Abstract
Human papillomavirus (HPV) E6 oncoproteins target many cellular proteins for ubiquitin-mediated proteasomal degradation. In the case of p53, this is mediated principally by the E6AP ubiquitin ligase. Several studies have reported that E6 can target certain of its substrates in an apparently E6AP-independent fashion and that several of these substrates vary in the degree to which they are degraded by E6 at different stages of malignancy. To more fully understand the regulation of the E6AP/E6 proteolytic targeting complex, we performed a mass spectroscopic analysis of HPV type 18 (HPV-18) E6 protein complexes and identified the HECT domain-containing ubiquitin ligase EDD as a new HPV-18 E6 binding partner. We show that EDD can interact independently with both E6 and E6AP. Furthermore, EDD appears to regulate E6AP expression levels independently of E6, and loss of EDD stimulates the proteolytic activity of the E6/E6AP complex. Conversely, higher levels of EDD expression protect a number of substrates from E6-induced degradation, partly as a consequence of lower levels of E6 and E6AP expression. Intriguingly, reduction in EDD expression levels in HPV-18-positive HeLa cells enhances cell resistance to apoptotic and growth arrest stimuli. These studies suggest that changes in the levels of EDD expression during different stages of the viral life cycle or during malignancy could have a profound effect upon the ability of E6 to target various substrates for proteolytic degradation and thereby directly influence the development of HPV-induced malignancy.
Human papillomaviruses (HPVs) are small double-stranded DNA viruses that cause hyperproliferative lesions in epithelial tissues, which can lead to malignancy. Persistent infection with high-risk HPV types such as 16 and 18 (HPV-16 and HPV-18, respectively) is the most important factor for cervical cancer development (46). The oncogenic activity of these HPV types is mediated by the joint action of two viral oncoproteins, E6 and E7. By interacting with cellular proteins that are involved in regulating cell cycle and apoptosis, these oncoproteins can induce cellular immortalization and transformation (26, 31). E7 interacts with a number of cellular proteins, with its targeting of the pRb family of pocket proteins for proteasome-mediated degradation being among the most important (3, 15). Major activities of the E6 oncoprotein include proteasome-mediated degradation of the p53 tumor suppressor (36) and of a number of cellular proteins containing PDZ domains (41). Thus, an important common feature of the high-risk HPV E6 and E7 proteins is their ability to utilize the proteasome machinery for efficient inactivation of their cellular targets. In the case of E7 this involves the Cul2 complex (19) while E6 is believed to function primarily through the E6AP ubiquitin ligase (20). E6AP was originally identified due to its requirement for E6-induced degradation of p53 (36). It is the prototype HECT domain-containing ubiquitin ligase (21) and plays a central role in many of E6's functions, albeit in some unexpected ways. Loss of E6AP appears to mimic loss of E6 in transcriptome analyses of HPV-16-containing cervical tumor-derived cell lines, suggesting that the effects of E6 upon the cellular transcriptome require E6AP (23). However, a number of studies have also shown various degrees of requirement for E6AP in E6's targeting of a number of substrates, including p53 and some PDZ domain-containing targets (6, 28). One apparent explanation for this is the recent observation that E6AP is required for high levels of E6 expression, with loss of E6AP resulting in enhanced proteasome-mediated degradation of HPV-18 E6 (43). Finally, a number of studies have also shown that p53 is degraded in vivo by E6 to various degrees, both within cervical lesions (7, 10, 24, 27) and in transgenic mouse models (33), suggesting that other mechanisms may modulate the E6/E6AP degradation activity. Indeed, a recent study showed that HPV-16 E6 interacts with the deubiquitinating enzyme USP15 (45), suggesting that E6 interacts with the ubiquitin proteasome machinery in multiple ways.
As part of our studies to more fully understand the regulation of E6 function, we performed proteomic analyses to identify additional interacting partners of HPV-18 E6. In this we identified the HECT domain-containing ligase EDD (5, 32) as a new interacting partner of HPV-18 E6. EDD has been linked to a variety of diseases, including cancer, and gives a neoplastic phenotype in knockout models in Drosophila (17, 30). We now show that EDD is important in the regulation of E6AP and, consequently, in the control of E6 levels and function.
MATERIALS AND METHODS
Cells and transfection.
All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). HEK293, (human embryonic kidney), H1299 (a p53-deficient [p53−/−] non-small-cell lung carcinoma cell line), HT1080 (fibrosarcoma), NIH 3T3 (mouse fibroblasts), E6AP−/− (mouse epithelial kidney cells), HeLa (HPV-18 positive), and CaSKi (HPV-16 positive) cells were transfected using calcium phosphate precipitation (29) or Lipofectamine 2000 (Invitrogen).
Plasmids.
Wild-type hemagglutinin (HA)-tagged HPV-18 E6 (HA-18E6) and untagged HPV-18 E6 (in pCDNA-3) have been described previously (12, 43) as have the following: pGWI-HA-Dlg (11), pCDNA3-HA-MAGI-2 (39), pRcCMV-EDD (8), pCDNA-p53 and pCDNA-Flag-p53 (34), and pCDNA3-E6AP and pCDNA3-E6AP C→A (catalitically inactive mutant) (4). For expression as glutathione S-transferase (GST) fusion proteins, the following were cloned into pGEX2T: HPV-18 E6, HPV-18 E6* (where E6* is a truncated form of the protein), HPV-16 E6, HPV-11 E6, and E6AP (4, 34, 35, 40). The GST HPV-18 E6 fusion protein with the mutation I130T [GST-18E6 (I130T)] was generated using a Gene Tailor Mutagenesis kit (Invitrogen). We gratefully thank Giannino Del Sal for providing HA-tagged ubiquitin.
Antibodies.
Mouse monoclonal antibodies against HPV-18 E6 (1:1,000; N terminus no. 399) were generated and generously provided by the Arbor Vita Corporation. The following antibodies were also used: anti-HA monoclonal antibody 12CA5 (Roche), anti-β-galactosidase (Promega), anti-Flag mouse monoclonal antibody M2 (Sigma), mouse anti-p53 DO-1 (Santa Cruz), rabbit anti-p53 (35), mouse anti-γ-tubulin (Sigma), rabbit anti-α-actin (Santa Cruz), mouse anti-E6AP (BD Transduction Labs), goat anti-EDD M-19 (Santa Cruz), and appropriate secondary antibodies conjugated to horseradish peroxidase (HRP; Dako), fluorescein, or rhodamine (Molecular Probes).
Fusion protein purification and in vitro binding assays.
GST-tagged fusion proteins were expressed and purified as described previously (40). Proteins were translated in vitro using a Promega TNT kit and radiolabeled with [35S]cysteine or [35S]methionine (Perkin Elmer). Equal amounts of in vitro translated proteins were added to GST fusion proteins bound to glutathione agarose (Sigma) and incubated for 1 h at 4°C. After extensive washing with phosphate-buffered saline (PBS) containing 0.25% NP-40, the bound proteins were analyzed by SDS-PAGE and autoradiography.
GST pulldowns using cellular extracts were performed by incubating GST fusion proteins immobilized on glutathione agarose with cells extracted in E1A buffer (25 mM HEPES, pH 7.0, 0.1% NP-40, 150 mM NaCl, plus protease inhibitor cocktail set I [Calbiochem]) for 1 h at 4°C on a rotating wheel. After extensive washing, the bound proteins were detected using SDS-PAGE and Western blotting.
Western blotting.
Total cellular extracts were prepared by directly lysing cells from 6-cm2 or 10-cm2 dishes in SDS lysis buffer, and protein detections were done as described previously (43).
Immunoprecipitation.
For coimmunoprecipitations and mass spectrometry pulldown experiments, HEK293 cells were transfected with the appropriate plasmids. After 24 h cells were extracted in either E1A or mass spectrometry lysis buffer (50 mM HEPES, pH 7.4 [at 4°C], 150 mM NaCl, 50 mM NaF, 1 mM EDTA, 0.25% NP-40), and extracts were incubated with anti-HA beads (Sigma) for 2 to 3 h on a rotating wheel at 4°C. The beads were then extensively washed, dried, and subjected to proteomic analysis (42).
Half-life experiments.
At 72 h posttransfection (see Fig. 6), cells were treated for different times, as indicated, with cycloheximide (50 μg/ml in dimethyl sulfoxide [DMSO]) to block protein synthesis. DMSO-treated cells were used as the control. Total cellular extracts were analyzed by Western blotting, and the intensity of the bands on the X-ray film was measured using the OptiQuant program. The standard deviation was calculated from three independent assays.
Immunofluorescence and microscopy.
Cells were stained and fixed for immunofluorescence as described previously (13). Secondary anti-rabbit or anti-mouse conjugated with fluorescein or rhodamine was used as appropriate (Molecular Probes); slides were analyzed using a Leica DMLB fluorescence microscope with a Leica photo camera (A01M871016), and the data were collected using a 100× objective oil immersion lens.
siRNA experiments.
For transient small interfering RNA (siRNA) experiments, HPV-positive HeLa and CaSKi cells and HPV-negative HT1080 cells were seeded in 6-cm2 dishes and transfected using Lipofectamine 2000 (Invitrogen) with the following siRNAs: siRNA against luciferase (Dharmacon) as a control, siRNA against HPV-18 E6/E7 (5′-CAUUUACCAGCCCGACGAG) (Dharmacon), siRNA against HPV-16 E6/E7 (5′-UUAAAUGACAGCUCAGAGG) (Dharmacon), siRNA against E6AP (Dharmacon and Santa Cruz), and siRNA against EDD (Santa Cruz and Dharmacon). For stable knockdown lines, HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) with nonspecific short hairpin RNA (shRNA; OriGene) as a control and shRNA against EDD (OriGene). Cells were maintained in medium containing 200 μg/ml puromycin. Cells negative for EDD were isolated, and EDD levels were verified by Western blot analysis.
Cell Synchronization and FACS analysis.
To assess G2/M phase arrest, HeLa cells were treated with 300 nM nocodazole (Sigma) for 18 h. To induce DNA damage-induced apoptosis, cells were treated with 10 μM etoposide (Sigma) for 12 h. Cells were harvested, and DNA content was assessed by propidium iodide staining and fluorescence-activated cell sorter (FACS) analysis as described previously (1).
RESULTS
EDD is a target of HPV-18 E6.
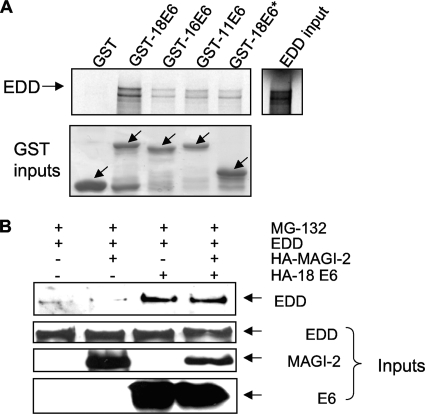
To identify other components of the proteolytic apparatus with which E6 might interact, HEK293 cells were transfected with a plasmid expressing HA-tagged HPV-18 E6. After 24 h, extracts were immunoprecipitated with anti-HA antibody, and the complexes were subjected to mass spectrometric analysis. Complexes were compared with pulldowns without E6 and all potentially contaminating hits were excluded. Although a number of novel potential binding partners were found, we focused our attention on those interactions that were potentially associated with the proteasome pathway. E6AP was easily detected in this analysis, and a large number of proteasome subunits were also identified, demonstrating a close association between E6 and the multiprotein complexes of the proteasome. In addition to these subunits, one other ubiquitin ligase was also coimmunoprecipitated with E6, and this was identified as a 300-kDa protein, a hyperplastic discs protein homolog, or EDD. This was intriguing because EDD is a putative tumor suppressor involved in DNA damage signaling (5) and has been shown to function as an E3 HECT domain-containing ubiquitin ligase (5). To verify whether EDD can complex with HPV-18 E6 in vitro and also to determine whether it could interact with E6 proteins from other HPV types, a series of GST pulldown assays were performed. EDD was translated in vitro and incubated with GST-18E6, GST-16E6, GST-11E6, GST-18E6*, or GST alone for control. The result in Fig. 1A shows that HPV-18 E6 binds to EDD much more strongly than either HPV-16 E6 or HPV-11 E6. Since HPV-18E6* binds much more weakly than the full-length HPV-18 E6, this suggests that the principal site of interaction between HPV-18 E6 and EDD is within the carboxy-terminal half of E6.
FIG. 1.
HPV-18 E6 protein binds to EDD in vitro and in vivo. (A) Radiolabeled in vitro translated EDD was incubated with GST, GST-18E6, GST-16E6, GST-11E6, and GST-18E6*. Bound proteins were assessed by autoradiography, and the input GST fusion proteins were visualized with Coomassie staining (lower panel). Input EDD (10%) is shown. (B) 293 cells were transfected with HA-tagged HPV-18 E6 (HA-18E6), EDD1, or HA-tagged MAGI-2, as indicated. After 24 h cells were incubated for 3 h with MG132 before being harvested, and cell extracts were immunoprecipitated with anti-HA antibody. Coprecipitating EDD was detected by Western blotting with anti-EDD antibody. The three lower panels show the protein inputs of EDD, MAGI-2, and HPV-18 E6.
We then proceeded to confirm that the interaction between E6 and EDD occurs in vivo. Cells were transfected with EDD, together with HA-18E6, HA-MAGI-2, or both, and pulldown assays were performed on the cell extracts. The results in Fig. 1B show that EDD is coimmunoprecipitated with HPV-18 E6, both in the presence and absence of HPV-18 E6's known substrate, MAGI-2, whereas no EDD was immunoprecipitated with MAGI-2 alone or when EDD was overexpressed alone.
EDD inhibits HPV-18 E6 degradation activity.
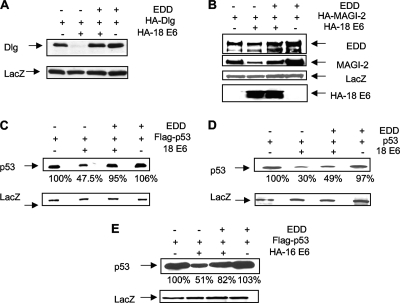
Since EDD has been shown to function as a ubiquitin ligase, it was of interest to investigate the potential role of EDD in E6's degradatory activities. To do this we analyzed whether EDD could affect the ability of E6 to degrade three known substrate proteins in vivo: Dlg, MAGI-2, and p53. In the first set of assays, 293 cells were transfected with Dlg or MAGI-2 in the presence and absence of HPV-18 E6 and exogenously added EDD. The results obtained in the experiment shown in Fig. 2 demonstrate that both Dlg (panel A) and MAGI-2 (panel B) are very efficiently degraded by E6 but that addition of EDD essentially abolishes the activity of E6 with respect to these two substrates.
FIG. 2.
EDD inhibits HPV-18 E6 degradation of Dlg, MAGI-2, and p53. 293 cells (A, B, C, and E) or p53 null H1299 cells (D) were transfected with HA-tagged Dlg, HA-MAGI-2, EDD1, and HPV-18 E6 (A, B, C, and D) or HPV-16 E6 (E), alone or in combination. After 24 h cells were harvested, and residual Dlg (A), MAGI-2 (B), and p53 (C, D, and E) were detected by Western blot analysis using either anti-HA antibody (A, B, and C) or anti-p53 antibody (D) or by anti-Flag where p53 was Flag tagged (C and E). The expression of ß-galactosidase (LacZ) was used as a control of transfection efficiency and loading (lower panels), and the percentage of p53 remaining in each track in panels C, D, and E is also shown.
We then performed a similar analysis on p53 in 293 and H1299 cells (Fig. 2C and D, respectively). As can be seen, p53 is also rescued from E6-induced degradation in the presence of exogenous EDD, albeit not quite as efficiently in H1299 cells. These effects appear to be specific for E6 since cotransfection of EDD alone with p53, Dlg, or MAGI-2 has no effect on their levels of expression. We also repeated the analysis with HPV-16 E6 and p53, and the results in Fig. 2E show that overexpressed EDD can also rescue p53 from HPV-16 E6-induced degradation. Taken together, these results demonstrate that the effects of EDD in rescuing MAGI-2, Dlg, and p53 from E6-induced degradation are most likely associated with a direct effect on overall E6 function; they are not restricted to a single target protein per se and are not due to a generalized indirect inhibitory effect on the proteasome pathway.
Endogenous EDD directly regulates HPV E6 activity in vivo.
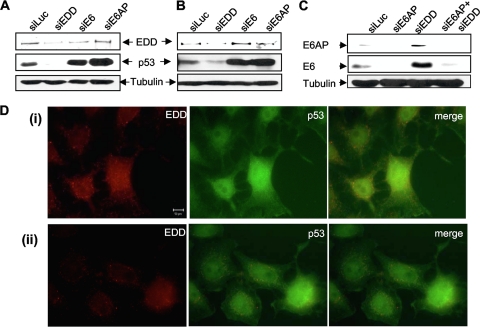
To investigate the role of EDD in the context of HPV E6 function in cervical tumor-derived cell lines, we performed siRNA EDD silencing in HPV-18-positive HeLa and HPV-16-positive CaSKi cells. Each cell line was transfected with siRNAs directed against EDD, HPV-18 E6 (HeLa), HPV-16 E6 (CaSKi), E6AP, or luciferase as a control. The levels of EDD and p53 expression were then analyzed by Western blotting after 72 h. The results in Fig. 3 show that siRNA directed against either E6 or E6AP results in a marked upregulation in the level of p53 expression (Fig. 3A and B), in agreement with previous studies (2, 22, 23, 43). In contrast, when EDD levels are depleted, there is a striking decrease in the levels of p53 expression in both the HeLa (Fig. 3A) and CaSKi (Fig. 3B) cells. These results show that EDD is a rate-determining factor in the ability of E6 to direct the degradation of p53 in cervical tumor-derived cell lines.
FIG. 3.
EDD knockdown enhances E6 activity. HeLa cells (A and C) and CaSKi cells (B) were transfected with siRNAs directed against luciferase (siLuc), EDD (siEDD), E6AP (siE6AP), or HPV-16 E6/E7 or HPV-18 E6/E7 (siE6), alone or in combination with siE6AP and siEDD. After 72 h cells were harvested, and the levels of EDD, p53, E6, and the tubulin loading control were detected by Western blotting. HeLa cells (D) were fixed and probed with goat anti-EDD and rabbit anti-p53 antibodies, followed by rhodamine-conjugated donkey anti-goat (red for EDD) and fluorescein isothiocyanate-conjugated donkey anti-rabbit (green for p53) antibodies. Two different fields are shown (i and ii).
Effects of siRNA EDD depletion on HPV E6 and E6AP in HeLa cells.
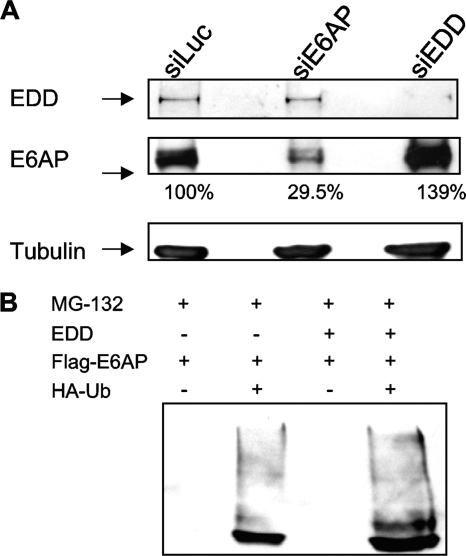
The above results demonstrate that EDD has a negative effect on the capacity of E6 to direct its substrates for degradation. To investigate the underlying mechanisms, we repeated the siRNA EDD knockdown assays in HeLa cells and assessed the effect upon E6AP and E6 expression levels. Since loss of E6AP can destabilize E6 (43), we also analyzed the effect on E6 levels of a double knockdown of both EDD and E6AP. At 72 h posttransfection, cells were harvested, and cellular lysates were analyzed by Western blotting for HPV-18 E6, E6AP, and tubulin. The results in Fig. 3C confirm that loss of E6AP destabilizes E6 (43). However, loss of EDD results in a striking increase in the levels of expression of both E6AP and E6. This suggests that EDD can directly affect the levels of E6 and E6AP expression and thereby influence the ability of E6 to direct the degradation of p53 in vivo. Knockdown of EDD in the context of cells lacking E6AP does not result in a significant alteration in the levels of E6 expression, suggesting that EDD does not directly affect E6.
These results suggest that changes in EDD expression levels might be expected to affect the ability of E6 to induce substrate protein degradation and might offer one possible explanation for the variable levels of p53 expression often seen by immunohistochemistry in cell lines and tumors (7, 10, 24, 27). To investigate this further, we performed immunofluorescence analysis of p53 and EDD in HPV-18-containing HeLa cells. The results shown in Fig. 3D demonstrate a clear concordance between the level of EDD expression and that of p53, with high EDD expression correlating with high p53 expression and vice versa.
EDD interacts independently with both E6 and E6AP.
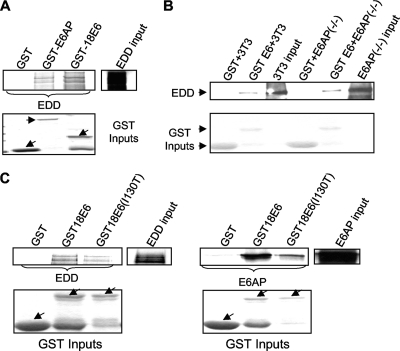
Since EDD was identified as an E6-interacting protein in a pulldown in which E6AP was also present, we proceeded to determine whether the ability of E6 to interact with EDD was in part dependent on E6AP and, further, whether E6AP itself could also potentially interact with EDD. To do this, EDD was in vitro translated and incubated with GST-18E6, GST-E6AP, and GST alone. The results in Fig. 4A show that EDD binds equally well to both HPV-18 E6 and to E6AP, demonstrating that EDD can interact with E6AP independently of E6.
FIG. 4.
E6 does not require E6AP to bind EDD. (A) In vitro translated EDD was incubated with GST, GST-E6AP, and GST-18E6. Bound proteins were assessed by autoradiography, and the input GST fusion proteins were visualized with Coomassie staining (lower panel). Input EDD (10%) is shown. (B) Extracts of E6AP−/− and 3T3 cells were incubated with GST and GST-18E6 fusion proteins for 2 h at 4°C. Bound proteins were assessed by Western blotting using EDD antibody. The EDD inputs from the cells are also shown. (C) In vitro translated EDD and E6AP were incubated with GST, GST-18E6, and GST-18E6 (I130T). Bound proteins were assessed by autoradiography, and the input GST fusion proteins were visualized with Coomassie staining (lower panel). GST-18 E6 (I130T) binding to EDD is 21% lower than that of wild-type GST-18E6, while GST-18E6 (I130T) binding to E6AP is 77% lower than that of wild-type GST-18E6. Arrows indicate GST fusion proteins.
To determine whether E6 could interact with EDD in the absence of E6AP, we analyzed the ability of a purified GST-E6 fusion protein to pull down EDD from E6AP-positive and E6AP null cells. The results in Fig. 4B show that E6 can bind to EDD in the absence of E6AP and, furthermore, that the presence of E6AP does not affect the amount of EDD bound to E6. We also made use of an E6 mutant (I130T), which has a reduced capacity to interact with E6AP (33). EDD and E6AP were in vitro translated and incubated with GST-18E6, GST-18E6 (I130T), or GST alone. The results, in Fig. 4C, show that wild-type HPV-18 E6 and the I130T mutant bind EDD similarly. In contrast, there is a much bigger difference in the capacity of the wild-type E6 and the I130T mutant to bind E6AP. This also supports the notion that E6 does not require E6AP for the EDD interaction and furthermore suggests that the binding sites for EDD and E6AP are distinct on E6.
EDD is a direct regulator of E6AP turnover.
Since loss of EDD increases E6AP levels in HPV-positive cells (Fig. 3C), we then investigated whether EDD could affect E6AP levels in cells that lack HPV sequences. To do this, HPV-negative HT1080 cells were transfected with siRNAs directed against EDD, E6AP, or luciferase. After 72 h the levels of both EDD and E6AP were analyzed by Western blotting. The results in Fig. 5A demonstrate that silencing of EDD in HT1080 cells results in a dramatic upregulation in the levels of E6AP. In contrast, ablation of E6AP expression appears to have no significant effect upon the levels of EDD, and this is consistent with the results in HPV-positive cells. These results suggest that EDD can directly affect the levels of expression of E6AP independently of the presence of HPV E6.
FIG. 5.
siEDD increases E6AP levels in HPV-negative cells. (A) HT1080 cells were transfected with siLuc, siE6AP, or siEDD. After 72 h cells were harvested, and levels of EDD, E6AP, and tubulin were determined by Western blotting. The numbers show the percentages of E6AP remaining. (B) 293 cells were cotransfected with EDD, Flag-E6AP, and HA-ubiquitin (HA-Ub), and after 24 h cells were treated for 3 h with the proteasome inhibitor MG132. The cells were then harvested, and complexes were immunoprecipitated with anti-HA-conjugated agarose beads. Complexes were then analyzed by Western blotting for E6AP using anti-Flag antibodies. Note the increased levels of mono- and polyubiquitinated forms of E6AP in the EDD-expressing cells.
Since EDD has been reported to possess ubiquitin ligase activity, we were next interested in determining whether overexpressed EDD can enhance the levels of E6AP ubiquitination. 293 cells were cotransfected with Flag-tagged E6AP, EDD, and HA-tagged ubiquitin expression plasmids. After 24 h the cells were harvested, and complexes were immunoprecipitated using anti-HA-conjugated agarose beads. HA-ubiquitin-bound E6AP was then detected by Western blotting with anti-Flag antibodies. The results in Fig. 5B show clear coimmunoprecipitation of E6AP with ubiquitin, and this is increased dramatically in the presence of exogenous EDD, suggesting that EDD can also enhance the levels of E6AP ubiquitination.
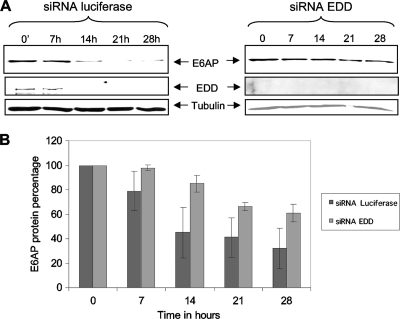
To verify whether loss of EDD could result in slower E6AP turnover, we performed studies to investigate changes in E6AP half-life in the presence and absence of EDD. Since the E6AP half-life is over 25 h in HPV-negative cells (22; V. Tomaić, personal observation) we used HPV-positive HeLa cells, where E6AP has a half-life of about 7 h (22). Cells were transfected with siRNA directed against either luciferase or EDD, and after 72 h they were treated with cycloheximide to block protein synthesis for different periods of time, after which the cells were harvested, and the levels of E6AP and EDD were determined by Western blotting. The results in Fig. 6A, together with the quantitation from multiple assays in Fig. 6B, show that in control cells E6AP has half-life of 7 to 14 h. In contrast, when EDD is depleted from the cells, the E6AP levels remain relatively stable until the 21-h time point, when they begin to decrease. These results show that silencing EDD in HPV-positive HeLa cells results in a dramatic decrease in E6AP protein turnover and provide an explanation for how changes in EDD levels might affect E6AP activity and, hence, the capacity of E6 to direct the degradation of a variety of different substrate proteins.
FIG. 6.
E6AP protein turnover is regulated by EDD. (A) HeLa cells were transfected with siLuc or siEDD. At 72 h posttransfection cells were treated with cycloheximide at different time points (0, 7 h, 14 h, 21 h, and 28 h). E6AP, EDD, and tubulin levels were detected by Western blotting. (B) Collated results from three independent experiments to measure residual E6AP protein levels, with band intensities determined using the OptiQuant quantification program. E6AP levels were normalized to 100% at time zero. Standard deviations are also shown.
Loss of EDD in HPV-positive cells enhances cell survival and aids evasion of checkpoint-activated cell growth arrest.
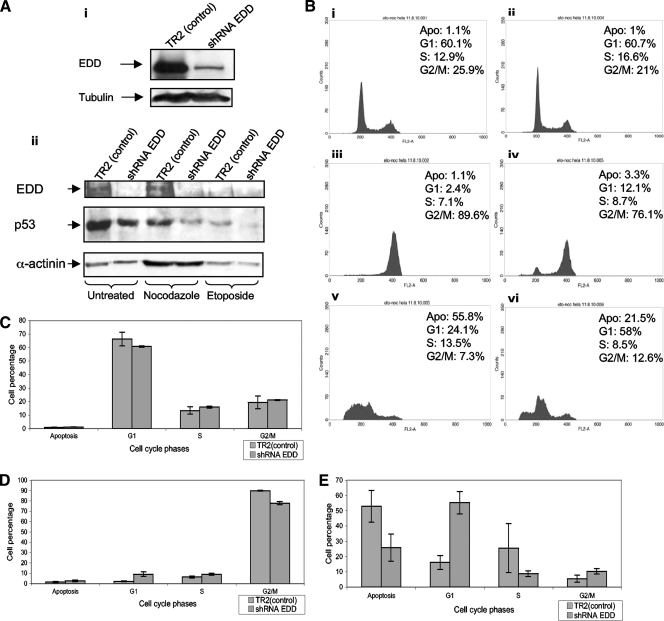
Since loss of EDD enhances the levels of E6/E6AP expression and consequent p53 degradation, we wanted to address the potential biological consequences for loss of EDD expression in HeLa cells. To do this, we generated HeLa cell lines stably expressing shRNAs against EDD. Cell clones were isolated, and the levels of EDD expression were verified by Western blotting. As can be seen from Fig. 7Ai, cells were obtained that had greatly reduced levels of EDD expression. We then analyzed the capacity of these cells to respond to different forms of checkpoint activation: G2/M arrest induced by microtubule disruption with nocodazole (44) and S phase arrest/apoptosis induced by etoposide-induced DNA damage (14). The results in Fig. 7B, together with the quantitations from multiple assays in Fig. 7C, D, and E, show that both the parental and EDD knockdown cells have relatively similar cell cycle profiles. However, upon treatment with nocodazole, the control cells show a complete G2/M arrest while the EDD knockdown cells show a small percentage of cells within the G1 and S phases. Likewise, following treatment with etoposide, the control cells exhibit a high level of apoptosis, as determined by sub-G1 DNA content, while the EDD knockdown cells have greatly reduced levels of apoptosis and a correspondingly higher number of cells within the G1 phase. To determine whether these differences in the response of the EDD knockdown cells were related to differences in the levels of p53 expression, Western blotting was also done to detect p53 and EDD in the control and EDD knockdown cells following exposure to etoposide and nocodazole. The results in Fig. 7Aii show significantly decreased levels of p53 in the EDD knockdown cells, further confirming the results of the transient siRNA experiments and suggesting that the differences in the apoptotic responses of these cells are due to lower levels of p53 expression. It is also interesting that there are lower levels of p53 expression in the control knockdown cells following treatment with nocodazole and etoposide, which is in agreement with previous studies showing enhanced degradation of p53 by E6 following induction of DNA damage response and activation of cell cycle checkpoint pathways (38). Intriguingly, there is also a significant decrease in the levels of EDD expression in the etoposide treated cells, which also occurs in cells that lack HPV sequences (V. Tomaić, personal observations), suggesting that DNA damage responses result in enhanced EDD turnover. Taken together, these results demonstrate, in the context of HPV-18-positive cervical cancer derived cells, that EDD has a potent tumor suppressor function, most likely through its capacity to affect the activity of E6.
FIG. 7.
EDD knockdown inhibits checkpoint-activated cell growth arrest and apoptosis. (A). HeLa cells stably transfected with shRNA EDD and with nonspecific shRNA TR2 (control) were harvested in SDS sample buffer, and residual EDD levels were assessed by Western blotting (i). The lower panel (ii) shows Western blots of EDD and p53 in the control and EDD-ablated cells growing asynchronously and postexposure to nocodazole and etoposide. (B) FACS analysis showing the cell cycle profiles of asynchronously growing cells (i and ii) and following nocodazole (iii and iv) and etoposide treatments (v and vi). Control cells are shown in panels i, iii, and v, with EDD knockdown cells in panels ii, iv and vi. Apo, apoptosis. (C, D, and E) Percentages of cells in each phase of the ell cycle in asynchronously growing cells (C) and following treatment with nocodazole (D) and etoposide (E) from at least three separate experiments; the standard deviations are also shown.
DISCUSSION
Using a proteomic approach, we have identified new components of the ubiquitin proteasome pathway with which HPV-18 E6 interacts. The integrity of the assay was confirmed by the detection of E6AP as a major interacting partner of E6. We also identified a large number of proteasome subunits, suggesting that E6 functions in close proximity to the proteasome complex. We also identified EDD, another HECT domain-containing ubiquitin ligase, as a novel interacting partner of E6. EDD was originally reported to play a critical role in coordinating the balance between cell cycle progression and differentiation (17), and EDD overexpression has been reported in several cancers, including those of ovary and breast, while truncating mutations have also been found in gastric and colon cancers (8, 30).
Although alteration in EDD function has been linked to cancer development, there is still little known about the biochemical activities of the protein, and few interacting partners have been reported. Perhaps most significantly from an HPV E6 standpoint, EDD is an E3 ubiquitin ligase HECT-domain containing protein (5), which has been shown to have the potential to function as an N-recognin in the N-end rule degradatory pathway (37). Furthermore, EDD has also been implicated in the regulation of several DNA damage response pathways. EDD interacts with and degrades topoisomerase II-binding protein (TopBP1), a protein associated with DNA damage response and cell cycle regulation (18). In addition, EDD was also reported to be required for optimal CHK2 Thr68 phosphorylation and kinase activity and for enhanced cell survival after DNA damage (16). More recent studies have suggested that EDD can form a ubiquitin ligase complex with DNA-damage binding protein 1 (DDB1) and Vpr-binding protein (VprBP), known as the EDVP (EDD, DDB1, VprBP) complex. This is dependent on a protein kinase, DYRK2, for its formation, and for the subsequent phosphorylation, ubiquitylation, and degradation of EDVP substrates (25), which are involved in regulating mitotic progression.
While we found that EDD was bound strongly by HPV-18 E6, only a weak interaction was found with HPV-16 and HPV-11 E6. This suggests that the interaction between E6 and EDD is restricted biochemically to HPV-18 E6, and it will be of interest to determine whether this association confers any unique characteristics to the function of HPV-18 E6. Potential alterations in DNA damage repair pathways are one obvious line of further investigation. It was technically possible that the interaction between E6 and EDD was indirect and that it might be being mediated by E6AP. However, using a non-E6AP binding mutant of E6 and pulldown assays using extracts from E6AP null cells, we were able to show that HPV-18 E6 can interact with EDD in the absence of E6AP.
Although EDD binding is primarily a feature of HPV-18 E6, it is clear that EDD plays a critical role in the normal function of both HPV-16 and HPV-18 E6. Thus, overexpression of EDD impairs the ability of HPV-18 E6 to direct the degradation of different substrates, and there is a correlation between the levels of p53 and EDD expression in HeLa cells, where high levels of EDD correlate with high levels of p53. This provides one possible explanation for the variable degrees to which p53 might be degraded in vivo under different physiological settings (7, 10, 24, 27) and raises the intriguing prospect that EDD might be a rate-limiting factor in the development of cervical cancer. In agreement with this, ablation of EDD expression in both HPV-16- and HPV-18-containing cervical tumor-derived cell lines results in enhanced degradation of p53. This would appear contradictory to the respective capacity of the two E6 proteins to bind EDD; however, the explanation for this is provided by E6AP, where loss of EDD results in a significant increase in the levels of E6AP expression. Based upon studies showing that E6AP is essential for the stability of E6 (43), it is not surprising that this increase in E6AP levels also results in an increase in the levels of HPV-18 E6. This provides a clear mechanistic explanation for the enhanced rates of p53 degradation in cells where EDD expression has been ablated. We also show that the reduction of EDD levels in HeLa cells has significant biological consequences. Previous studies had shown that loss of EDD resulted in an impaired G2/M checkpoint arrest in response to DNA damage in HeLa cells (16). Our studies also show a reduced G2/M checkpoint activation in response to microtubule destabilization induced by nocodazole treatment. Most importantly, we also observed significantly reduced levels of apoptosis in cells expressing reduced levels of EDD following induction of DNA damage with etoposide; again, this is consistent with the ability of E6 to enhance p53 degradation. Intriguingly, we found that EDD levels were also lower following treatment with etoposide. This does not seem to be an HPV-related phenomenon since we also observed similar effects in an HPV-negative context. Obviously, further studies are warranted to investigate these aspects further. However, taken together these results would suggest that, in the context of HPV infection, EDD is a potent tumor suppressor protein.
We also showed that EDD can regulate the levels of E6AP expression in the absence of HPV. In addition, in HeLa cells, where E6AP is turned over more rapidly, partly in response to E6 (22), loss of EDD induces a striking extension of the E6AP half-life from 7 h to 14 h. Whether this degradation of E6AP by EDD is dependent upon EDD's ubiquitination activity remains to be determined. However, E6AP lacks sequences that would suggest susceptibility to the N-end rule pathway of degradation; nonetheless, in transient overexpression assays, we detect significantly increased levels of E6AP ubiquitination when EDD is present. Obviously, further studies are required to elucidate more fully the mechanism by which EDD controls E6AP protein levels. However, this does suggest a possible route for restoring E6AP expression levels under conditions where the E6AP protein expression has been lost in certain disease syndromes (9).
In summary, we have identified a novel mechanism for regulating the activity of the HPV E6/E6AP ubiquitin ligase complex, whereby alterations in the levels of EDD expression may have a significant impact upon the ability of E6/E6AP to direct the degradation of several cellular substrates, p53 in particular, and thereby affect the capacity of HPV-infected cells to undergo apoptosis or to enter G2/M arrest in response to various inhibitory stimuli.
Acknowledgments
The anti-18 E6 (N terminus no. 399) antibody was generated and kindly provided by the Arbor Vita Corporation. We are also very grateful to Michelle Henderson and Colin Watts for providing us with the EDD expression plasmid.
This work was supported in part by research grants to L.B. from the Associazione Italiana per la Ricerca sul Cancro, the Association for International Cancer Research, and the Telethon Foundation of Italy, grant number GGP10006.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1.Banks, L., S. C. Barnett, and T. Crook. 1990. HPV-16 E7 functions at the G1 to S phase transition in the cell cycle. Oncogene 5:833-837. [PubMed] [Google Scholar]
- 2.Beer-Romero, P., S. Glass, and M. Rolfe. 1997. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene 14:595-602. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 4.Brimer, N., C. Lyons, and S. B. Vande Pol. 2007. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 358:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callaghan, M. J., et al. 1998. Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene 17:3479-3491. [DOI] [PubMed] [Google Scholar]
- 6.Camus, S., et al. 2007. Ubiquitin-independent degradation of p53 mediated by high-risk human papillomavirus protein E6. Oncogene 26:4059-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavatorta, A. L., et al. 2004. Differential expression of the human homologue of drosophila discs large oncosuppressor in histologic samples from human papillomavirus-associated lesions as a marker for progression to malignancy. Int. J. Cancer 111:373-380. [DOI] [PubMed] [Google Scholar]
- 8.Clancy, J. L., et al. 2003. EDD, the human orthologue of the hyperplastic discs tumour suppressor gene, is amplified and overexpressed in cancer. Oncogene 22:5070-5081. [DOI] [PubMed] [Google Scholar]
- 9.Clayton-Smith, J., and L. Laan. 2003. Angelman syndrome: a review of the clinical and genetic aspects. J. Med. Genet. 40:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, K., C. S. Herrington, M. F. Evans, K. C. Gatter, and J. O. McGee. 1993. p53 antigen in cervical condylomata, intraepithelial neoplasia, and carcinoma: relationship to HPV infection and integration. J. Pathol. 171:27-34. [DOI] [PubMed] [Google Scholar]
- 11.Gardiol, D., S. Galizzi, and L. Banks. 2002. Mutational analysis of the discs large tumour suppressor identifies domains responsible for human papillomavirus type 18 E6-mediated degradation. J. Gen. Virol. 83:283-289. [DOI] [PubMed] [Google Scholar]
- 12.Gardiol, D., et al. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 13.Grm, H. S., P. Massimi, N. Gammoh, and L. Banks. 2005. Crosstalk between the human papillomavirus E2 transcriptional activator and the E6 oncoprotein. Oncogene 24:5149-5164. [DOI] [PubMed] [Google Scholar]
- 14.Hande, K. R. 1998. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer 34:1514-1521. [DOI] [PubMed] [Google Scholar]
- 15.Helt, A. M., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson, M. J., et al. 2006. EDD mediates DNA damage-induced activation of CHK2. J. Biol. Chem. 281:39990-40000. [DOI] [PubMed] [Google Scholar]
- 17.Henderson, M. J., et al. 2002. EDD, the human hyperplastic discs protein, has a role in progesterone receptor coactivation and potential involvement in DNA damage response. J. Biol. Chem. 277:26468-26478. [DOI] [PubMed] [Google Scholar]
- 18.Honda, Y., et al. 2002. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J. Biol. Chem. 277:3599-3605. [DOI] [PubMed] [Google Scholar]
- 19.Huh, K., et al. 2007. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 81:9737-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 13:4918-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, W. H., S. L. Beaudenon, A. L. Talis, J. M. Huibregtse, and P. M. Howley. 2000. Human papillomavirus type 16 E6 induces self-ubiquitination of the E6AP ubiquitin-protein ligase. J. Virol. 74:6408-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley, M. L., K. E. Keiger, C. J. Lee, and J. M. Huibregtse. 2005. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J. Virol. 79:3737-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lie, A. K., S. Skarsvag, H. Skomedal, O. A. Haugen, and R. Holm. 1999. Expression of p53, MDM2, and p21 proteins in high-grade cervical intraepithelial neoplasia and relationship to human papillomavirus infection. Int. J. Gynecol. Pathol. 18:5-11. [DOI] [PubMed] [Google Scholar]
- 25.Maddika, S., and J. Chen. 2009. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11:409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 27.Mantovani, F., and L. Banks. 1999. Inhibition of E6 induced degradation of p53 is not sufficient for stabilization of p53 protein in cervical tumour derived cell lines. Oncogene 18:3309-3315. [DOI] [PubMed] [Google Scholar]
- 28.Massimi, P., A. Shai, P. Lambert, and L. Banks. 2008. HPV E6 degradation of p53 and PDZ containing substrates in an E6AP null background. Oncogene 27:1800-1804. [DOI] [PubMed] [Google Scholar]
- 29.Matlashewski, G., et al. 1987. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. EMBO J. 6:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori, Y., et al. 2002. Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res. 62:3641-3645. [PubMed] [Google Scholar]
- 31.Münger, K., et al. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 32.Munoz, M. A., et al. 2007. The E3 ubiquitin ligase EDD regulates S-phase and G2/M DNA damage checkpoints. Cell Cycle 6:3070-3077. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, M., et al. 2002. A mutant of human papillomavirus type 16 E6 deficient in binding α-helix partners displays reduced oncogenic potential in vivo. J. Virol. 76:13039-13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pim, D., P. Massimi, and L. Banks. 1997. Alternatively spliced HPV-18 E6* protein inhibits E6-mediated degradation of p53 and suppresses transformed cell growth. Oncogene 15:257-264. [DOI] [PubMed] [Google Scholar]
- 35.Pim, D., A. Storey, M. Thomas, P. Massimi, and L. Banks. 1994. Mutational analysis of HPV-18 E6 identifies domains required for p53 degradation in vitro, abolition of p53 transactivation in vivo and immortalisation of primary BMK cells. Oncogene 9:1869-1876. [PubMed] [Google Scholar]
- 36.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 37.Tasaki, T., et al. 2005. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25:7120-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas, J. T., and L. A. Laimins. 1998. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J. Virol. 72:1131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, M., et al. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, M., G. Matlashewski, D. Pim, and L. Banks. 1996. Induction of apoptosis by p53 is independent of its oligomeric state and can be abolished by HPV-18 E6 through ubiquitin mediated degradation. Oncogene 13:265-273. [PubMed] [Google Scholar]
- 41.Thomas, M., et al. 2008. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 27:7018-7030. [DOI] [PubMed] [Google Scholar]
- 42.Tomaić, V., et al. 2009. Human and primate tumour viruses use PDZ binding as an evolutionarily conserved mechanism of targeting cell polarity regulators. Oncogene 28:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomaić, V., D. Pim, and L. Banks. 2009. The stability of the human papillomavirus E6 oncoprotein is E6AP dependent. Virology 393:7-10. [DOI] [PubMed] [Google Scholar]
- 44.Vasquez, R. J., B. Howell, A. M. Yvon, P. Wadsworth, and L. Cassimeris. 1997. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell 8:973-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vos, R. M., J. Altreuter, E. A. White, and P. M. Howley. 2009. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J. Virol. 83:8885-8892.19553310 [Google Scholar]
- 46.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]