:30-41. [DOI] [PubMed] [Google Scholar]
- 25.Legrain, P., and L. Selig. 2000. Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett. 480:32-36. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., G. S. Baillie, and M. D. Houslay. 2009. Mdm2 directs the ubiquitination of β-arrestin-sequestered cAMP phosphodiesterase-4D5. J. Biol. Chem. 284:16170-16182. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Li, X., et al. 2009. A scanning peptide array approach uncovers association sites within the JNK/βarrestin signalling complex. FEBS Lett. 583:3310-3316. [DOI] [PubMed] [Google Scholar]
- 28.Lu, Q., F. M. Longo, H. Zhou, S. M. Massa, and Y. H. Chen. 2009. Signaling through Rho GTPase pathway as viable drug target. Curr. Med. Chem. 16:1355-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ménétrey, J., et al. 2007. Structural basis for ARF1-mediated recruitment of ARHGAP21 to Golgi membranes. EMBO J. 26:1953-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng, D., et al. 2009. MEK1 binds directly to βarrestin1, influencing both its phosphorylation by ERK and the timing of its isoprenaline-stimulated internalization. J. Biol. Chem. 284:11425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meshki, J., et al. 2009. Neurokinin 1 receptor mediates membrane blebbing in HEK293 cells through a Rho/Rho-associated coiled-coil kinase-dependent mechanism. J. Biol. Chem. 284:9280-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, A., et al. 2003. Vessel- and vasoconstrictor-dependent role of rho/rho-kinase in renal microvascular tone. J. Vasc. Res. 40:244-251. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopal, S., K. Rajagopal, and R. J. Lefkowitz. 2010. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 9:373-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan, M. J., S. P. Didion, S. Mathur, F. M. Faraci, and C. D. Sigmund. 2004. Angiotensin II-induced vascular dysfunction is mediated by the AT1A receptor in mice. Hypertension 43:1074-1079. [DOI] [PubMed] [Google Scholar]
- 35.Shenoy, S. K., and R. J. Lefkowitz. 2005. Seven-transmembrane receptor signaling through β-arrestin. Sci. STKE 2005:cm10. [DOI] [PubMed] [Google Scholar]
- 36.Smith, K. J., et al. 2007. 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, β-arrestin and RACK1. Cell. Signal. 19:2612-2624. [DOI] [PubMed] [Google Scholar]
- 37.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 38.Urcan, E., et al. 2010. Real-time xCELLigence impedance analysis of the cytotoxicity of dental composite components on human gingival fibroblasts. Dent. Mater. 26:51-58. [DOI] [PubMed] [Google Scholar]
- 39.Vincent, S., and J. Settleman. 1999. Inhibition of RhoGAP activity is sufficient for the induction of Rho-mediated actin reorganization. Eur. J. Cell Biol. 78:539-548. [DOI] [PubMed] [Google Scholar]
- 40.Violin, J. D., and R. J. Lefkowitz. 2007. β-Arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol. Sci. 28:416-422. [DOI] [PubMed] [Google Scholar]
- 41.Vistejnova, L., et al. 2009. The comparison of impedance-based method of cell proliferation monitoring with commonly used metabolic-based techniques. Neuro Endocrinol. Lett. 30(Suppl. 1):121-127. [PubMed] [Google Scholar]
- 42.Wei, H., et al. 2003. Independent β-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc. Natl. Acad. Sci. U. S. A. 100:10782-10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, N., et al. 2006. Real-time monitoring of morphological changes in living cells by electronic cell sensor arrays: an approach to study G protein-coupled receptors. Anal. Chem. 78:35-43. [DOI] [PubMed] [Google Scholar]