Abstract
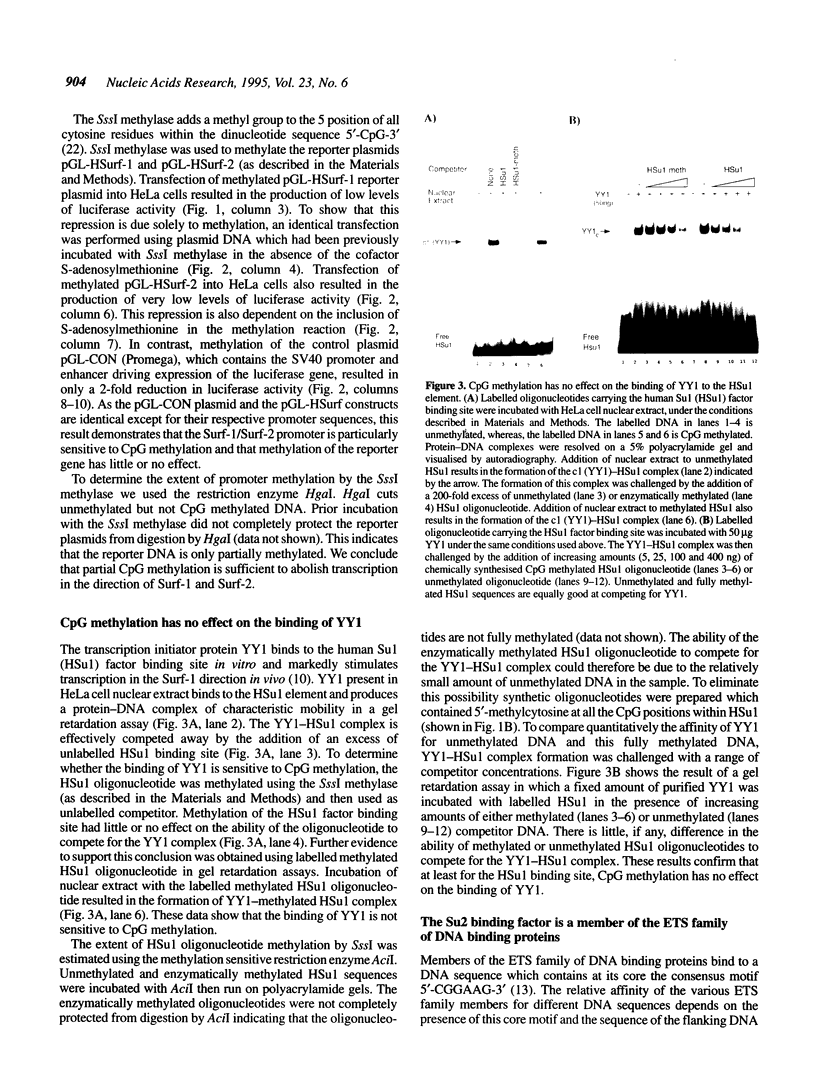
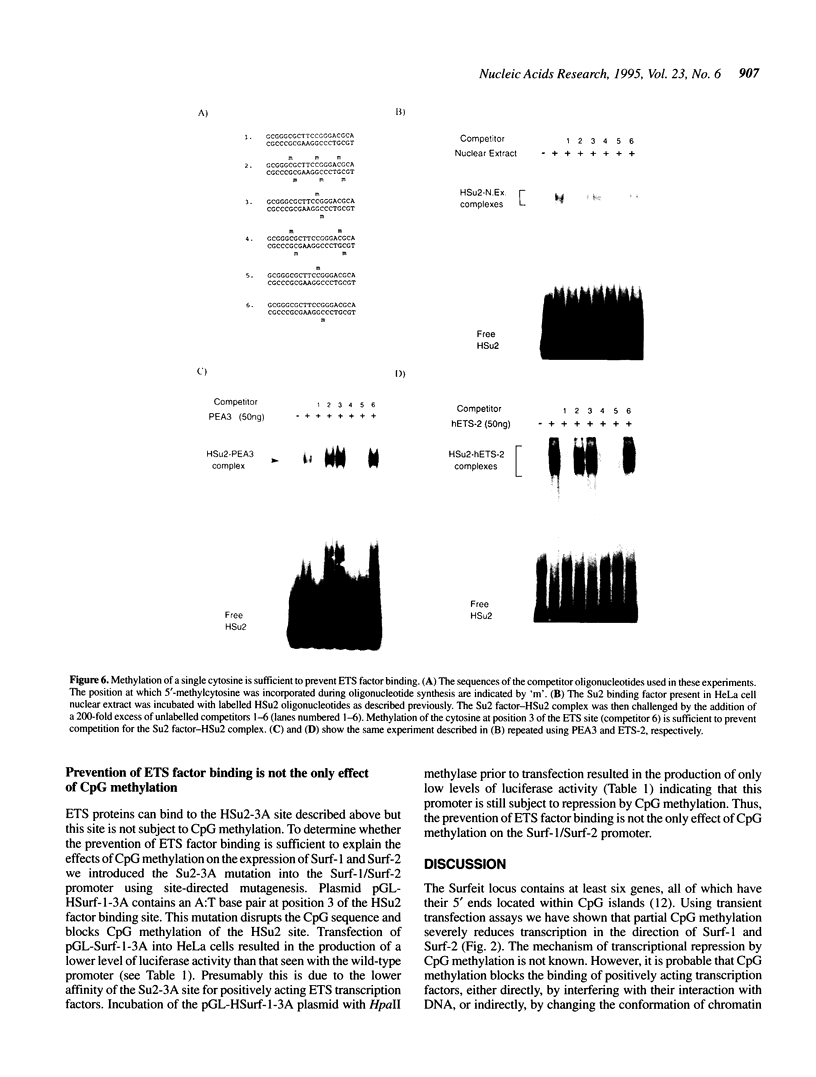
The divergently transcribed Surf-1 and Surf-2 housekeeping genes are separated by a bi-directional, TATA-less promoter which lies within a CpG-rich island. Here we show that CpG methylation severely reduces transcription in the direction of both Surf-1 and Surf-2. Previous work has identified three promoter elements (Su1, Su2 and Su3) which are conserved between the human and mouse Surf-1/Surf-2 promoters. These elements bind transcription factors present in human and mouse cell nuclear extracts in vitro and mutations which prevent factor binding also reduce promoter activity in vivo. Transcription initiation factor YY1 binds to the Su1 site and stimulates transcription in the direction of Surf-1 and, to a lesser extent, Surf-2. Here we show that members of the ETS family of transcription factors bind to the Su2 site. Although the Su1 factor binding site contains three CpG dinucleotides, the binding of YY1 is not affected by CpG methylation. In contrast, CpG methylation abolishes the binding of ETS proteins to the Su2 site; methylation of a single cytosine, at position 3 of the consensus ETS site, is sufficient to prevent factor binding. This direct effect on the binding of ETS proteins is, however, not in itself sufficient to explain the repression of this promoter by CpG methylation. A mutation of the Su2 site which removes the sequence CpG, but which does not prevent ETS factor binding, fails to relieve this promoter from repression by CpG methylation.
Full text
PDF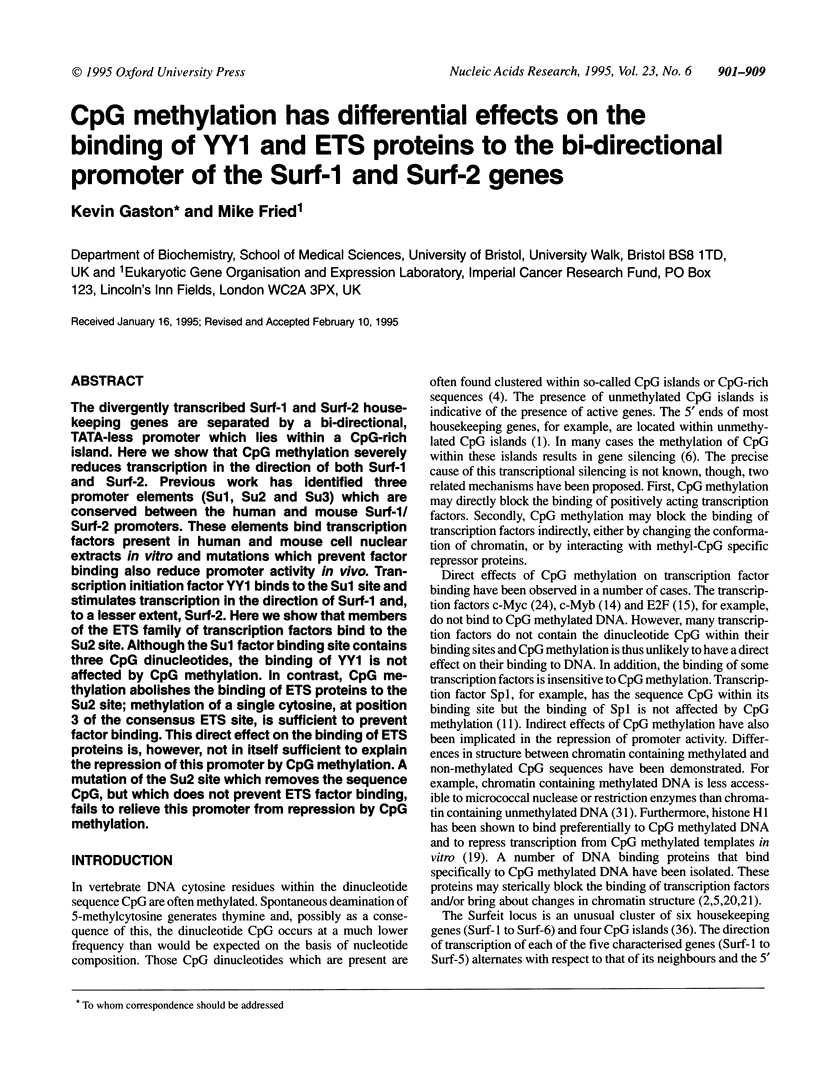





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Antequera F., Macleod D., Bird A. P. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989 Aug 11;58(3):509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- Bhat N. K., Fisher R. J., Fujiwara S., Ascione R., Papas T. S. Temporal and tissue-specific expression of mouse ets genes. Proc Natl Acad Sci U S A. 1987 May;84(10):3161–3165. doi: 10.1073/pnas.84.10.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Dalton S., Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992 Feb 7;68(3):597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- Fisher R. J., Mavrothalassitis G., Kondoh A., Papas T. S. High-affinity DNA-protein interactions of the cellular ETS1 protein: the determination of the ETS binding motif. Oncogene. 1991 Dec;6(12):2249–2254. [PubMed] [Google Scholar]
- Gaston K., Fried M. YY1 is involved in the regulation of the bi-directional promoter of the Surf-1 and Surf-2 genes. FEBS Lett. 1994 Jun 27;347(2-3):289–294. doi: 10.1016/0014-5793(94)00567-2. [DOI] [PubMed] [Google Scholar]
- Gaston K., Kolb A., Busby S. Binding of the Escherichia coli cyclic AMP receptor protein to DNA fragments containing consensus nucleotide sequences. Biochem J. 1989 Jul 15;261(2):649–653. doi: 10.1042/bj2610649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C., Fried M. The mouse surfeit locus contains a cluster of six genes associated with four CpG-rich islands in 32 kilobases of genomic DNA. Mol Cell Biol. 1990 Feb;10(2):605–614. doi: 10.1128/mcb.10.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höller M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988 Sep;2(9):1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Karim F. D., Urness L. D., Thummel C. S., Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A., Gunther C. V., Nye J. A. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990 Sep;4(9):1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H. Methylation-sensitive DNA binding by v-myb and c-myb proteins. Oncogene. 1993 Jan;8(1):111–115. [PubMed] [Google Scholar]
- Kovesdi I., Reichel R., Nevins J. R. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Galvin K. M., Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard A. C., Fried M. The bidirectional promoter of the divergently transcribed mouse Surf-1 and Surf-2 genes. Mol Cell Biol. 1991 Mar;11(3):1281–1294. doi: 10.1128/mcb.11.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard A., Gaston K., Fried M. The Surf-1 and Surf-2 genes and their essential bidirectional promoter elements are conserved between mouse and human. DNA Cell Biol. 1994 Nov;13(11):1117–1126. doi: 10.1089/dna.1994.13.1117. [DOI] [PubMed] [Google Scholar]
- Levine A., Yeivin A., Ben-Asher E., Aloni Y., Razin A. Histone H1-mediated inhibition of transcription initiation of methylated templates in vitro. J Biol Chem. 1993 Oct 15;268(29):21754–21759. [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., Bird A. P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992 Oct 11;20(19):5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Nur I., Szyf M., Razin A., Glaser G., Rottem S., Razin S. Procaryotic and eucaryotic traits of DNA methylation in spiroplasmas (mycoplasmas). J Bacteriol. 1985 Oct;164(1):19–24. doi: 10.1128/jb.164.1.19-24.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992 Jun;6(6):975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K., Park K., Atchison M. L., Howe C. C. The intracisternal A-particle upstream element interacts with transcription factor YY1 to activate transcription: pleiotropic effects of YY1 on distinct DNA promoter elements. Mol Cell Biol. 1993 Nov;13(11):6621–6628. doi: 10.1128/mcb.13.11.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E., Lewis B., Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993 Sep 30;365(6445):462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- Seto E., Shi Y., Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991 Nov 21;354(6350):241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- Shi Y., Seto E., Chang L. S., Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991 Oct 18;67(2):377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Hahn S. L., Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993 Jan 15;211(1-2):7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Kerckaert J. P., Wasylyk B. A novel modulator domain of Ets transcription factors. Genes Dev. 1992 Jun;6(6):965–974. doi: 10.1101/gad.6.6.965. [DOI] [PubMed] [Google Scholar]
- Watson D. K., McWilliams M. J., Lapis P., Lautenberger J. A., Schweinfest C. W., Papas T. S. Mammalian ets-1 and ets-2 genes encode highly conserved proteins. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7862–7866. doi: 10.1073/pnas.85.21.7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman A. G., Sassone-Corsi P., Grundström T., Zenke M., Chambon P. Stimulation of in vitro transcription from the SV40 early promoter by the enhancer involves a specific trans-acting factor. EMBO J. 1984 Dec 20;3(13):3129–3133. doi: 10.1002/j.1460-2075.1984.tb02269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. J., Fried M. The MES-1 murine enhancer element is closely associated with the heterogeneous 5' ends of two divergent transcription units. Mol Cell Biol. 1986 Dec;6(12):4558–4569. doi: 10.1128/mcb.6.12.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- Woods D. B., Ghysdael J., Owen M. J. Identification of nucleotide preferences in DNA sequences recognised specifically by c-Ets-1 protein. Nucleic Acids Res. 1992 Feb 25;20(4):699–704. doi: 10.1093/nar/20.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J. H., Cowie A., Lachance P., Hassell J. A. Molecular cloning and characterization of PEA3, a new member of the Ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev. 1992 Mar;6(3):481–496. doi: 10.1101/gad.6.3.481. [DOI] [PubMed] [Google Scholar]








