Abstract
After fixation in the human genome, human endogenous retroviruses (HERVs) are bona fide cellular genes despite their exogenous origin. To be able to spread within the germ line and the early embryo, the ancient retroviral promoters must have adapted to the requirements for expression in these cell types. We describe that in contrast to the case for current exogenous retroviruses, which replicate in specific somatic cells, the long terminal repeat (LTR) of the human endogenous retrovirus HERV-K acts as a TATA- and initiator element-independent promoter with a variable transcription start site. We present evidence that the HERV-K LTR is regulated by the transcription factors Sp1 and Sp3. Mutating specific GC boxes, which are binding sites for Sp proteins, and knocking down Sp1 and Sp3 by use of small interfering RNA (siRNA) significantly reduced the promoter activity. Binding of Sp1 and Sp3 to the promoter region was confirmed using electrophoretic mobility shift assays (EMSAs) and chromatin immunoprecipitation (ChIP). Our data explain why certain HERV-K proviruses have lost promoter competence. Since vertebrate promoters lacking canonical core promoter elements are common but poorly studied, understanding the HERV-K promoter not only will provide insight into the regulation of endogenous retroviruses but also can serve as a paradigm for understanding the regulation of this class of cellular genes.
Human endogenous retroviruses (HERVs) bear witness that during primate/human evolution exogenous retroviruses have repetitively infected and colonized the germ lines of their respective hosts. HERV sequences constitute approximately 8% of the human genome. However, all present-day proviral loci in the human lineage are rendered noninfectious by mutations and deletions, probably through genetic drift and—given the mutagenic potential of retroviruses—selection for replication-incompetent proviruses. In addition, detailed in silico analyses showed that several HERV proviruses are already inactivated during the primary infection cycle by an APOBEC3G cytosine deaminase, an antiretroviral gene which leaves specific mutation marks within the proviral DNA (17, 33). The diverse effects of the HERV load on the human genome, either beneficial or detrimental, have been summarized in several comprehensive reviews (5, 13, 30, 46, 51, 55).
Among the HERVs, the betaretrovirus subgroup HERV-K/HML-2 (HERV-K), is unique in the respect that several proviruses have retained open reading frames for some, if not all, retroviral proteins and are still able to form retrovirus-like particles (7, 36, 44, 45). HERV-K is a complex retrovirus carrying, inter alia, the rec gene, encoding a nuclear RNA export adapter essential for the expression of the viral proteins (37). Although not interchangeable, rec is a functional homologue of the HIV rev, human T-cell leukemia virus (HTLV) rex, and mouse mammary tumor virus (MMTV) rem genes. rec has been deleted in most of the HERV-K elements (type 1 proviruses) but is present in the majority of the youngest human-specific integrants (type 2 proviruses) (11, 18). Although they are repressed in somatic tissues, HERV-K proteins are reexpressed in certain tumors, most prominently germ cell tumors (GCTs) and melanomas. Such expression can induce T- and B-cell immune responses (21, 22, 56). We found that in melanoma patients, anti-HERV-K antibodies are inversely correlated with survival time and thus are not only a diagnostic marker but also a prognostic marker for disease progression (21). Similar results were described for patients suffering from germ cell tumors (27). It has been postulated that expression of HERV-K proteins may directly contribute to malignant transformation in melanomas (49, 58). In germ cell tumors, the Rec protein potentially influences the onset and/or progression of the disease owing to its interaction with the tumor suppressor proteins PLZF and TLZF, which in turn leads to the activation of proto-oncogenes such as c-myc (26, 55).
In the present study, we aimed to understand how the expression of HERV-K is regulated mechanistically in these tumor cells. Transcription of exogenous retroviruses, which infect somatic cells, is usually initiated by a TATA box motif and other core promoter elements. Regulation of expression occurs in a tissue-specific manner, by ubiquitous as well as tissue-specific or even virally encoded transcription factors. Little is known yet about the transcriptional regulation of HERV-K proviruses. In somatic tissues, their expression seems to be tightly repressed, and it is generally supposed that epigenetic mechanisms play a major role. For human CD4+ cells, high-throughput sequencing data displayed enrichment of HERV-K elements in chromatin carrying the inactivating histone marks H3K9 and H3K79me3 (12). Similarly, transcription of murine endogenous retroviruses (muERVs) is silenced already, early during embryogenesis, by epigenetic modifiers which induce extensive CpG methylation of the proviral DNA and/or establish repressive histone marks associated with heterochromatin (8, 43, 63). Deletion of the H3K9 methyltransferase ESET or the retroviral restriction factor TRIM28, which binds to ESET, reactivated muERV expression (43). Carcinogenesis is known to perturb gene regulation and to alter epigenetic marks and thus could lead to reexpression of silenced genes. Indeed, in germ cell tumor and melanoma cell lines, the amount of HERV-K transcription was found to be correlated inversely with the density of CpG methylation in certain proviral promoters (62). In those cell lines, it was possible to enhance the amount of HERV-K transcripts with 5-azacytidine (5′-Aza), an inhibitor of DNA methyltransferases (20). However, the fact that the completely unmethylated promoter of the youngest fixed HERV-K provirus was transcriptionally silent in T47D cells argues that in addition to epigenetic mediators, transcription factors play an important role in HERV-K expression (32). In the study presented here, we characterize the HERV-K promoter and its regulation in more detail. In agreement with previously published data (29), we observed a number of slightly dispersed transcription start sites (TSS) in addition to a major TSS. We demonstrate that in contrast to exogenous retrovirus long terminal repeats (LTRs), the HERV-K LTR is not regulated by canonical core promoter elements. We present evidence that, instead, the HERV-K promoter is dependent on Sp1 and Sp3 proteins, and we identify essential binding sites for the Sp transcription factors. Our data explain why certain HERV-K proviruses have lost promoter competence. Transcriptionally active endogenous retroviruses seem to be regulated similarly to genes expressed in pluripotent stem cells, by epigenetic DNA and histone modifications as well as specific transcription factors. Tumors which mirror, at least in part, such a genetic and cellular environment may support their reexpression, although additional specific activating or repressing factors have to be postulated, as HERV-K proteins are expressed in some tumor types and not in others.
MATERIALS AND METHODS
Plasmids.
HERV-K LTRpck30 and LTR21 were cloned into the HindIII restriction site of pGL3_basic by use of pSVOAL vectors containing various HERV-K LTRs (unpublished data; see reference 28). To generate the 3′-deletion constructs, LTRpck30 was transferred to the HindIII site of the vector pBluescript II KS (Stratagene). The sequence pck30_30dG was cut from the pBS-pck30 plasmid with KpnI and SmaI and ligated into pGL3 cut with the same enzymes. For the pGL3_pck30dH construct, the restriction enzymes KpnI and BamHI were used, for pGL3_pck30dI KpnI and BclI were used, and for pGL3_pck30dK KpnI and NsiI were used; after generation of blunt ends, the fragments were cloned into pGL3_basic digested with KpnI. To produce pGL3_pck30dA, a part of the LTR was cut out with Bsu36I and EcoRI. After religation of the vector, the plasmid was digested with HindIII and the LTR sequence was cloned back into the HindIII-digested acceptor plasmid pGL3_basic. The mutated constructs were generated with a QuikChange II site-directed mutagenesis kit (Stratagene). All HERV-K-specific primers used in this publication are listed below. pGL3_pck30meth was methylated for 1 h at 37°C, using the SssI methylase (NEB) and S-adenosylmethionine. The methylated DNA was purified by agarose gel electrophoresis and eluted with a gel extraction kit (Qiagen).
Primers.
All primer sequences are given in the 5′-to-3′ direction. Primers used to create mutation constructs were as follows: Sp_1*_FW, AAACCACCTTAGGGC; Sp_1*_RV, AGCCCTAAGGTGGTTT; Sp_2*_FW, ACCTGCAGGCAGCAAT; Sp_2*_RV, ATTGCTGCCTGCAGGT; Sp_3*_FW, CACATCTCCCTCGAGA; Sp_3*_RV, TCTCGAGGGAGATGTG; Sp_4*_FW, GGCTGGTGGGATCCTCCA; Sp_4*_RV, TGGAGGATCCCACCAGCC; pck30dT_FW, GGAATGTCTCGGTTTTTTTCCCGATTGTATGC; pck30dT_RV, GCATACAATCGGGAAAAAAACCGAGACATTCC; Inr_FW, CTCTTCGAGAAACACCCACAGATGAAAAAAAAATACTAAGGGAACTCAGAGGCTG; and Inr_RV, CAGCCTCTGAGTTCCCTTAGTATTTTTTTTTCATCTGTGGGTGTTTCTCGAAGAG. Primers used for PCR investigations after rapid amplification of cDNA ends (RACE) were as follows: B2_new, GAAAAGCCTCCACGTTGGGCACCA; Lys1,2, GCCCCAGGTTGGGCGCCA; MKr17, CGGGCGCAACTGCAACTCCGATAAATAACG, and Env-sense, CCAGACTCCCAGACTATAACCTGTG. Primers used to generate electrophoretic mobility shift assay (EMSA) probes were as follows: pck30-1_FW, AATTGTCTTGTGACCCTGACACATCCCCCTCTTCGAGAAACACCCACAGATGATCAGTAAATACTA; pck30-1_RV, TAGTATTTACTGATCATCTGTGGGTGTTTCTCGAAGAGGGGGATGTGTCAGGGTCACAAGACAATT; LTR21_FW, AATTGTCTTGTGACCCTGACACATCTCCCTCTAGGAGAAACACCCACAGATGATCAGTAAATACTA; and LTR21_RV, TAGTATTTACTGATCATCTGTGGGTGTTTCTCCTAGAGGGAGATGTGTCAGGGTCACAAGACAATT. Primers used to analyze chromatin immunoprecipitation (ChIP) fragments were as follows: ChIP-LTR-FW, ATGTTTGTCTGCTGACCCTCTCC; ChIP-Env-FW, GTGCACAAGTGAGTCCAGCTGTT; and ChIP-Env-RV, CATATAAGGGGGCTTTACGCAA.
Luciferase reporter assays.
Luciferase assays were performed using a dual-luciferase assay system (Promega) according to the manufacturer's instructions. A total of 0.5 × 106 cells were seeded in 6-well plates and incubated for 1 or 2 (GH cells) days. Cells were transfected with 0.9 μg of the respective pGL-vector (encoding firefly luciferase) and 0.1 μg of the phRL vector (encoding Renilla luciferase driven by a thymidine kinase promoter), using the GeneJuice transfection reagent (Merck) according to the recommendations of the supplier. Relative light units (RLU) were measured in a microplate luminometer (MicroLumat Plus LB96V; Berthold). For normalization, firefly luciferase light units were divided by Renilla luciferase light units. For luciferase assays with Sp protein-ablated cells, we applied 150 ng small interfering RNA (siRNA), using the HiPerfect transfection reagent (see “Sp protein immunodetection and knockdown”). Twenty-four hours after siRNA transfection, cells were transfected with luciferase constructs by using GeneJuice as described above. After 24 h, luciferase activity was assayed using the dual-luciferase reagents and protocol.
5′- and 3′-RACE.
Poly(A)-selected RNA was isolated with a PolyATract mRNA isolation system (Promega). The TSS and termination sites of HERV-K were identified using a GeneRacer kit with SuperScript III reverse transcriptase (RT) (Invitrogen) according to the manufacturer's instructions. For cDNA synthesis and for the first PCR primer, B2new was used, and nested PCR was performed with Lys1,2. Reaction products were analyzed by agarose gel electrophoresis, purified using a Minelute gel extraction kit (Qiagen), cloned into the pGEM-T Easy vector (Promega), and sequenced. The TSS of pGL3_pck30 was determined by transfecting GH cells with vector DNA. The luciferase-specific primer MKr17 was used for cDNA synthesis and PCR amplification. For the 3′-RACE approaches, cDNA synthesis was performed using a GeneRacer oligo(dT) primer, GeneRacer 3′ primer, and Env sense primer for PCRs.
Sp protein immunodetection and knockdown.
Cells were lysed in triple lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% desoxycholate) containing Complete protease inhibitor cocktail (Roche). The lysates were cleared by centrifugation. Total protein was determined by Bio-Rad protein assay. Thirty micrograms of each sample was mixed with SDS loading dye. After boiling, samples were subjected to gel electrophoresis as previously described (21). Membranes were incubated in blocking buffer (phosphate-buffered saline containing 0.1% Tween and 4% nonfat milk) overnight at 4°C. Sp1 (Pep2) and Sp3 (D-20) antibodies from Santa Cruz and a beta-actin antibody (AC74; Sigma) were used. All antibodies were incubated for 1 h in blocking buffer. Antibody binding was visualized using ECL chemiluminescence detection solution (Amersham).
Translation of Sp1 and Sp3 was silenced with the siRNA duplexes Hs_SP1_1_HP and Hs_SP3_1_HP (Qiagen), respectively. siRNA against the HIV capsid, with the target sequence 5′-AAGATTGTACTGAGAGACAGG-3′, was used as an unrelated control. Applying the HiPerfect transfection reagent (Qiagen) according to the manufacturer's protocol, cells were transfected with 150 or 300 ng siRNA per 0.5 × 106 cells. Forty-eight hours after transfection, cells were harvested for detection of Sp1 and Sp3 proteins by immunodetection.
Cell lines, ChIP, and EMSA.
The human teratocarcinoma cell line GH was established and grown as described previously (36). The cell line Mel-C9 was subcloned from UKRV Mel 2 by N. Fuchs (unpublished data). UKRV Mel 2 and HCl Mel 19 are human melanoma cell lines kindly provided by D. Schadendorf (University Hospital, Essen, Germany). All melanoma lines were grown in RPMI 1640 supplemented with 10% fetal calf serum.
A ChIP-IT Express kit (Active Motif) was used according to the supplied protocol. The samples were lysed and subjected to enzymatic shearing. DNA digestion was performed overnight at 37°C, using 15 U each of the restriction enzymes HindIII, NspI, and AflIII in buffer 2 containing bovine serum albumin (BSA) (New England Biolabs). Two micrograms of specific antibody for Sp1 (Pep2) or Sp3 (D-20) (Santa Cruz) or of beta-actin antibody (AC74; Sigma) was used for each immunoprecipitation, with overnight incubation. After elution of the digested chromatin, reversion of cross-linking, and proteinase K treatment, immunoprecipitated DNA was detected by PCR, using 5 μl eluate as a template. For amplification of the HERV-K LTR, the ChIP-LTR-FW and B2_new primers were used. PCR was performed with Taq polymerase (Applied Biosystems) and the following cycle conditions: 1 cycle of 94°C for 5 min; 36 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 40 s; and 1 cycle of 72°C for 10 min. Primers targeting the HERV-K env gene were used as a control (ChIP-Env-FW and ChIP-Env-RV). PCR conditions were the same as those for the LTR sequence, except that the annealing temperature was decreased to 55°C. Fractionated protein extracts for EMSA were prepared as described previously (28). The probe was generated by annealing of the complementary oligonucleotides pck30-1_FW and pck30-1_RV at equimolar concentrations. The double-stranded oligonucleotides were 5′-end labeled using T4 polynucleotide kinase (NEB) and [γ-32P]ATP (3,000 Ci/mmol; Amersham).
Statistics.
The hypothesis of a significant deviation from 100% (pGL3_pck30 vector) for TATA box mutants, initiator element (Inr) mutants, and siRNA was tested for each cell line and group separately by means of a t test, using log percentages. For GC boxes and 3′ deletions, analysis of variance (ANOVA) was performed, with an additional random factor day included in the model accounting for the high variability between some experiments. For statistically significant results, the following convention was used: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Analysis was performed with SAS/STAT software, version 9.2 (SAS System for Windows).
RESULTS
HERV-K LTR promoter activity is cell type dependent and is inactivated by point mutations and CpG methylation.
To characterize the promoters of endogenous HERV-K proviruses, we chose two LTRs, LTRpck30 and LTR21, from a selection of HERV-K promoter sequences previously isolated and analyzed in our laboratory. These two LTRs differ by 5.6% of their nucleotides (Fig. 1). LTRpck30 was chosen because we had previously identified and extensively studied the Rec-responsive element (RcRE) sequence of this LTR (39, 40). To obtain an active promoter, we had combined in LTRpck30 the 3′ U3/R and 5′ R/U5 regions of a HERV-K cDNA clone generated from HERV-K mRNA isolated from the GCT cell line GH (39). An NCBI Blast search (http://www.ncbi.nlm.nih.gov) identified the HERV-K provirus 108 on chromosome 7 as the source of LTRpck30. HERV-K 108 is human specific (6), and its LTR was recently confirmed to be an active promoter in luciferase reporter assays (62). LTR21 was amplified by PCR, together with a variety of other LTR sequences, from genomic DNA isolated from GH cells (unpublished data). To avoid the amplification of 3′-LTRs and solitary LTRs, we chose a sense primer located in the beginning of the U3 region and an antisense primer spanning the primer binding site (PBS). The LTR21 sequence is most similar to the 5′-LTR of HERV-K 110/18 (48) on chromosome 1, differing by only 9 point mutations. This divergence may reflect single-nucleotide polymorphisms similar to those described for two other HERV-K proviruses (24) or may have resulted from PCR amplification. HERV-K 18 is evolutionarily older, as it is also present in most hominoids (6).
FIG. 1.
Nucleotide alignment of HERV-K LTRs. Identical nucleotides are indicated by dots, and variations are shown by the one-letter code for nucleotides. The insertion/deletion of a nucleotide is depicted by a dash. GC boxes 1 to 4 are boxed, and the RcRE sequence is marked by a black line. The binding sequence for the yin yang transcription factor (YY1) is shown in gray. TSS, transcription start site; TTS, transcription termination site.
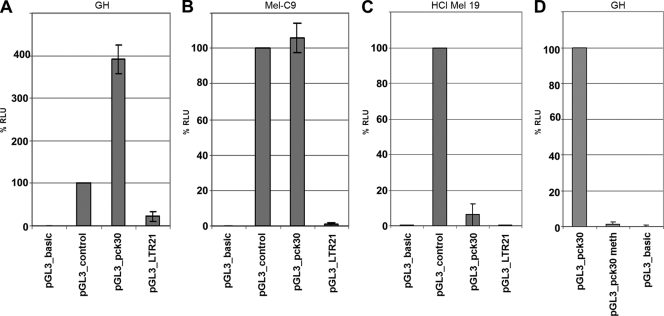
We cloned the LTRs into the promoterless luciferase reporter vector pGL3_basic to analyze their promoter strengths in various transiently transfected cell lines, using a dual-luciferase assay system (Fig. 2). In contrast to LTRpck30, LTR21 showed only very weak activity in all cell lines, indicating that the LTRs of certain HERV-K proviruses have been inactivated by point mutations during the course of evolution. Importantly, LTRpck30 was active in the cell lines expressing HERV-K proteins, the GCT line GH and the melanoma line Mel-C9, but not in the HERV-K-negative melanoma line HCl Mel 19. In GH cells, LTRpck30 was even significantly more active than the simian virus 40 (SV40) control promoter. In agreement with a previous publication describing that reactivation of HERV-K Gag expression in Raji cells by treatment with 5′-Aza and trichostatin A failed (20), we could not reactivate HERV-K transcription in HCl Mel 19 cells by using the same agents. These findings indicate that the erasure of inactivating epigenetic marks by chemical agents or during carcinogenesis is insufficient to reactivate HERV-K transcription if the necessary cell line-specific transcription factors are lacking. Conversely, when we transfected GH cells with pGL3_pck30 DNA treated with the methyltransferase SssI to generate methylated CpG dinucleotides, we observed a complete loss of the luciferase activity (Fig. 2D). This result confirms previous data (62) showing that HERV-K promoters are silenced by epigenetic modifications despite the presence of transcription factors which principally support HERV-K transcription. Thus, both epigenetic and transcription factors regulate HERV-K expression.
FIG. 2.
HERV-K LTR reporter plasmids analyzed with a dual-luciferase reporter system. GH (germ cell tumor) (A), Mel-C9 (melanoma) (B), and HCl Mel 19 (melanoma) (C) cells were transfected with pGL3_pck30, pGL3_LTR21, pGL3_basic (promoterless), or pGL3_control (SV40 promoter) DNA. (D) GH cells were also transfected with pGL3_pck30 DNA treated with the CpG dinucleotide-methylating enzyme SssI (pGL3_pck30meth). % RLU are shown as means ± standard deviations (SD) for six (A to C) or three (D) individual transfections. LTR21 was inactive in all cell lines (A to C, 4th bars), and LTRpck30 was active in GH and Mel-C9 cells (A and B, 3rd bars) but was silenced upon methylation of CpG (D, 2nd bar).
The HERV-K LTR acts as a TATA-less promoter, and analysis by 5′- and 3′-RACE defines the authentic U3, R, and U5 regions.
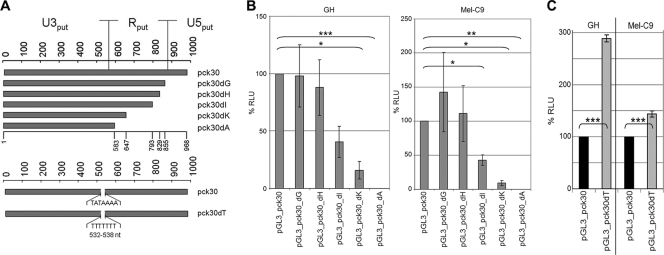
In retrovirus LTRs, the U3 region usually harbors the core promoter elements. Based on the hypothesis that transcription is initiated by a putative TATA box motif at nucleotide positions 532 to 538, the structural parts of the HERV-K LTR were previously predicted by computational comparisons (48). To identify the HERV-K core promoter experimentally, we generated a series of consecutive 3′-deletion LTR constructs (Fig. 3 A) and tested their activity in GH and Mel-C9 cells (Fig. 3B). Deletion of up to 139 nucleotides (nt) (pGL3_pcK30dG and pGL3_pcK30dH) had no statistically significant effect. Thus, we could not confirm the existence of a negative regulatory element within this region (15). Further 3′ deletions, however, significantly decreased the promoter activity in both cell lines. pGL3_pck30dI produced approximately 40% of the relative luciferase counts measured with pGL3_pck30, and pGL3_pck30dK produced only about 20% of the relative counts. No promoter activity was detected for pGL3_pck30dA. These results were unexpected, as the pGL3_pck30dA construct still included the predicted U3 region and the putative core promoter. Therefore, we analyzed whether the TATA box was at all functional. The TATAAAA motif was changed to a heptamer of thymidines, generating the construct pGL3_pck30dT. This mutation did not decrease promoter function in transiently transfected GH and Mel-C9 cells. Instead, we observed significantly increased transcriptional activity (Fig. 3C), indicating that mutating the motif may have abolished the binding of a transcriptional repressor or may have enabled the binding of an enhancing factor. All constructs tested remained inactive in HCl Mel 19 cells (data not shown). Based on these experimental results, we hypothesized that the U3 region comprised nt 1 to 793 within LTRpck30dI.
FIG. 3.
The HERV-K LTR is a TATA-less promoter with a core promoter different from the predicted one. (A) Putative HERV-K LTR structure predicted in the literature and 3′-deletion constructs. The putative TATA box motif is indicated. (B) Promoter competence of the 3′-deletion constructs was determined in GH and Mel-C9 cells and compared to that of the pGL3_pck30 promoter. % RLU are shown as means ± SD for nine individual transfections. The 3′ deletions within the putative R region induced significant losses of promoter activity (pGL3_pcK30dI and -dK). No promoter activity was seen with the predicted U3 core promoter region (pGL_pck30dA). (C) When the TATA motif was mutated to TTTTTTT (pck30dT), the promoter function was significantly enhanced in GH and Mel-C9 cells.
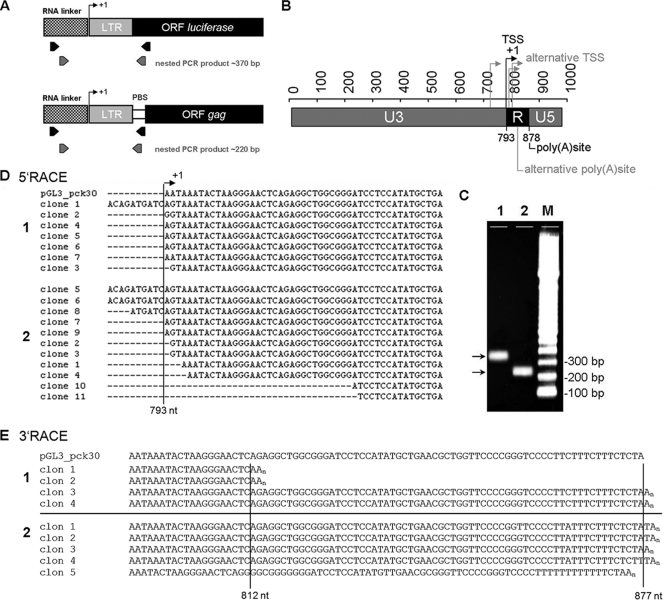
To determine the exact TSS of the HERV-K promoter by 5′-RACE, we used two complementary approaches. For consistency with the luciferase promoter assays, the TSS from the promoter construct pGL3_pck30 was defined using RNA from transfected GH cells and downstream primers located within the luciferase gene. To identify the TSS of HERV-K proviruses reactivated in germ cell tumors and melanomas, we used RNAs from untransfected cells and downstream primers spanning the PBS (Fig. 4 A). The resulting amplification products were cloned and sequenced (Fig. 4C and D). We identified a major TSS at nt 793 and observed that the TSS of the pGL3_pck30 plasmid was identical to the major TSS of HERV-K proviruses expressed in GH and Mel-C9 cells. Some RACE clones, especially those from Mel-C9 cells, showed slightly different TSS, ranging from up to 10 nt upstream to approximately 30 nt downstream of the major TSS. This variation in TSS is a feature often observed in TATA-less promoters. The TSS defines the boundary between the U3 and R regions (Fig. 4B). The data precisely corroborate the results obtained in the transient reporter assay using 3′-deletion mutants. The pGL3_pck30dI construct, which exerts a moderate activity that can be expected for a viral core promoter, indeed harbors the U3 region as defined by 5′-RACE. Thus, the U3 core promoter region is 233 nt longer than the predicted one. The boundary of the R and U5 regions is defined by the polyadenylation site in the 3′-LTR. Our group previously published that the polyadenylation site of HERV-K proviruses expressed in GH cells is located at nt 877, 84 nt downstream from the polyadenylation signal AATAAA (36). This polyadenylation site was confirmed in Mel-C9 cells by 3′-RACE (Fig. 4E), using PCR primers targeting a region within the HERV-K env gene. A second polyadenylation site, at position 812, only 19 nt downstream from the polyadenylation signal, was also observed in Mel-C9 cells (Fig. 4E). In summary, the U3 region—the core promoter—comprises nt 1 to 792, and the R region spans nt 793 to 876 (a total of 84 nt), starting with the polyadenylation signal. The U5 region spans nt 877 to 968 (a total of 92 nt). Transcription start and termination sites are more precisely regulated in germ cells than in melanoma cells.
FIG. 4.
Definition of HERV-K LTR structure by 5′- and 3′-RACE. (A) Locations of the PCR primers used for amplification in 5′-RACE experiments. For endogenous transcripts, the reverse primer binds to the HERV-K PBS, whereas for amplification of cDNAs from pGL3_pck30 transfectants, the reverse primer binds within the luciferase gene. (B) Schematic presentation of redefined U3, R, and U5 regions. +1, major TSS at nt 793 in the 5′-LTR. The transcription termination site in the 3′-LTR is also plotted. Alternative TSS and termination sites are indicated in a light shade. (C) 5′-RACE nested PCR products of 370 bp for transcripts from the transfected plasmid pGL3_pck30 (lane 1) and of 220 bp for endogenous HERV-K transcripts (lane 2). (D) Sequences of clones from endogenous transcripts of GH (1) and Mel-C9 (2) cells in comparison to the 5′-RACE product obtained from the transfected pGL3_pck30 plasmid. The first bases transcribed from the pGL3_pck30 plasmid are AAT; transcription of endogenous proviruses started with AGT. This difference resulted from the LTRpck30 construct which combined the U3 region of the HERV-K 108 3′-LTR with the R/U5 region of its 5′-LTR. (E) 3′-RACE clones from Mel-C9 (1) and GH (2) cells, with the major polyadenylation site at nt 877 and a second polyadenylation site at nt 812 compared to the sequence of the pGL3_pck30 plasmid.
The HERV-K LTR does not possess a functional Inr, but Sp1 and Sp3 proteins mediate promoter activity.
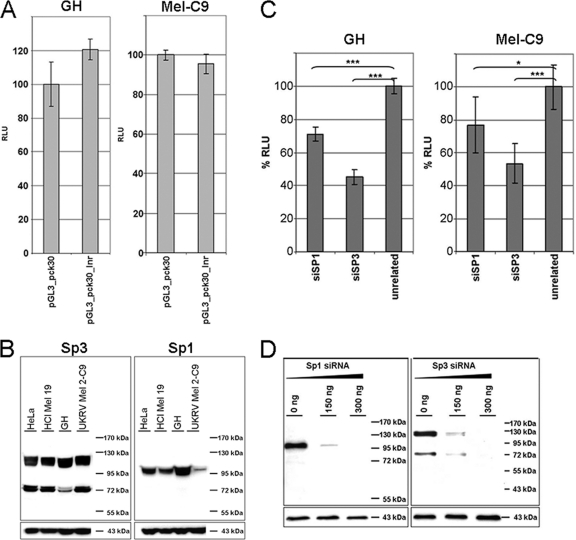
In TATA-less promoters, transcription may be initiated at an Inr (57). We found the sequence TCAGTAA at the major HERV-K TSS, which differs by only the last two nucleotides from the consensus sequence of Inr motifs [YYA+N(T/A)YY] (23). When this putative HERV-K Inr sequence was mutated to a heptamer of adenines, generating the reporter construct pGL3_pck30_Inr, no decrease in promoter activity was observed (Fig. 5 A). Thus, HERV-K transcription initiation in GH and Mel-C9 cells is not mediated by either an initiator or a TATA motif. It appears that HERV-K endogenization induced a switch from conventional LTR regulation by core promoter elements to an alternative form of regulation.
FIG. 5.
The HERV-K LTR does not possess a functional initiator element, but silencing of Sp1 and Sp3 reduces pGL3_pck30 promoter activity. (A) HERV-K transcription initiation is not mediated by an initiator element (Inr). The putative Inr wild-type sequence, TCAGTAA, was changed into a heptamer of adenines in the LTR reporter pGL3_pck30_Inr construct. Luciferase activities of pGL3_pck30 (wild type) and the pGL3_pck30_Inr mutant were determined in GH and Mel-C9 cells at 24 h posttransfection. The promoter activity of LTRpck30 was set to 100% for each cell line. The results are means ± SD for six individual experiments. (B) Immunoblot analysis of endogenous Sp1 and Sp3 protein levels in HeLa, HCl Mel 19, GH, and Mel-C9 cell lines. Fifty micrograms of cell lysate was used. Actin served as a loading control. (C) GH and Mel-C9 cells were cotransfected with pGL3_pck30 DNA and 150 ng specific siRNA or with 150 ng unrelated siRNA. % RLU are given as means ± SD for three individual transfections. The mock level was set to 100%. HERV-K LTR activity was significantly reduced by Sp1 as well as Sp3 ablation. (D) Sp1 or Sp3 knockdown reduced HERV-K promoter activity. Mel-C9 and GH cells were transfected with 150 or 300 ng of either Sp1- or Sp3-specific siRNA. Immunoblot analysis using 30 μg of protein revealed that the amounts of Sp1 and Sp3 proteins were significantly reduced by 150 ng of the respective siRNA at 48 h posttransfection. Actin was used to control for equal loading of cell lysates.
Transcription initiation of TATA-independent cellular promoters can be mediated by the ubiquitous transcription factors Sp1 and Sp3. Sp1, for instance, has been shown to tether the transcription preinitiation complex to a TATA-less promoter by interacting with a component of TFIID (60), a complex composed of the TATA binding protein (TBP) and TBP-associated factors. We determined the levels of Sp1 and Sp3 proteins in the cells used in this study by immunoblot analysis (Fig. 5B). The amount of Sp1 protein was largest in GH cells and was very small in Mel-C9 cells. Four isoforms of Sp3 exist, with two long isoforms harboring two activation domains and two shorter isoforms with only one activation domain (reviewed recently in reference 34). Both melanoma lines expressed similar levels of all Sp3 isoforms. In GH cells, however, the amounts of both short Sp3 isoforms were reduced significantly.
To elucidate the roles of Sp1 and Sp3 in HERV-K promoter function, we first analyzed if ablation of Sp1 or Sp3 by RNA interference reduced HERV-K LTR activity in luciferase reporter assays. The functionality of the siRNA (Qiagen) directed against Sp1 or Sp3 was confirmed by Western blot analysis. A total of 150 ng of siRNA effectively knocked down the Sp1 or Sp3 protein level in Mel-C9 and GH cells, as shown for Mel-C9 cells in Fig. 5D. Subsequently, the luciferase reporter plasmid pGL3_pck30 was cotransfected with 150 ng siRNA into GH or Mel-C9 cells and compared to transfections with unrelated siRNA. Upon Sp1 or Sp3 knockdown, we observed a significant but not complete reduction in promoter activity in both cell lines (Fig. 5C). Thus, both Sp transcription factors mediate the transcriptional activity of the HERV-K LTR and may, at least partly, substitute for each other.
In vivo and in vitro binding of Sp1 and Sp3 proteins to the HERV-K LTR.
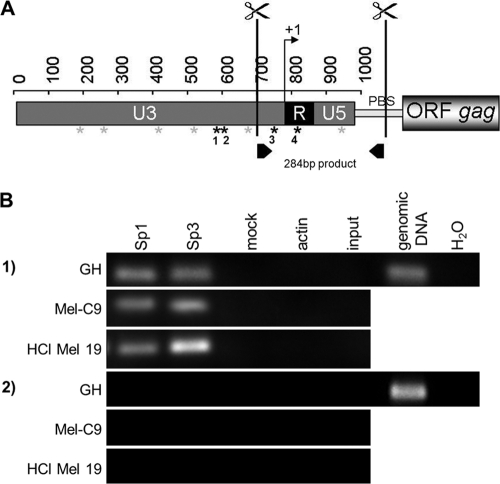
HERV-K LTR inspection using the Transcription Element Search System (TESS), an Internet tool for predicting transcription factor binding sites in DNA sequences (http://www.cbil.upenn.edu/cgi-bin/tess/tess), suggested 10 putative binding sites for the transcription factors Sp1 and Sp3. Six of these GC boxes have identical sequences in LTRpck30 and LTR21 (Fig. 6 A, gray asterisks), but four GC boxes differ between LTRpck30 and LTR21 (Fig. 1, boxed areas, and Fig. 6A, black asterisks). Two of these boxes are located approximately 30 nt upstream and downstream of the TSS. The other two are clustered in the region from nt 570 to 600.
FIG. 6.
Sp1 and Sp3 bind to HERV-K LTRs in vivo. (A) ChIP assays were performed, with cutting of proviral HERV-K LTRs at the indicated sites. Putative binding sites for Sp1/Sp3 are depicted by asterisks; black asterisks indicate sequence differences between LTRpck30 and LTR21 in four Sp1/Sp3 binding motifs. (B) Immunoprecipitation was performed with antibodies directed against Sp1, Sp3, or actin. Specific PCR primers excluding amplification of products from solitary LTRs were used to analyze the precipitates (see panel A; PBS denotes the primer binding site), as well as env-specific primers. PCRs on digested and cross-linked fragments before the immunoprecipitation step (input) and on mock IPs served as negative controls, as well as PCR without a template (H2O). Genomic DNA from GH cells was amplified as a positive control. ChIP with anti-Sp1 and anti-Sp3 antibodies yielded PCR amplicons with LTR-specific primers but not env-specific primers.
To illustrate that the transcription factors Sp1 and Sp3 indeed bind to the promoters of endogenous HERV-K proviruses, we performed ChIP assays using GH, Mel-C9, and HCl Mel 19 cells. Focusing on the putative GC boxes in the vicinity of the TSS, we designed a strategy that favors analyses of proviral 5′-LTRs over proviral 3′-LTRs and solitary LTRs. After formaldehyde cross-linking and cell lysis, the DNA was digested with two restriction enzymes cutting most proviral HERV-K 5′-LTRs at positions 710 (U3) and 1078 (5′-UTR) to generate a fragment of optimal size for ChIP (Fig. 6A). The protein-DNA complexes were subjected to immunoprecipitation, using anti-Sp1 or anti-Sp3 antibody as well as anti-actin antibody as a control. Cross-linking was reversed and the precipitated DNA purified. The region spanning nt 710 to 995 was then amplified by PCR. For comparison, a coding region of similar size was analyzed for the HERV-K env gene (nt 7301 to 7615). For all cell lines, amplicons of the 284-bp LTR fragment were detected in the DNAs isolated from precipitations with anti-Sp1 and anti-Sp3 antibodies (Fig. 6B, panel 1). Sequencing confirmed that the amplicons were indeed derived from HERV-K proviruses (data not shown). In contrast, the primer pair specific for HERV-K env sequences never yielded any PCR product (Fig. 6B, panel 2). No PCR signal was detected for precipitations with anti-actin antibodies or using the cross-linked and digested protein-DNA complexes (mock IP and input control), whereas a genomic template yielded the expected positive results. In the setting of the ChIP protocol used here, a positive signal should occur only after IP enrichment, explaining why the input control was negative. With HCl Mel 19 cells, the anti-Sp3 ChIP generated a stronger PCR signal than those with GH and Mel-C9 cells. This finding suggests that Sp3 may contribute to the silencing of HERV-K proviruses in HCl Mel 19 cells, as Sp3 can enhance or repress transcription, depending on posttranslational modifications and/or the presence or absence of coactivators and corepressors. Although we could demonstrate by ChIP that the transcription factors Sp1 and Sp3 bind to the examined promoter region of HERV-K proviruses in vivo, the approach was not suitable to determine which of the GC boxes was functional.
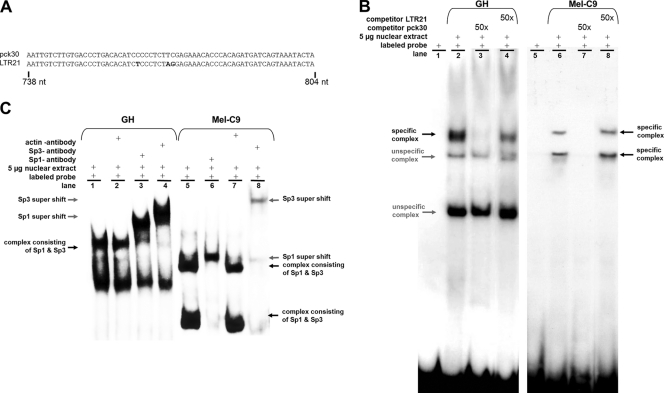
We hypothesized that GC boxes 1 to 4, which are mutated in the inactive LTR21, are functional in LTRpck30. We first analyzed GC box 3 because it is located only 36 nt upstream of the TSS and because its deletion significantly reduced the HERV-K promoter activity (the 3′-deletion construct pck30dk had only 10 to 20% of the wild-type activity [Fig. 3B, 5th bars]). To investigate whether this GC box indeed constitutes a binding site for the transcription factors Sp1 and Sp3, we performed gel retardation analyses using PCR-generated amplicons comprising the sequences depicted in Fig. 7 A. Incubation of the labeled LTRpck30 probe with nuclear extracts isolated from GH and Mel-C9 cells revealed several DNA-protein complexes (Fig. 7B). The high-molecular-mass complexes marked by bold arrows indicate specific binding of proteins to the GC box, since they could be competed with a 50-fold molar excess of the unlabeled LTRpck30 probe but not with the unlabeled LTR21 probe. Nuclear extracts from GH cells formed a single specific DNA-protein complex; nuclear extracts from Mel-C9 cells induced two shifts of the probe. To demonstrate the presence of Sp1 or Sp3 within the specific DNA-protein complexes, we performed supershift experiments using Sp1- and Sp3-specific antibodies and anti-actin antibody (negative control). All of the specific complexes observed with GH and Mel-C9 nuclear extracts shifted in the presence of Sp1 as well as Sp3 antibody (Fig. 7C). No supershift was induced with anti-actin antibody. These assays demonstrate that both Sp1 and Sp3 bind to the examined GC box, probably as a heteromer. The significance of the two specific Sp1/Sp3-containing complexes formed with Mel-C9 nuclear extracts remains elusive. They may contain different additional proteins or different Sp3 isoforms, or they may represent a different stoichiometry of Sp1 and Sp3 within the DNA-protein complex.
FIG. 7.
In vitro binding of Sp1 and Sp3 proteins to the GC box upstream of the TSS. (A) HERV-K LTR sequences generated by PCR as probes for EMSA. Nucleotide differences are depicted in bold. (B) Nuclear extracts of GH cells or Mel-C9 cells were incubated with the 32P-labeled LTRpck30 probe. Protein-probe complexes were separated by polyacrylamide gel electrophoresis and detected by autoradiography. A 50-fold molar excess of unlabeled LTRpck30 probe competed with the specific complexes (lane 3), but the unlabeled LTR21 probe did not (lane 4). (C) When anti-Sp1 or anti-Sp3 antibody was added to the EMSA reaction mixtures, the respective specific probe-protein complexes (black arrows) were shifted to complexes of higher molecular mass (supershift; gray arrows). Incubation with an anti-actin antibody did not induce a supershift.
Three GC boxes mediate transcriptional activity of the HERV-K LTR.
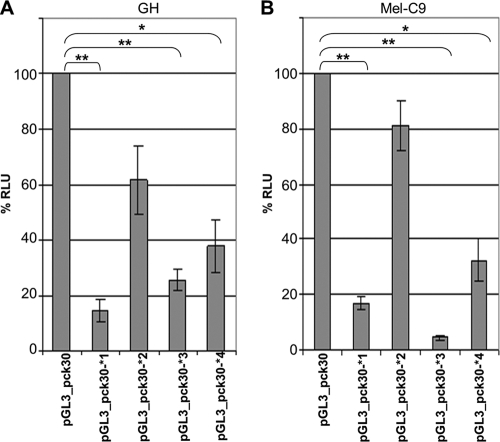
In addition to GC box 3, there are three putative Sp1/Sp3 binding sites which differ between LTRpck30 and LTR21 (Fig. 6A). Deletion of GC box 4 (pGL3_pck30dI) led to significantly reduced promoter activity (Fig. 3B). Deletion of both GC boxes in the vicinity of the TSS (pGL3_pck30dK) further reduced the activity. The pGL3_pck30dA construct, in which three of the GC boxes were missing, had no promoter activity (Fig. 3B). These findings indicated that the GC boxes might have additive effects. To analyze whether individually mutating the four Sp1/Sp3 binding sites would produce similar results to those for the deletions, we introduced the respective GC box motifs from the LTR21 sequence (Fig. 1) into the pGL3_pck30 backbone and measured the effects on the LTR promoter in GH and Mel-C9 cells, using a dual-luciferase reporter assay. The pGL3_pck30*4 construct induced a significant loss of luciferase activity in both cell types (Fig. 8). Mutating GC box 3 (pGL3_pck30*3 construct) had an even more severe effect. This result correlates with the finding that the respective LTR21 sequence did not bind Sp proteins in the EMSA experiments (Fig. 7B, no competition). Both GC box mutants corroborated the data obtained with the respective deletion mutants. Mutation of GC box 2 (pGL3_pck30*2) had only a moderate effect, while mutation of GC box 1 (pGL3_pck30*1) again evoked a drastic reduction of promoter activity. In summary, we have shown by deletion and mutation that the HERV-K promoter is regulated by GC boxes which are binding sites for Sp1 and Sp3. Mutation of GC box 3 had the strongest inactivating effect in Mel-C9 cells, and mutation of GC box 1 had the strongest inactivating effect in GH cells. This observation may indicate that Sp1 and Sp3 interact with coactivators which differ between cell lines and tissues to regulate the HERV-K promoter activity. Alternatively, other LTR regions with transcription factor binding sites may differentially modulate HERV-K promoter activity, depending on the cellular context.
FIG. 8.
Mutation of the four GC boxes affects HERV-K promoter activity in GH and Mel-C9 cells. GC boxes 1 to 4, depicted by black asterisks in Fig. 5A, were individually mutated to the respective LTR21 sequence. The promoter competence of the mutants was determined in GH and Mel-C9 cells and compared to that of the wild-type pGL3_pck30 promoter. % RLU are shown as means ± SD for nine individual transfections. Every mutation evoked a significant loss of luciferase activity. Mutation of GC box 2 had the slightest effect.
We achieved a partial (40% of LTRpck30 activity) reactivation of the LTR21 promoter by exchanging nt 580 to 988 (containing GC boxes 2 to 4) with the respective LTRpck30 sequence. Replacing nt 238 to 988 (containing all four CG boxes) yielded no improvement, and replacing sequences containing only GC box 1 or GC box 4 had no significant effect. We obtained even better promoter activity (80% of LTRpck30 activity) with a chimera harboring a functional YY1 binding site and three intact GC boxes (nt 1 to 283 of LTRpck30 combined with nt 284 to 580 of LTR21 and nt 581 to 988 of LTRpck30) (data not shown). The data corroborate our previous observation that YY1 is an important enhancer of HERV-K promoter activity (28). In our previous publication, we reported that mutation of the YY1 binding site induced a 50% reduction in the relative light units measured in luciferase reporter assays. In summary, these findings indicate that three GC boxes are essential for HERV-K promoter activity, but they also indicate that other transcription factor binding sites, such as the YY1 binding site, positively or negatively regulate HERV-K transcription.
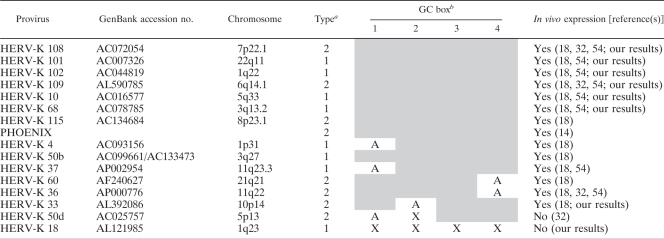
In Table 1, we have compiled data on all proviruses which have been found to be expressed in cell lines or tissues by us (unpublished data) or others (references 18, 32, and 54 and references therein). All transcriptionally active proviruses harbor at least three of the LTRpck30 GC box motifs. Proviruses that are not expressed carry two or more mutant GC boxes displaying either the respective LTR21 motif or a slightly different one. These in vivo data strongly support our finding that the HERV-K promoter is regulated by binding of Sp1 and Sp3 proteins to three essential GC boxes in the vicinity of the transcription start site. Thus, HERV-K proviruses have been inactivated not only by mutations or deletions within the open reading frames but also by direct hits of sensitive motifs in the viral promoter.
TABLE 1.
Activity of LTRs in reporter assays and/or expression of HERV-K transcripts in germ cell tumors and melanomas correlates with the presence of at least three intact GC boxes in the respective proviral promoterc
Presence (2) or absence (1) of the nuclear export adaptor gene rec.
Shading indicates that GC box motifs 1 to 4 are identical to the respective LTRpck30 sequences, “X” indicates that GC box motifs 1 to 4 are identical to the respective inactivating signatures of the LTR21 sequences, and “A” indicates that the GC box motif carries another point mutation.
Only full-length LTR sequences were included in the analysis.
DISCUSSION
The retroviral LTR structure, composed of the U3, R, and U5 regions, is unique owing to its specific assembly during the retroviral replication cycle. The U3 region usually harbors the principal core promoter elements as well as important transcriptional enhancers. Using 5′- and 3′-RACE, we determined the HERV-K LTR composition (Fig. 4) and found that it is significantly different from a previous prediction based on computational comparisons (48). In particular, the U3 region, with a total length of 792 nt, is considerably longer than estimated. This is in agreement with results obtained by studying the TSS within solitary HERV-K LTRs and proviral HERV-K LTRs expressed in human testicular parenchyma specimens (29). Embedded within this region is the RcRE, involved in the Rec-mediated nuclear export of viral transcripts (39). Several contemporary complex retroviruses also display long U3 regions harboring regulatory elements or genes, e.g., the human and simian T-cell leukemia virus U3 regions encode Rex-responsive elements with a function similar to that of the RcRE (2). Unusually, the TSS coincides with the transcription termination signal. We previously published that the termination signal and the termination site are 84 nt apart (36), the actual length of the R region as determined by our RACE approaches.
Furthermore, we have demonstrated that the HERV-K LTR acts as a TATA-independent promoter, despite the presence of a putative TATA box (Fig. 3C). Likewise, a putative initiator motif (61) spanning the transcription start site was not functional (Fig. 5A). No other putative canonical core promoter motifs (25), neither a downstream promoter element nor upstream or downstream TFIIB recognition elements, could be predicted at the expected distances in the HERV-K LTR. Thus, the HERV-K LTR belongs to a class of human promoters which assemble transcription preinitiation complexes independently of any core promoter elements. Such promoters are poorly understood mechanistically but represent the majority of vertebrate promoters and are usually rich in G and C nucleotides (3, 57).
Transcription at TATA-independent promoters may be initiated by transcription factors of the SP/KLF family. In our study, we have shown by RNA interference (RNAi) that the Sp1 and Sp3 proteins mediate HERV-K transcription (Fig. 5C and D) and that both transcription factors bind to promoter sequences in vivo and in vitro (Fig. 6 and 7). The transcription factor Sp1 is involved in the activation of many housekeeping and pluripotency genes and contributes to regulatory processes such as chromatin remodeling and the protection of methylation-free CpG islands, predominantly during early embryogenesis and in germ cells (9, 16, 38). Importantly, these are the cells in which endogenous retroviruses must be able to replicate in order to spread within a host genome.
Using mutation and deletion constructs, we studied four Sp binding sites (GC boxes) which positively influence HERV-K promoter activity (Fig. 3 and 8). Our analyses suggested that LTR21 was inactivated due to point mutations within the relevant GC boxes. The C-to-T conversion present in GC box 4 as well as the G-to-A conversion in GC box 1 may have resulted in the course of evolution from the occasionally occurring deamination of 5-methylcytosine to thymidine, an indication that the provirus may have been silenced transcriptionally by CpG methylation after integration. In contrast, GC box 2 may have been mutated initially during the integration of the provirus into its host, as it displays a plus-strand GGG-to-AGG change, a signature typical for the action of the APOBEC3G deaminase during the first-strand synthesis of retroviruses. All proviruses described as being expressed, in the literature or by us (Table 1), contain at least three functional GC boxes in the 5′ LTR. If two or more of these boxes are mutated, the affected LTR is not promoter competent.
Interestingly, GC boxes 1 and 3 are 189 nt apart, GC boxes 1 and 4 are 245 nt apart, and GC boxes 2 and 4 are 213 nt apart, distances covered by a nucleosome (146 nt) and the spacing between two nucleosomes (30 to 50 nt; in some instances, 10 to 114 nt). In order to initiate transcription, regulatory factors must compete with and disassemble the preexisting nucleosomes at promoter regions, especially at the TSS. Histone binding factors such as nucleoplasmin and chromatin remodeling enzymes can increase the binding of transcription factors such as Sp1 to nucleosomal DNA, which in turn can initiate nucleosomal disassembly (10). In such a setting, one could envision that binding of Sp1 and Sp3 proteins to two adjacent nucleosomes upstream of the HERV-K TSS (e.g., GC boxes 1 and 3) could create a nucleosome-free region to initiate transcription. This is supported by the fact that the regions upstream and downstream of the TSS were accessible to restriction enzyme cleavage in our ChIP experiments (Fig. 6), which is only possible if they were not obscured by nucleosomes.
Our data add to findings that the transcription of several other groups of HERVs, as well as muERVs related to HERV-K, is not initiated at a TATA motif present in the LTR but is mediated by Sp1 or Sp3 protein binding to several GC boxes in the vicinity of the TSS, although the involvement of an Inr has often been postulated (31, 41, 59). This suggests that the potential to infect and replicate in pluripotent embryonic stem cells and germ cells requires switching the transcriptional regulation of a retrovirus from a TATA-dependent to a TATA-independent mechanism. Endogenous retrovirus promoters obviously adapt to the environment present in early embryonic and germ cells, with the precise parameter allowing ERV expression only slowly being understood. TATA-dependent cellular genes and exogenous retroviruses are generally regulated not only by ubiquitous transcription factors such as Sp1 but also by a variety of cell-type-specific transcription factors regulated by various signaling pathways. Early after zygote formation, such specialized inducible factors are not yet present, whereas regulators such as Sp1 are already in place. Sp1 is involved, inter alia, in the transcription of the TATA-less pluripotency Oct3/4 gene (50) and the TATA-less Dnmt3 promoters (1). Sp1 and Sp3 also regulate their own TATA-less promoters in a positive-feedback loop (35, 47).
Recently, it was published that multiple epigenetic modifiers, such as chromatin remodeling factors, polycomb repressor complexes, and DNA methyltransferases, are involved in silencing endogenous retroviral elements and that different factors may exert differential effects between stem cell types. We have shown that a fully methylated HERV-K promoter loses its promoter competence. Using methylation-sensitive PCR and bisulfite sequencing analyses, a recent publication showed a generally higher level of proviral promoter methylation in melanoma cell lines which did not express HERV-K (62). Similar results have been shown for HERV-K expression in germ cell tumors (32) and for HERV-E promoter activity in the placenta (52). Murine ERVs are also silenced by DNA hypermethylation (42), but they display an unexpected variability, especially high interindividual variances in their DNA methylation patterns (53). Such stochastic individual variances may explain why HERV-K reactivation was observed in only a percentage of melanoma patients (21). The high prevalence of HERV-K reactivation in germ cell tumors may result from the general DNA hypomethylation typical for embryonic stem cells and germ cells.
Although our data revealed an essential role of the transcription factors Sp1 and Sp3 in the regulation of the HERV-K LTR, our observation that the unmethylated LTRpck30 was active in Mel-C9 but not HCl Mel 19 cells argues for the presence of additional tissue-specific transcription factors to achieve full promoter competence or silence. We have previously shown that the transcription factor YY1 (which is equally expressed in both melanoma lines) positively influences HERV-K LTR activity. On the other hand, YY1 can function as a polycomb repressor protein in vivo, requiring, e.g., the corepressor CtBP for transcriptional repression (4). It has also been shown that activated YY1 counteracts Sp1 by interfering with the general transcription machinery (19). Further experiments are needed to decipher the epigenetic modifiers involved in HERV-K expression and silencing and to identify further potential tissue-specific transcription factors.
In summary, we have shown that the LTRs of HERV-K proviruses are TATA-independent promoters regulated by three GC boxes which are binding sites for the transcription factors Sp1 and Sp3. Together with coactivators or enhancers, Sp1 and Sp3 might mediate the formation of a nucleosome-free active chromatin structure at the TSS and enable transcription.
Acknowledgments
We express our gratitude to H. Schmitz for excellent technical assistance, to D. Schadendorf (University Hospital, Essen, Germany) for supplying melanoma cell lines, and to B. Schnierle and G. Schumann (Paul Ehrlich Institut, Langen, Germany), R. Marschalek (Pharmaceutical Biology Frankfurt, Frankfurt, Germany), and A. Loewer (Harvard Medical School, Boston, MA) for helpful discussions.
Footnotes
Published ahead of print on 19 January 2011.
REFERENCES
- 1.Aapola, U., K. Maenpaa, A. Kaipia, and P. Peterson. 2004. Epigenetic modifications affect Dnmt3L expression. Biochem. J. 380:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, Y. F., G. M. Gilmartin, S. M. Hanly, J. R. Nevins, and W. C. Greene. 1991. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell 64:727-737. [DOI] [PubMed] [Google Scholar]
- 3.Albert, T. K., K. Grote, S. Boeing, and M. Meisterernst. 2010. Basal core promoters control the equilibrium between negative cofactor 2 and preinitiation complexes in human cells. Genome Biol. 11:R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atchison, L., A. Ghias, F. Wilkinson, N. Bonini, and M. L. Atchison. 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 22:1347-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101(Suppl. 2):14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbulescu, M., et al. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 7.Boller, K., et al. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196:349-353. [DOI] [PubMed] [Google Scholar]
- 8.Bourc'his, D., and T. H. Bestor. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431:96-99. [DOI] [PubMed] [Google Scholar]
- 9.Brandeis, M., et al.1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., B. Li, and J. L. Workman. 1994. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 13:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costas, J. 2001. Evolutionary dynamics of the human endogenous retrovirus family HERV-K inferred from full-length proviral genomes. J. Mol. Evol. 53:237-243. [DOI] [PubMed] [Google Scholar]
- 12.Day, D. S., L. J. Luquette, P. J. Park, and P. V. Kharchenko. 2010. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol. 11:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Parseval, N., and T. Heidmann. 2005. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet. Genome Res. 110:318-332. [DOI] [PubMed] [Google Scholar]
- 14.Dewannieux, M., et al.2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domansky, A. N., et al. 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472:191-195. [DOI] [PubMed] [Google Scholar]
- 16.Ellis, J., et al. 1996. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human beta-globin locus control region. EMBO J. 15:562-568. [PMC free article] [PubMed] [Google Scholar]
- 17.Esnault, C., S. Priet, D. Ribet, O. Heidmann, and T. Heidmann. 2008. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology 5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flockerzi, A., et al. 2008. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics 9:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galvin, K. M., and Y. Shi. 1997. Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 17:3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotzinger, N., M. Sauter, K. Roemer, and N. Mueller-Lantzsch. 1996. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. J. Gen. Virol. 77:2983-2990. [DOI] [PubMed] [Google Scholar]
- 21.Hahn, S., et al. 2008. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res. Hum. Retroviruses 24:717-723. [DOI] [PubMed] [Google Scholar]
- 22.Humer, J., et al. 2006. Identification of a melanoma marker derived from melanoma-associated endogenous retroviruses. Cancer Res. 66:1658-1663. [DOI] [PubMed] [Google Scholar]
- 23.Javahery, R., A. Khachi, K. Lo, B. Zenzie-Gregory, and S. T. Smale. 1994. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 14:116-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jha, A. R., et al. 2009. Cross-sectional dating of novel haplotypes of HERV-K 113 and HERV-K 115 indicate these proviruses originated in Africa before Homo sapiens. Mol. Biol. Evol. 26:2617-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juven-Gershon, T., J. Y. Hsu, J. W. Theisen, and J. T. Kadonaga. 2008. The RNA polymerase II core promoter—the gateway to transcription. Curr. Opin. Cell Biol. 20:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann, S., et al. 2010. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J. Gen. Virol. 91:1494-1502. [DOI] [PubMed] [Google Scholar]
- 27.Kleiman, A., et al. 2004. HERV-K(HML-2) Gag/Env antibodies as indicator for therapy effect in patients with germ cell tumors. Int. J. Cancer 110:459-461. [DOI] [PubMed] [Google Scholar]
- 28.Knossl, M., R. Lower, and J. Lower. 1999. Expression of the human endogenous retrovirus HTDV/HERV-K is enhanced by cellular transcription factor YY1. J. Virol. 73:1254-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovalskaya, E., A. Buzdin, E. Gogvadze, T. Vinogradova, and E. Sverdlov. 2006. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology 346:373-378. [DOI] [PubMed] [Google Scholar]
- 30.Kurth, R., and N. Bannert. 2010. Beneficial and detrimental effects of human endogenous retroviruses. Int. J. Cancer 126:306-314. [DOI] [PubMed] [Google Scholar]
- 31.La Mantia, G., et al. 1992. Identification of regulatory elements within the minimal promoter region of the human endogenous ERV9 proviruses: accurate transcription initiation is controlled by an Inr-like element. Nucleic Acids Res. 20:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavie, L., M. Kitova, E. Maldener, E. Meese, and J. Mayer. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79:876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, Y. N., M. H. Malim, and P. D. Bieniasz. 2008. Hypermutation of an ancient human retrovirus by APOBEC3G. J. Virol. 82:8762-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, L., and J. R. Davie. 2010. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann. Anat. 192:275-283. [DOI] [PubMed] [Google Scholar]
- 35.Lou, Z., V. M. Maher, and J. J. McCormick. 2005. Identification of the promoter of human transcription factor Sp3 and evidence of the role of factors Sp1 and Sp3 in the expression of Sp3 protein. Gene 351:51-59. [DOI] [PubMed] [Google Scholar]
- 36.Lower, R., et al. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. U. S. A. 90:4480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lower, R., R. R. Tonjes, C. Korbmacher, R. Kurth, and J. Lower. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macleod, D., J. Charlton, J. Mullins, and A. P. Bird. 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8:2282-2292. [DOI] [PubMed] [Google Scholar]
- 39.Magin, C., R. Lower, and J. Lower. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magin-Lachmann, C., et al. 2001. Rec (formerly Corf) function requires interaction with a complex, folded RNA structure within its responsive element rather than binding to a discrete specific binding site. J. Virol. 75:10359-10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maksakova, I. A., and D. L. Mager. 2005. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J. Virol. 79:13865-13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maksakova, I. A., Y. Zhang, and D. L. Mager. 2009. Preferential epigenetic suppression of the autonomous MusD over the nonautonomous ETn mouse retrotransposons. Mol. Cell. Biol. 29:2456-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsui, T., et al. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927-931. [DOI] [PubMed] [Google Scholar]
- 44.Mayer, J., E. Meese, and N. Mueller-Lantzsch. 1997. Chromosomal assignment of human endogenous retrovirus K (HERV-K) env open reading frames. Cytogenet. Cell Genet. 79:157-161. [DOI] [PubMed] [Google Scholar]
- 45.Mayer, J., E. Meese, and N. Mueller-Lantzsch. 1997. Multiple human endogenous retrovirus (HERV-K) loci with gag open reading frames in the human genome. Cytogenet. Cell Genet. 78:1-5. [DOI] [PubMed] [Google Scholar]
- 46.Medstrand, P., et al. 2005. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet. Genome Res. 110:342-352. [DOI] [PubMed] [Google Scholar]
- 47.Nicolas, M., V. Noe, K. B. Jensen, and C. J. Ciudad. 2001. Cloning and characterization of the 5′-flanking region of the human transcription factor Sp1 gene. J. Biol. Chem. 276:22126-22132. [DOI] [PubMed] [Google Scholar]
- 48.Ono, M. 1986. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J. Virol. 58:937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oricchio, E., et al. 2007. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene 26:4226-4233. [DOI] [PubMed] [Google Scholar]
- 50.Pikarsky, E., H. Sharir, E. Ben Shushan, and Y. Bergman. 1994. Retinoic acid represses Oct-3/4 gene expression through several retinoic acid-responsive elements located in the promoter-enhancer region. Mol. Cell. Biol. 14:1026-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prudhomme, S., B. Bonnaud, and F. Mallet. 2005. Endogenous retroviruses and animal reproduction. Cytogenet. Genome Res. 110:353-364. [DOI] [PubMed] [Google Scholar]
- 52.Reiss, D., Y. Zhang, and D. L. Mager. 2007. Widely variable endogenous retroviral methylation levels in human placenta. Nucleic Acids Res. 35:4743-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reiss, D., Y. Zhang, A. Rouhi, M. Reuter, and D. L. Mager. 2010. Variable DNA methylation of transposable elements: the case study of mouse early transposons. Epigenetics 5:68-79. [DOI] [PubMed] [Google Scholar]
- 54.Ruprecht, K., et al. 2008. Human endogenous retrovirus family HERV-K(HML-2) RNA transcripts are selectively packaged into retroviral particles produced by the human germ cell tumor line Tera-1 and originate mainly from a provirus on chromosome 22q11.21. J. Virol. 82:10008-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruprecht, K., J. Mayer, M. Sauter, K. Roemer, and N. Mueller-Lantzsch. 2008. Endogenous retroviruses and cancer. Cell. Mol. Life Sci. 65:3366-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauter, M., et al. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saxonov, S., P. Berg, and D. L. Brutlag. 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 103:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serafino, A., et al. 2009. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 315:849-862. [DOI] [PubMed] [Google Scholar]
- 59.Sjottem, E., S. Anderssen, and T. Johansen. 1996. The promoter activity of long terminal repeats of the HERV-H family of human retrovirus-like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3′ to the TATA box. J. Virol. 70:188-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smale, S. T. 1997. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim. Biophys. Acta 1351:73-88. [DOI] [PubMed] [Google Scholar]
- 61.Smale, S. T., and D. Baltimore. 1989. The “initiator” as a transcription control element. Cell 57:103-113. [DOI] [PubMed] [Google Scholar]
- 62.Stengel, S., U. Fiebig, R. Kurth, and J. Denner. 2010. Regulation of human endogenous retrovirus-K expression in melanomas by CpG methylation. Genes Chromosomes Cancer 49:401-411. [DOI] [PubMed] [Google Scholar]
- 63.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]