Abstract
The L* protein encoded by Theiler's murine encephalomyelitis virus (TMEV) is a unique example of a picornaviral protein encoded by an alternative open reading frame. This protein is an important determinant of TMEV persistence in the mouse central nervous system. We showed that in infected cells, L* is partitioned between the cytosol and the mitochondria. In mitochondria, L* is anchored in the outer membrane and exposed to the cytosol. The targeting of L* to mitochondria is independent of other viral components and likely depends on a conformational signal. L* targeting to mitochondria might involve chaperones of the Hsp70 family, as these proteins are shown to interact.
The use of overlapping open reading frames (ORFs) enables viruses to limit their genome sizes, not only by expressing multiple proteins from a unique nucleotide sequence but also by sparing the need for additional regulatory elements. Theiler's murine encephalomyelitis virus (TMEV), or Theiler's virus, is a neurotropic picornavirus responsible for either acute or persistent and demyelinating infections of the mouse central nervous system (CNS) (3). The L* protein encoded by this virus is the sole protein encoded by an alternative ORF within the picornavirus family (19). L* was found to play an important role in the establishment of persistent CNS infections by TMEV (4, 7, 25). In vitro, L* facilitates viral infection of macrophages (17, 23, 24), possibly through antiapoptotic activity (7, 9). A role for L* in the infection of macrophages fits the observations that macrophages bear the major viral load during persistent infection of the CNS and that L* is expressed in these cells in vivo (2, 13). The exact mechanism by which L* facilitates viral persistence and infection of macrophages is still unclear. Subcellular localization of the protein in infected cells might give a hint regarding protein function. Our work aimed at extending previous data that suggested an association between L* and microtubules (16).
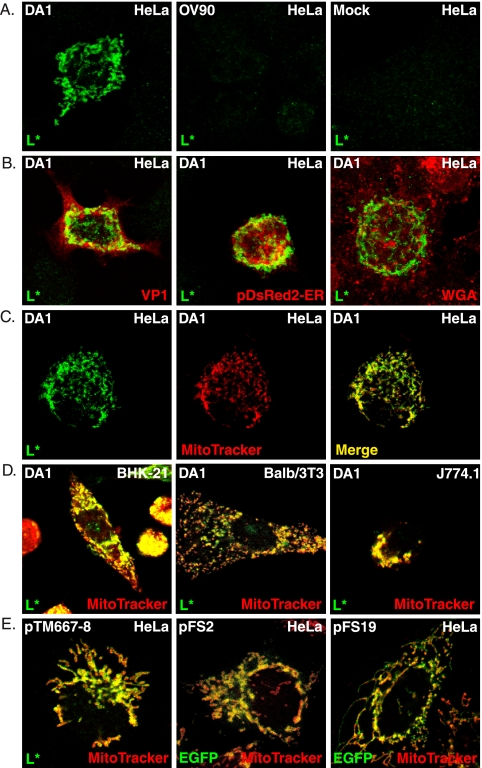
Subcellular localization of the TMEV-encoded L* protein was analyzed in HeLa cells infected for 24 h with 5 PFU per cell of the wild-type TMEV DA1 strain or with the OV90 mutant virus, in which AUG codons 1, 5, and 41 of L* are mutated to ACG and which therefore expresses minimal amounts of L* (25). Cells were processed for indirect immunofluorescence as described previously (18). Two primary anti-L* rabbit polyclonal antibodies were used for immunodetection, one kindly provided by Y. Ohara (16) and the other produced in our laboratory by immunization of a rabbit with peptides CNPRETPLHLTRVTPSPQVT and CALFAQPLTLLPDLNI coupled to N-ethylmaleimide-activated keyhole limpet hemocyanin or to ovalbumin (Pierce). The antiserum (UC172) was adsorbed by serial passage on fixed, permeabilized HeLa, BALB/3T3, and BHK-21 cells. The secondary antibodies were anti-rabbit antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen). As expected, no L* protein was detected in cells infected with the OV90 mutant or in mock-infected cells. Surprisingly, when either primary antibody was used, the fluorescence pattern observed in DA1-infected cells was a punctated and reticulated cytoplasmic pattern that did not match the previously reported association of L* to microtubules (Fig. 1A).
FIG. 1.
L* protein is targeted to mitochondria. Localization of L* was examined by confocal microscopy in infected (A to D) or transfected (E) cells, using the adsorbed polyclonal rabbit antiserum UC172. (A) L* was detected in a reticular and punctated pattern in HeLa cells infected with DA1 but not in mock-infected cells or in cells infected with the AUG-to-ACG mutant OV90. (B) Merged images showing the lack of colocalization of L* with the VP1 capsid protein of TMEV (monoclonal antibody F12B3), the endoplasmic reticulum detected after transfection of the pDsRed2-ER vector (Clontech), or the ER-plus-Golgi compartment detected using Alexa Fluor 594 wheat germ agglutinin (WGA) (Invitrogen). (C to E) Mitochondria were detected by the addition of Mito Tracker red CMXRos dye (Invitrogen) for 30 min before cells were fixed and L* was processed for immunodetection. (C) Single and merged images showing L* and mitochondria detected in infected HeLa cells. (D) Merged images showing mitochondrial localization of L* in infected BHK-21, BALB/3T3, and J774.1 cells. (E) Merged images showing colocalization of L* or EGFP with Mito Tracker in cells transfected with plasmids expressing L* or EGFP-L* fusions under the control of the cytomegalovirus promoter: pTM667-8, L* alone; pFS2, EGFP-L*; and pFS19, EGFP-L*GDVII.
Confocal microscopy was used to identify the structures with which L* associated in infected cells. L* did not colocalize with viral protein VP1, with the endoplasmic reticulum (ER), or with glycosylated proteins of the secretory pathway (including the Golgi network) (Fig. 1B) but clearly colocalized with mitochondria detected using Mito Tracker (Fig. 1C). The targeting of L* to mitochondria was confirmed by colocalization of L* with the F1β subunit of the F1F0 ATP-synthase (data not shown).
L* also colocalized with mitochondria in infected BHK-21, BALB/3T3, and L929 cells, as well as in the macrophage cell lines RAW264.7 and J774.1. L* was detected in mitochondria from 4 and 6 h postinfection in BHK-21 and HeLa cells, respectively (Fig. 1D and data not shown).
To examine whether the mitochondrial localization of L* depended on viral replication or on virus-encoded proteins, HeLa cells were transfected with plasmids expressing either L* alone (pTM667-8) or N- and C-terminal fusion proteins made with L* and the enhanced green fluorescent protein (EGFP). The untagged L* protein expressed from pTM667-8 displayed mitochondrial localization irrespective of its expression level. When EGFP-L* or L*-EGFP was expressed from the pFS2 or pFS5 plasmid, respectively, two different patterns appeared. In cells displaying high expression levels, EGFP fluorescence was detected either as perinuclear aggregates or throughout the cell (data not shown). These aberrant patterns were interpreted as resulting from L* overexpression. In cells with intermediate or low levels of expression, EGFP colocalized with Mito Tracker for both N-terminal and C-terminal fusions (Fig. 1E and data not shown). L* is among the most divergent proteins of the Theiler's virus strains with neurovirulent and persistent phenotypes (14). Furthermore, a polymorphism in L* (L93S) occurring between 2 molecular clones of the persistent strain DA was found to affect the phosphorylation and activity of L* (21). Nevertheless, fusion proteins formed between EGFP and either L* from the neurovirulent TMEV strain GDVII (EGFP-L*GDVII) or L* from the L93S mutant of strain DA1 (EGFP-L*DAI-L93S) also colocalized with mitochondria when they were expressed from transfected plasmids (Fig. 1E and data not shown). Thus, ectopically expressed L* proteins of persistent and neurovirulent TMEV strains are targeted to mitochondria in the absence of other viral components.
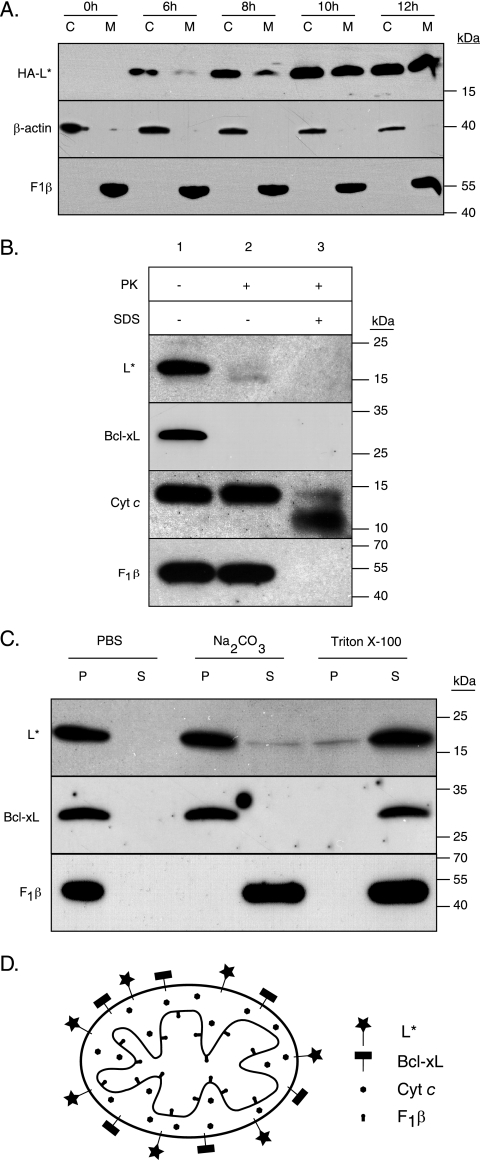
Mitochondrial targeting of L* was confirmed by fractionation experiments. L929 cells infected with FS97, expressing a hemagglutinin epitope-tagged L* (HA-L*), were fractionated using digitonin (8). After centrifugation for 10 min at 12,000 × g, the pellet containing mitochondria and the soluble fraction were suspended in Laemmli buffer, and SDS-PAGE was performed. Detection of β-actin and F1β confirmed the separation of cytoplasmic and mitochondrial fractions. L* was partitioned into both cytoplasmic and mitochondrial fractions. L* first appeared in the cytoplasmic fraction and increased over time in the mitochondrial fraction (Fig. 2A).
FIG. 2.
L* is anchored in the mitochondrial outer membrane and exposed to the cytosol. (A) Western blot analysis of cytosolic (C) and mitochondrial (M) extracts prepared at the indicated time points after infection of L929 cells with virus FS97. This virus is a DA1 derivative bearing capsid mutations from KJ6 that adapt the virus to grow on L929 cells (11) and expressing an N-terminally HA-tagged L*. (B) Mitochondria prepared from HeLa cells expressing L* from a retroviral construct were treated with PBS (lane 1), proteinase K (2.5 μg per ml for 1 h at 30°C) (lane 2), or 0.1% SDS and proteinase K (lane 3). Degradation of L*, Bcl-xL, cytochrome c, and F1β was analyzed by immunoblotting. (C) Western blot of pellet (P) and soluble (S) fractions of mitochondria isolated as described above and treated with PBS, Na2CO3, or Triton X-100. (D) Cartoon showing the localization of the mitochondrial proteins used as markers and the localization of L* as deduced from the results. Primary antibodies used for Western blotting were anti-HA (MMS101P; Covance), anti-β-actin (A5441; Sigma), anti-F1β (A-21351; Invitrogen), anti-cytochrome c (sc-13156; Santa Cruz), and anti-Bcl-xL (sc-7195; Santa Cruz).
To map L* at the mitochondrial level, we generated HeLa cells stably expressing L*, using retroviral vectors. Mitochondria were prepared from these cells according to the procedure described by Seth et al. (20) and were subjected to a mild proteinase K treatment to digest surface-exposed proteins. Treated and untreated mitochondria were analyzed by immunoblotting to assess the degradation of L* and specific mitochondrial proteins. Cytochrome c and F1β were resistant to the treatment (Fig. 2B, lane 2), confirming that proteinase K did not access the intermembrane space or the mitochondrial matrix. Cytochrome c and F1β were not, however, intrinsically resistant to proteinase K, as these proteins were degraded when mitochondria were solubilized with SDS prior to the proteinase K treatment (Fig. 2B, lane 3). L* and Bcl-xL, a tail-anchored protein of the mitochondrial outer membrane, were degraded by proteinase K, suggesting that L*, like Bcl-xL, is exposed at the cytosolic face of the mitochondrial outer membrane.
Next, we investigated the mode of association of L* with the mitochondrial membrane, using alkaline extraction. This method allows discrimination between loosely associated perimembrane proteins and transmembrane proteins (6). Mitochondrial fractions isolated from HeLa cells stably expressing L* were incubated for 1 h on ice with either phosphate-buffered saline (PBS; negative control), 0.1 M Na2CO3 (pH 11.5), or 1% Triton X-100 (positive control) and were subjected to ultracentrifugation to separate membrane fractions from proteins released in the supernatant.
After treatment with PBS, L*, F1β, and Bcl-xL proteins sedimented in the pellet (Fig. 2C). After treatment with Na2CO3 (pH 11.5), F1β was detected in the supernatant, showing the efficacy of the alkaline extraction. In contrast, L* and Bcl-xL were detected predominantly in the pellet, suggesting that L* is membrane anchored, as is Bcl-xL (12). As expected, after solubilization of membranes with 1% Triton X-100, these proteins were released and detected in the supernatant. Taken together, these findings provide evidence that L* protein is anchored in the mitochondrial outer membrane, where it is exposed to the cytosol.
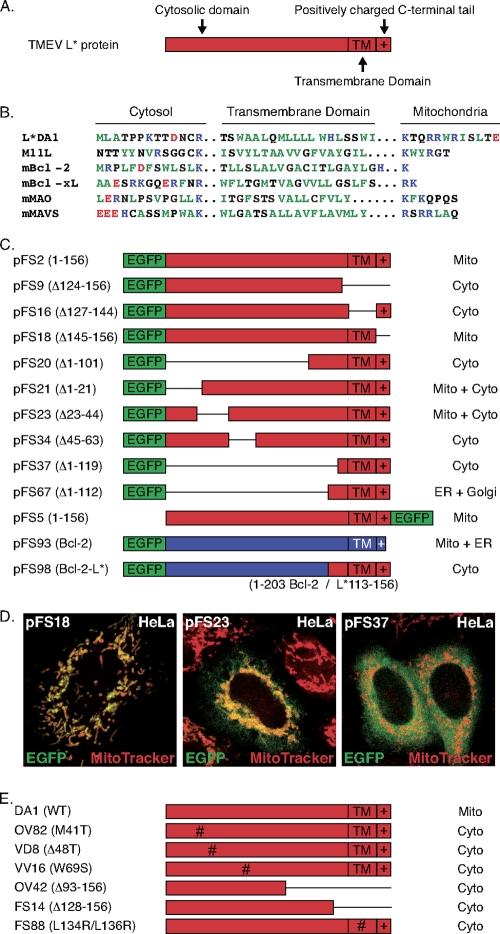
Database searches failed to detect significant similarities between L* and other proteins. However, L* sequence analysis revealed a C-terminal motif made of a previously identified putative transmembrane domain (4) flanked by basic residues and followed by a positively charged tail, typical of tail-anchored proteins such as Bcl-2 (15). To test whether this was the signal targeting L* to the mitochondrial membrane, deletion mutants were made from the EGFP-L* construct. As shown in Fig. 3, no discrete internal targeting signal could be identified using this approach. Deletion of the 22 N-terminal amino acids or of residues 23 to 44 only partly suppressed mitochondrial targeting. In contrast, internal deletions, including the deletion of the hydrophobic domain, blocked mitochondrial targeting (Fig. 3C and D). Also, point mutations or deletions introduced in the viral backbone affected the mitochondrial targeting of L* in infected cells (Fig. 3E). More surprisingly, deletion of the positively charged C-terminal end of the protein did not affect mitochondrial localization. This finding is in contrast to what is known of well-characterized tail-anchored proteins, whose tail ablation prevents mitochondrial targeting (10, 15, 22). Also, substitution of the putative L* tail anchor domain for that of Bcl-2 failed to target the Bcl-2 protein to mitochondria (Fig. 3C and D). Thus, our data suggest that L* targeting to mitochondria involves a noncanonical conformational signal.
FIG. 3.
The mitochondrial targeting signal of L* is noncanonical and conformational. (A) L* domains defined from sequence analysis include a putative Bcl-2-like tail-anchoring domain, which is made of a hydrophobic domain (TM) preceded by positively charged residues and followed by a C-terminal positive tail (indicated by a plus sign [+]). (B) Sequence alignment of the L* C-terminal domain and characterized tail-anchoring domains from other proteins. Hydrophobic residues are shown in dark green, basic residues are shown in blue, and acidic residues are shown in red. (C) In-frame deletions (symbolized by lines) were introduced in plasmid pFS2, which expresses the EGFP-L* fusion protein. The extents of the deletions present in the constructs are indicated beside the plasmid names (left column). pFS5 expresses an L*-EGFP fusion. pFS93 expresses an EGFP-Bcl-2 fusion. In pFS98, the tail-anchoring domain of Bcl-2 was replaced by the C-terminal end of L* (Bcl-2 amino acids 1 to 203 joined to L* amino acids 113 to 156). The right column indicates the subcellular localization defined by confocal microscopy after transfection of the constructs in HeLa cells. The expression and stability of the fusion proteins in transfected cells were confirmed by Western blotting (not shown). (D) Representative merged confocal images showing detection by EGFP and Mito Tracker of the indicated EGFP-L* mutants. (E) L* protein expressed from the 6 DA1 virus derivatives carrying the indicated amino acid substitutions or deletions (Δ) failed to show mitochondrial localization after infection of HeLa cells. The expression and stability of these proteins in infected cells were checked by Western blotting (not shown).
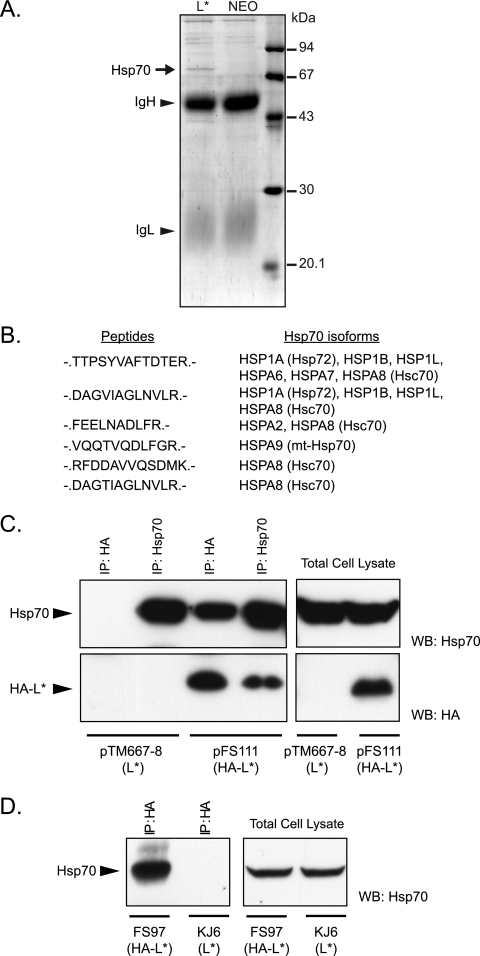
We hypothesized that L* could be targeted to mitochondria through interaction with a mitochondrial partner. Therefore, L* was immunoprecipitated from HeLa cells constitutively expressing L* from a retroviral construct. SDS-PAGE of immunoprecipitated extracts showed a clear band for a 70-kDa protein that coimmunoprecipitated with L*. Mass spectrometric (5) analysis revealed that this protein belongs to the Hsp70 family (Fig. 4). Hsp70 is known as a “sticky protein.” Therefore, interaction was further confirmed by detection of L* in Hsp70 immunoprecipitates and, in a more physiological setting, by coimmunoprecipitation of L* and Hsp70 from L929 cells infected with an HA-tagged L* protein-expressing virus (Fig. 4C and D).
FIG. 4.
L* interacts with chaperones of the Hsp70 family. (A) Coomassie blue G-250-stained SDS-polyacrylamide gel of proteins immunoprecipitated from HeLa cells stably expressing L* or from HeLa cells carrying an empty retroviral vector (NEO), using anti-L* polyclonal antiserum UC172 and protein A-Sepharose (P-3391; Sigma). Bands corresponding to immunoglobulin heavy (IgH) and light (IgL) chains are shown. (B) The band corresponding to Hsp70, visible on the gel (arrow), was cut, digested in-gel with trypsin, and analyzed by tandem mass spectrometry. Sequences of peptides of the Hsp70 family and the Hsp70 isoforms that were identified are shown. (C) Coimmunoprecipitation of L* and Hsp70 from 293T cells transfected with plasmid pTM667-8, expressing L*, or with plasmid pFS111, expressing an N-terminally HA-tagged L*. Detection by Western blotting of Hsp70 (upper panel) or HA-L* (lower panel) in the indicated immunoprecipitates (IP) (left panel) or in the corresponding total cell lysates (right panel). (D) Detection of Hsp70 after immunoprecipitation of HA-tagged L* from L929 cells infected for 16 h with virus FS97 (HA-L*) or with virus KJ6 (wt L*). Detection of Hsp70 by Western blotting in HA-L*-immunoprecipitated extracts (left panel) and in total cell lysates (right panel). WB, Western blot.
Our data contrast with previous findings that show an association of L* with microtubules (16). Using imaging and fractionation experiments, we observed that L* partitioned as a cytoplasmic and mitochondrial protein. L* was first detected in the cytoplasm and accumulated in the mitochondrial fraction over time. Sensitivity to mild proteinase K treatment and resistance to stripping by alkaline treatment suggest that L* is anchored in the outer membrane of the mitochondria, facing the cell cytosol. This localization fits with the antiapoptotic role previously reported for L* (7, 9), since it would bring L* in close vicinity to the pro- and antiapoptotic factors of the Bcl-2/Bax family. L* mitochondrial targeting appears to involve a noncanonical conformational signal. Interaction with Hsp70 might either promote mitochondrial targeting or protect hydrophobic domains of L* prior to membrane insertion or association with another partner. It is noteworthy that Hsp70 was reported to participate in the membrane targeting of some tail-anchored proteins (1).
Acknowledgments
We thank Françoise Bontemps (de Duve Institute, University of Louvain) for the gift of anti-cytochrome c antibody.
F.S. was the recipient of an FSR fellowship (University of Louvain). D.V. is a Collaborateur Logistique of the FRS-FNRS. This work was supported by FSR (University of Louvain), ARC (Communauté Française de Belgique), and FRSM (Fonds National de la Recherche Médicale convention 3.4576.08 and crédit aux chercheurs).
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1.Abell, B. M., C. Rabu, P. Leznicki, J. C. Young, and S. High. 2007. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell Sci. 120:1743-1751. [DOI] [PubMed] [Google Scholar]
- 2.Asakura, K., H. Murayama, T. Himeda, and Y. Ohara. 2007. Expression of L* protein of Theiler's murine encephalomyelitis virus in the chronic phase of infection. J. Gen. Virol. 88:2268-2274. [DOI] [PubMed] [Google Scholar]
- 3.Brahic, M., J. F. Bureau, and T. Michiels. 2005. The genetics of the persistent infection and demyelinating disease caused by Theiler's virus. Annu. Rev. Microbiol. 59:279-298. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H. H., W. P. Kong, L. Zhang, P. L. Ward, and R. P. Roos. 1995. A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nat. Med. 1:927-931. [DOI] [PubMed] [Google Scholar]
- 5.Drozak, J., M. Veiga-da-Cunha, D. Vertommen, V. Stroobant, and E. Van Schaftingen. 2010. Molecular identification of carnosine synthase as ATP-grasp domain-containing protein 1 (ATPGD1). J. Biol. Chem. 285:9346-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghadge, G. D., L. Ma, S. Sato, J. Kim, and R. P. Roos. 1998. A protein critical for a Theiler's virus-induced immune system-mediated demyelinating disease has a cell type-specific antiapoptotic effect and a key role in virus persistence. J. Virol. 72:8605-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heibein, J. A., M. Barry, B. Motyka, and R. C. Bleackley. 1999. Granzyme B-induced loss of mitochondrial inner membrane potential (ΔΨm) and cytochrome c release are caspase independent. J. Immunol. 163:4683-4693. [PubMed] [Google Scholar]
- 9.Himeda, T., Y. Ohara, K. Asakura, Y. Kontani, and M. Sawada. 2005. A lentiviral expression system demonstrates that L* protein of Theiler's murine encephalomyelitis virus (TMEV) has an anti-apoptotic effect in a macrophage cell line. Microb. Pathog. 38:201-207. [DOI] [PubMed] [Google Scholar]
- 10.Horie, C., H. Suzuki, M. Sakaguchi, and K. Mihara. 2002. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell 13:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jnaoui, K., and T. Michiels. 1998. Adaptation of Theiler's virus to L929 cells: mutations in the putative receptor binding site on the capsid map to neutralization sites and modulate viral persistence. Virology 244:397-404. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann, T., et al. 2003. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipton, H. L., G. Twaddle, and M. L. Jelachich. 1995. The predominant virus antigen burden is present in macrophages in Theiler's murine encephalomyelitis virus-induced demyelinating disease. J. Virol. 69:2525-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michiels, T., N. Jarousse, and M. Brahic. 1995. Analysis of the leader and capsid coding regions of persistent and neurovirulent strains of Theiler's virus. Virology 214:550-558. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen, M., D. G. Millar, V. W. Yong, S. J. Korsmeyer, and G. C. Shore. 1993. Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J. Biol. Chem. 268:25265-25268. [PubMed] [Google Scholar]
- 16.Obuchi, M., T. Odagiri, K. Asakura, and Y. Ohara. 2001. Association of L* protein of Theiler's murine encephalomyelitis virus with microtubules in infected cells. Virology 289:95-102. [DOI] [PubMed] [Google Scholar]
- 17.Obuchi, M., et al. 2000. L* protein of Theiler's murine encephalomyelitis virus is required for virus growth in a murine macrophage-like cell line. J. Virol. 74:4898-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul, S., and T. Michiels. 2006. Cardiovirus leader proteins are functionally interchangeable and have evolved to adapt to virus replication fitness. J. Gen. Virol. 87:1237-1246. [DOI] [PubMed] [Google Scholar]
- 19.Roos, R. P., W. P. Kong, and B. L. Semler. 1989. Polyprotein processing of Theiler's murine encephalomyelitis virus. J. Virol. 63:5344-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 21.Stavrou, S., et al. 2010. Theiler's murine encephalomyelitis virus L* amino acid position 93 is important for virus persistence and virus-induced demyelination. J. Virol. 84:1348-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart, T. L., S. T. Wasilenko, and M. Barry. 2005. Vaccinia virus F1L protein is a tail-anchored protein that functions at the mitochondria to inhibit apoptosis. J. Virol. 79:1084-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takata, H., et al. 1998. L* protein of the DA strain of Theiler's murine encephalomyelitis virus is important for virus growth in a murine macrophage-like cell line. J. Virol. 72:4950-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Eyll, O., and T. Michiels. 2000. Influence of the Theiler's virus L* protein on macrophage infection, viral persistence, and neurovirulence. J. Virol. 74:9071-9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Eyll, O., and T. Michiels. 2002. Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence. J. Virol. 76:10665-10673. [DOI] [PMC free article] [PubMed] [Google Scholar]