Abstract
While group B coxsackieviruses (CVB) use the coxsackievirus and adenovirus receptor (CAR) as the receptor through which they infect susceptible cells, some CVB strains are known for their acquired capacity to bind other molecules. The CVB3/RD strain that emerged from a CVB3/Nancy population sequentially passaged in the CAR-poor RD cell line binds decay-accelerating factor (DAF) (CD55) and CAR. A new strain, CVB3/RDVa, has been isolated from RD cells chronically infected with CVB3/RD and binds multiple molecules in addition to DAF and CAR. The capsid proteins of CVB3/RD differ from those of CVB3/28, a cloned strain that binds only CAR, by only four amino acids, including a glutamate/glutamine dimorphism in the DAF-binding region of the capsid. The capsid proteins of CVB3/RD and CVB3/RDVa differ by seven amino acids. The ability of CVB3/RDVa to bind ligands in addition to CAR and DAF may be attributed to lysine residues near the icosahedral 5-fold axes of symmetry. Considered with differences in the stability of the CVB3 strains, these traits suggest that in vitro selection in a CAR-limited environment selects for virus populations that can associate with molecules on the cell surface and survive until CAR becomes available to support infection.
Some strains of group B coxsackieviruses (CVB) bind ligands in addition to the coxsackievirus and adenovirus receptor (CAR), including decay-accelerating factor (DAF) (CD55), nucleolin, and heparan sulfate (1, 8, 23). These promiscuous strains were selected by passage of CVB serotype 3 (CVB3) in cells that expressed little CAR and in which the parental CVB3 strain produced limited cytopathic effect (CPE) (e.g., RD cells and fibroblasts [18, 19]). The results were initially interpreted as evidence that CVB3 strains could evolve to utilize new receptors, but subsequent studies showed that DAF-binding CVB3/RD still requires CAR to infect cells (14, 21). Infection of human cells by a strain called CVB3/Nancy-PD, which binds heparan sulfate, can be blocked by antibody against CAR, indicating that it, too, still uses CAR to infect HeLa cells (19). While DAF binding may facilitate access of CVB3/RD to CAR sequestered in tight junctions of polarized epithelial cells (6), the ability to bind DAF apparently confers no such advantage to CVB3 during infection of HeLa cells (16). These results all indicate that growth of CVB3 on nonpolar, CAR-poor cells selects for variants that are able to bind additional cell surface ligands, but in vitro evolution has not replaced CAR as the necessary and sufficient receptor that mediates infection. The selective advantage of CVB3 that can bind ligands in addition to the receptor that mediates infection has not thus far been satisfactorily explained.
The molecularly cloned strain CVB3/28 (22) is representative of CVB3 strains that cause complete CPE in cultures of HeLa cells, but not in cultures of RD cells (5), and do not agglutinate red blood cells (RBC). Hemagglutination is characteristic of DAF-binding enteroviruses (17). CVB3/RD is the prototypical laboratory DAF-binding, hemagglutinating CVB3 strain selected by serial passage on RD cells. CVB3/RD causes CPE in both HeLa and RD cell cultures yet requires CAR for infection (14, 21). CVB3/RDVa is a strain that emerged from a carrier culture of RD cells originally inoculated with CVB3/RD. Comparison of these three strains reveals that competition among variants in the quasispecies in the RD cell culture environment where CAR is limiting selects for strains with additional capacity for binding to the cell surface and increased stability that allows them to survive until CAR becomes available.
MATERIALS AND METHODS
Cells and viruses.
HeLa and RD (CCL-136) cells were obtained from the American Type Culture Collection (Manassas, VA). RDt3 cells are RD cells that express CAR with a truncated cytoplasmic domain (2, 7). The cells were maintained at 37°C with 6% CO2 and 94% air in DMEM-10, comprised of Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, glutamine (0.9 mM), penicillin-streptomycin (90 units/ml and 90 μg/ml, respectively), and gentamicin (67 μg/ml), all from Invitrogen (Carlsbad, CA).
CVB3/RD (18) was obtained at passage three from the laboratory of M. G. Rossmann (Purdue University) and was subsequently passaged only in RD cells for these studies. CVB3/28 was used from a third passage stock following initial transfection of HeLa cells with the infectious cDNA plasmid (22). RD cells inoculated with CVB3/RD (5.1 × 107 50% tissue culture infective doses [TCID50] of virus added per T75 flask of 50 to 80% confluent RD cells) became chronically infected. The medium was replaced every 2 or 3 days, and the cells were subcultured before reaching confluence. The medium was collected after observing unexpected cell loss at about 4 months postinoculation, and a stock of virus called CVB3/RDVa, was prepared by propagation in RD cells.
Virus quantitation, preparation, and molecular analysis.
Virus titers were determined on both HeLa and RD cells and expressed as TCID50/ml. The titers of CVB3/28 and CVB3/RD were determined from the analyses on HeLa cells; the titer of CVB3/RDVa was taken from the analysis on RD cells. After removal of cellular debris by centrifugation, virus was isolated from cell culture lysates by shaking with an equal volume of chloroform and centrifugation through 30% sucrose, 1 mM MgCl2, 0.02 M Tris, and 1 M NaCl (pH 7.5) into approximately 200 μl of glycerol (7). Tris was used at 0.25 M with 0.5 M NaCl (pH 7.6) during isolation of radiolabeled virus. Viruses were resuspended in 0.1 M NaCl at 2 × 109 TCID50/ml, aliquoted, and stored at −80°C.
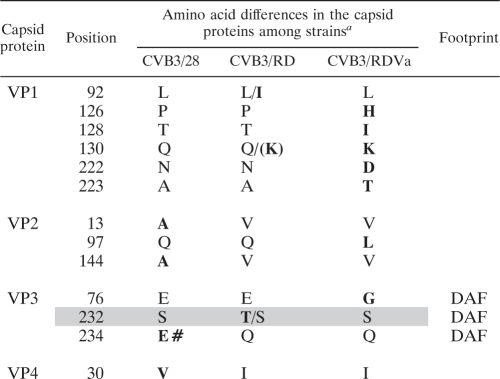
Viral RNA was isolated from virus stocks (ZR viral RNA kit; Zymo Research, Orange, CA). Approximately 10 ng of eluted RNA was reverse transcribed in a mixture containing 6 pmol each of 4 primers (primers GN, ID12, ID14, and DI-5; Table 1) using the Improm II reverse transcription system (Promega Corp., Madison, WI) with 3 mM MgCl2 and 0.5 mM each deoxynucleoside triphosphate (dNTP). cDNA was amplified using Go-Taq polymerase (Promega Corp., Madison, WI) with 2.5 mM MgCl2 and 0.2 mM (each) dNTP and 0.45 μM concentrations of the primers E3Rev and ID8. The 2.8-kb product of this PCR was purified on spin columns (DNA Clean & Concentrator; Zymo Research, Orange, CA) and directly sequenced with the primers E3Rev, Puff3, Puff5, FN, FL4, C, EN, ID3, and ID8 with an Applied Biosystems (ABI) 3730 48-capillary electrophoresis DNA analyzer (University of Nebraska Medical Center [UNMC] High-Throughput DNA Sequencing and Genotyping Core Facility). Both strands were sequenced in the capsid protein-coding region. As this sequence was derived from a population of viral RNA molecules, 100 ng of each cDNA population was cloned into pCR2.1-TOPO (TOPO TA cloning kit; Invitrogen Life Science, Carlsbad, CA) and sequenced as described before with the primers M13Forward, M13Reverse, Puff3, Puff5, FN, FL4, C, EN, and ID3. These sequences were used to confirm the sequence at sites in which the population had variation present. Sequences were aligned using Vector NTI Advance 11 (access to the Invitrogen Life Science software provided by University of Nebraska Medical Center Genetic Sequence Analysis Core). Sites of sequence variation were determined, and the sequence was aligned with the CVB3/28 sequence (GenBank accession no. AY752944.2; with Gln at position 264 of VP1 based on reanalyzed sequence data) to locate the open reading frame. The amino acid sequences of the capsid proteins were inferred from the open reading frame and aligned with the amino acid sequence of CVB3/28.
TABLE 1.
Primers used for sequence analysis
| Primer | Sequence (5′-3′) | Genomic locationa | Primer anneals to strand |
|---|---|---|---|
| E3Rev | GGAACCGACTACTTTGGGTGTCCGTG | 537-562 | − |
| Puff5 | ATGCAGTACCACTACTTAGG | 1235-1254 | − |
| Puff3 | ATGTTATCCATAGGTACACT | 1574-1593 | + |
| C | CTAGGAGAGATCTTGAAC | 2036-2053 | − |
| EN | CCATGCTTGGTACTCATGT | 2181-2199 | − |
| FN | GACAGAGAARTCATTGCATGC | 2384-2404 | + |
| FL4 | TGTGACGTGTGACCTGTCTC | 2555-2574 | + |
| ID3 | CACGCCACGTTAAGAACTACC | 2601-2621 | − |
| ID8 | ACTGCCCCTGATTGTTGTCC | 3305-3324 | + |
| ID12 | CACAGGAGATCACGGATGTC | 3701-3720 | + |
| GN | CCTGTTCCATTGCATCATCTTC | 3725-3746 | + |
| ID14 | TCCTCACCACAATTACTAAG | 3901-3920 | + |
| DI-5 | GGCAGTCACAGTGATCAGG | 3931-3949 | + |
Genomic location based on the sequence deposited in GenBank under accession no. AY752944.
Single-step growth curves.
For single-step growth curves, cells were seeded in multiple 24-well plates at 20,000 cells/well. Approximately 24 h after the cultures were seeded, virus was added to the cells at a multiplicity of infection (MOI) of about 25 TCID50 per cell. One hour after the cells were inoculated, the wells were rinsed three times with DMEM-10 and returned to the incubator with 1 ml of fresh medium per well. The plates were removed from the incubator at timed intervals and placed at −80°C until all time points had been sampled. The plates were subjected to three freeze/thaw cycles, cellular debris was removed by centrifugation for 3 min at 13,000 rpm in a microcentrifuge, and the titer of virus was determined. Except for the analysis of growth in RDt3 cells (where the titers of the viruses were determined on RDt3 cells), the titer of each CVB3 strain was determined on the cell line (HeLa or RD) most susceptible to CPE.
Protection by anti-CAR Rmcb monoclonal antibody and by soluble CAR (sCAR).
Monoclonal antibody Rmcb against human CAR (12) was used to assess the role of CAR in infection by the different virus strains. HeLa and RD cells were seeded into wells of 96-well plates at 6,000 cells per well. The next day, dilutions of pure Rmcb IgG in DMEM-10 were added to the wells to provide the indicated final concentrations. The plates were returned to the incubator for 30 min prior to adding CVB to the cells in the wells at an MOI of ∼10. About 24 h after inoculation, the cells in the wells were examined for CPE, fixed, and stained. Stained plates were digitally imaged and integrated for stain intensity.
sCAR was expressed in Raji cells from pEF-ECAR and purified from spent medium by immunoaffinity chromatography on Rmcb-AffiGel (Bio-Rad). The pEF-ECAR-coding DNA was generated and cloned (GenScript USA Inc., Piscataway, NJ) in the pEF-myc vector (Invitrogen). The coding sequence included the CAR signal sequence and the extracellular amino acids through serine 233. The C-terminal FLAG antigen in the coding sequence was not detectable in the secreted protein, indicating that the carboxyl terminus of the protein has been modified. The wells of 96-well plates were seeded with HeLa cells (6,000/well) or RD cells (8,000/well) 1 day before inoculation. Viruses were mixed with dilutions of sCAR in DMEM-10 and placed at 37°C. After about 1.5 h, the medium was removed from the HeLa and RD cells and replaced with 100 μl of the CVB3-sCAR samples (MOI of ∼10). Infections were allowed to proceed for 24 and 48 h prior to fixing and staining the surviving cells.
Virus overlay blots.
Cell lysates and virus overlay blots were prepared as previously described (3), using 10 to 15% SDS-polyacrylamide gels (13). Each lane was loaded with 50 μg of cell protein in Laemmli loading buffer (without reducing agents and without heating). The blots (on Immobilon-P) were blocked overnight with Tris-buffered saline (TBS) containing 0.1% Tween 20 plus 6% milk. CVB3 (35S labeled) was diluted in 10 ml of DMEM containing 20% fetal calf serum (FCS) (100,000 cpm/ml) and incubated with the blots on a rotating mixer at room temperature for 2 h. The blots were washed with TBS containing Tween 20 (150 ml/wash per blot) twice for 8 min and twice for 15 min and then dried overnight between filter papers pressed between glass or Plexiglas plates. Dried blots were placed on film at −80°C for detection of bound virus. CAR and DAF were localized on Western blots of the cell lysates using monoclonal antibody E1.2D3 (4) and anti-CD55 (Santa Cruz Biotechnology), respectively.
Hemagglutination.
Red blood cells (RBC) (type A negative) were prepared from fresh human blood samples collected with citrate anticoagulant. RBC were collected by centrifugation (15 min at 800 rpm in a Sorvall RT6000B with H1000B rotor) and washed five times with Dulbecco's phosphate-buffered saline (DPBS; Invitrogen). Washed RBC were resuspended at 1% (vol/vol) in DPBS with bovine serum albumin (BSA) (Sigma) at 10 mg/ml and dispensed (50 μl) into conical-bottom wells of a 96-well plate. The viruses were diluted in DPBS containing BSA, and 50-μl aliquots were added to wells containing RBC. Samples were mixed and allowed to stand undisturbed at room temperature for 3 to 4 h before photography.
Virus decay kinetics.
To assess virus stability under tissue culture conditions, purified virus (ca. 2 × 108 to 2 × 109 infectious virus in 100 μl) was added to 5 ml DMEM-10 and placed in a tissue culture incubator at 37°C and 6% CO2. In one of the experiments, the medium was preequilibrated overnight prior to adding virus. Aliquots (200 μl) were periodically removed and stored at −80°C until assayed for infectious virus.
Digital images.
Digital images were captured with a Canon 10D camera or an Epson Perfection V200 scanner and processed in Photoshop.
Nucleotide sequence accession numbers.
The nucleotide sequences of CVB3/28, CVB3/RD, and CVB3/RDVa were deposited in GenBank under accession numbers AY752944.2, HQ157560, and HQ164111, respectively.
RESULTS
Selection for CVB3/RDVa in chronically infected RD cells.
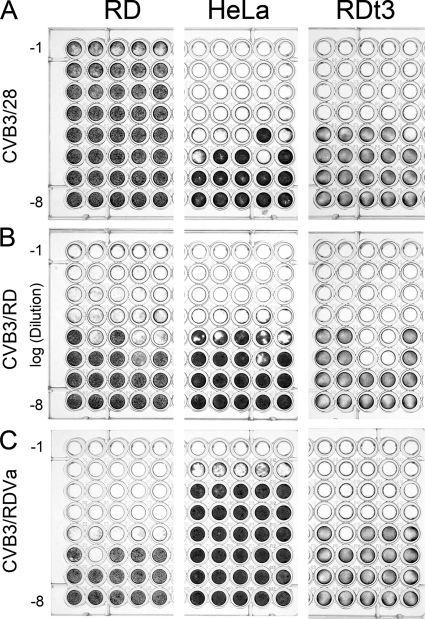
RD cells that survived the initial infection by CVB3/RD continued to proliferate and produce virus. This carrier culture was continued in anticipation that continuous pressure from infectious virus would further deplete the RD cell population of CAR, thus providing an environment in which the CVB3 quasispecies might evolve to use a new receptor. After 4 months of regular growth, the culture experienced an unexpected sharp decrease in cell density. When supernatant from this culture was tested on both HeLa and RD cells, the virus caused notably less extensive CPE on HeLa cells than on RD cells, unlike CVB3/RD, which produces similar degrees of CPE on HeLa and RD cells (Fig. 1). This phenotypically distinct CVB3 population was called CVB3/RDVa. For comparison of all three virus strains, the titers of CVB3/RDVa, CVB3/RD, and CVB3/28 were determined on HeLa, RD, and RDt3 cells. After the assay, the wells were examined for microscopic CPE for determination of virus titers, and the remaining cells were stained to demonstrate overt cytopathic effects (Fig. 1). As expected (5), CVB3/28 caused little CPE on RD cells compared to the CPE on HeLa cells (Fig. 1A), but the cytopathic phenotype was fully apparent on RDt3 cells. CVB3/RD caused similar CPE on all three cell lines (e.g., Fig. 1B shows nearly complete CPE in wells at dilutions to 1/104 and partial CPE at 1/105). Unlike either CVB3/28 or the parental CVB3/RD strain, CVB3/RDVa produced greater CPE on RD and RDt3 cells than on HeLa cells (Fig. 1C). The concentrations of CVB3/28, CVB3/RD, and CVB3/RDVa were 2.8 × 107, 2.1 × 107, and 1.2 × 105 TCID50/ml, respectively, when assayed on HeLa cells. Analysis of the same dilutions on RD cells gave concentrations of 2.1 × 105, 8.9 × 106, and 2.1 × 107 TCID50/ml. Clearly, the concentrations of infectious CVB3/28 were underestimated on RD cells, and the concentrations of infectious CVB3/RDVa were underestimated on HeLa cells. The concentrations of infectious CVB3/RD determined on HeLa cells were marginally higher than on RD cells in this experiment.
FIG. 1.
The three CVB3 strains differ in CPE on HeLa and RD cells. Serial 10-fold dilutions (10−1 to 10−8) of CVB3 strains were added to wells containing RD, HeLa, or RDt3 cells in a typical TCID50 assay format. (A) CVB3/28 shows little overt CPE on RD cells compared to the complete CPE on HeLa and RDt3 cells. (B) CVB3/RD causes comparable degrees of CPE on RD, HeLa, and RDt3 cells. (C) CVB3/RDVa causes extensive CPE on RD and RDt3 cells at dilutions where HeLa cells appear unaffected. The cells were fixed with ethanol-acetic acid (75:25) and stained with crystal violet (0.05% in 20% methanol) approximately 72 h after inoculation.
CVB3/RD and CVB3/RDVa agglutinate human red blood cells.
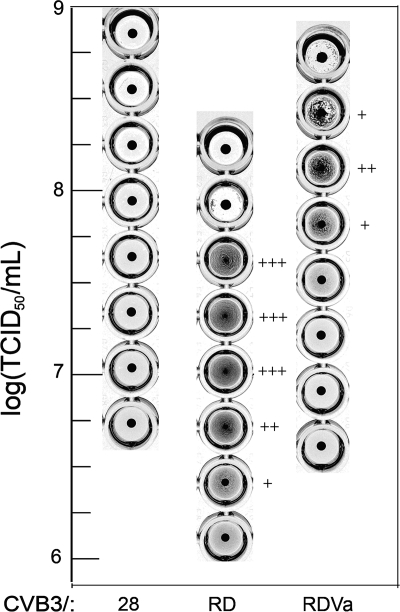
The CVB3/RDVa strain retained the ability to agglutinate red blood cells (Fig. 2). Red blood cell agglutination by CVB3/RDVa occurred at a higher virus concentration than by CVB3/RD, and the agglutinating concentration range was narrower (Fig. 2), suggesting that there might be some qualitative differences in the interaction between viruses and red blood cells. Even so, these results confirmed that the DAF-binding region of the parental CVB3/RD strain remained substantially intact in CVB3/RDVa. CVB3/28 did not agglutinate red blood cells.
FIG. 2.
CVB3/RD and CVB3/RDVa agglutinate human RBC. The degree of agglutination in wells is indicated by the number of plus symbols as follows: +, weak agglutination; ++, moderate agglutination; +++, strong agglutination. Wells in which all of the red blood cells have settled into a button at the bottom of the well are negative. CVB3/28 produced no hemagglutination.
CVB3/RDVa uses CAR to infect cells.
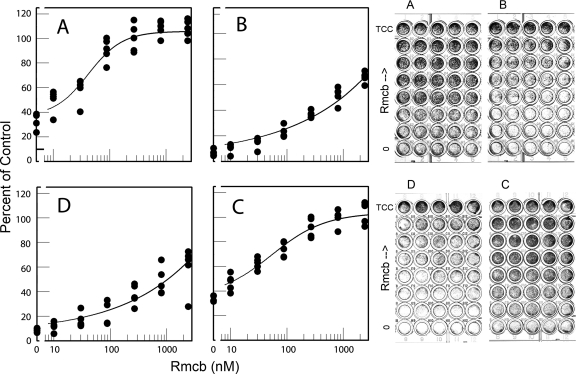
Capsid protein amino acids that map to the CAR-binding footprint (11) are conserved in CVB3/28 and CVB3/RD, both of which are known to use CAR for infection (5). CVB3/RDVa caused more CPE in the low-CAR RD cells than in the high-CAR HeLa cells, which raised questions as to its use of CAR to infect cells. Monoclonal antibody Rmcb, which binds CAR and inhibits infection by CAR-binding CVB strains (12), was added at various dilutions to block CAR on HeLa and RD cells. Subsequent challenge with the CVB3 strains showed that the Rmcb monoclonal antibody attenuated CPE caused by all three CVB strains on both cell lines, although to different degrees (Fig. 3). Rmcb near 1,000 nM protected HeLa cells from CVB3/28 and protected RD cells from CVB3/RD (Fig. 3A and C). In contrast, 1,000 nM Rmcb reduced CPE by about 50% for HeLa cells inoculated with CVB3/RD and RD cells inoculated with CVB3/RDVa. The inhibition curves are consistent with Rmcb acting as a competitive inhibitor of virus binding to receptor, with reduced efficacy if the virus concentration at the cell surface is increased (Fig. 3B and D).
FIG. 3.
Antibody to receptor blocks infection. Rmcb, a monoclonal antibody against CAR, protects HeLa cells (A and B) and RD cells (C and D) against virus strains CVB3/28 (A), CVB3/RD (B and C), and CVB3/RDVa (D) to different extents. Each row on a plate contains five replicate wells. The cells were fixed and stained about 24 h after inoculation. Digital images of the stained plates were integrated, and the results for the plates shown to the right of the figure were graphed and are shown to the left of the figure. TCC is the tissue culture control.
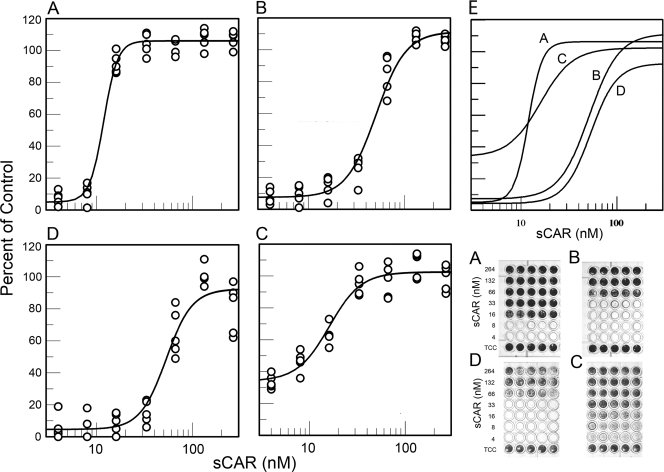
Soluble CAR (sCAR) has been shown to protect cells from infection by CAR-binding CVB and can convert the virions to A-particles (9, 14). sCAR inhibited infection by CVB3/28, CVB3/RD, and CVB3/RDVa, but the concentration of sCAR required to achieve complete protection varied among the three virus strains (Fig. 4). HeLa cells were well protected from CVB3/28 by 16 to 33 nM sCAR, which was well below the sCAR concentration required to achieve equivalent HeLa cell protection from CVB3/RD (Fig. 4A and B). The dose-response curve for sCAR protection of RD cells from CVB3/RDVa (Fig. 4D) was similar to that for protection of HeLa cells from CVB3/RD (Fig. 4B). Curiously, comparison of the 50% inhibition points indicated that sCAR was more effective for protecting RD cells from CVB3/RD than for protecting HeLa cells from the same strain (Fig. 4B and C). The preliminary interpretation of this result is that the virus is not fully inactivated and competitive inhibition between CAR and sCAR is significant in these experiments. The sCAR provides a more effective competitive blockade on RD cells (low CAR, low DAF) than on HeLa cells (high CAR, high DAF) (Fig. 5) (e.g., see reference 21). In this interpretation, the differences in sCAR required to achieve 50% inhibition can be attributed to differences in local virus concentration on the cell surface and competition for CAR present in markedly different cell surface densities.
FIG. 4.
Soluble receptor protects against infection. (A to D) Soluble CAR protects HeLa cells (A and B) and RD cells (C and D) from CVB3/28 (A), CVB3/RD (B and C), and CVB3/RDVa (D). The experiment shown was ended at 48 h. (E) Composite of the fitted curves in panels A to D. Each row of the stained plates contains five replicate wells. Digital images of the stained plates were integrated, and the results graphed. TCC is the tissue culture control.
FIG. 5.
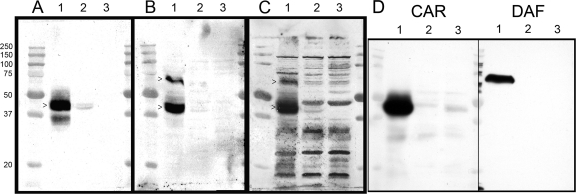
Virus overlay blots reveal differences in ligand binding among the CVB3 strains. (A to C) Lysates of HeLa cells (lanes 1), RD cells (lanes 2), and cells surviving in the RD carrier culture (lanes 3) probed with radiolabeled CVB3/28 (A), CVB3/RD (B), or CVB3/RDVa (C) reveal distinctive binding patterns for each of the three virus strains. The Mr (×10−3) molecular size markers are shown to the left of panel A. >, bands that correspond to DAF or CAR. (D) Lysates were blotted and probed with antibodies against CAR and DAF. Each lane was loaded with 50 μg of cell protein.
CVB3/RDVa binds multiple ligands in addition to CAR and DAF.
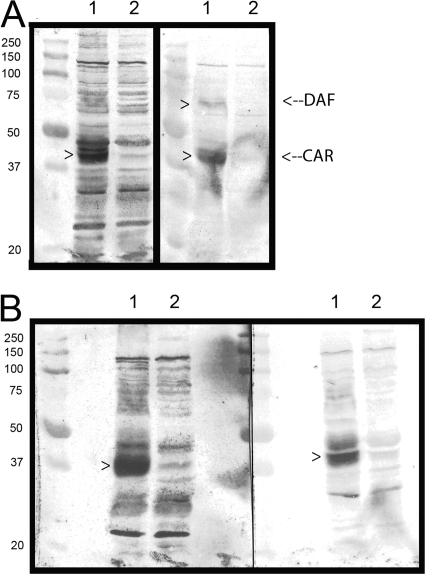
The phenotypic differences among the three CVB3 strains became more apparent when radiolabeled viruses were used to probe blots of cell lysates (Fig. 5). As expected from previous results (5), CVB3/28 detected a strong CAR presence in the HeLa cell lysate with a much weaker presence in the RD cell lysate. CVB3/RD detected bands corresponding to both CAR and DAF. However, CVB3/RDVa bound multiple bands with comparable signals in both HeLa and RD lanes, in addition to bands corresponding to CAR and DAF. No CAR was detected in the lysate from the carrier culture by any of the CVB strains (Fig. 5, lanes 3), consistent with the ongoing infection and loss of CAR-positive cells. CVB3/RDVa binding to bands other than DAF and CAR was reduced or eliminated by blocking the blot with histones (Fig. 6A). In a complementary manner, the addition of heparin to the CVB3/RDVa probe solution also reduced the intensity of, or eliminated binding to, many of the additional bands (Fig. 6B). These data confirm that CAR is bound by all three CVB3 strains and that the hemagglutinating CVB3 strains also bind DAF and reveal that CVB3/RDVa has acquired the additional capacity to bind multiple macromolecules present in both HeLa and RD cell lysates, most likely via charge-mediated interactions.
FIG. 6.
Ionic macromolecules reduce promiscuous binding of CVB3/RDVa on overlay blots. Blots of HeLa cell lysates (lanes 1) and RD cell lysates (lanes 2) were prepared and probed with radiolabeled CVB3/RDVa. The blots were prepared as described for Fig. 5. In panel A, the blocking solution for the blot on the right included lysine-rich histone (calf thymus [ICN]) at 1 mg/ml, and the buffer used for diluting the radiolabeled CVB3/RDVa included histone at 1.4 mg/ml. In panel B, the blot on the right was probed with radiolabeled CVB3/RDVa in solution supplemented with heparin (Sigma) at 2 mg/ml (preincubated 20 min at room temperature). The left-hand blots in both panels A and B were prepared and probed as described for Fig. 5.
Virus growth in HeLa, RD, and RDt3 cells.
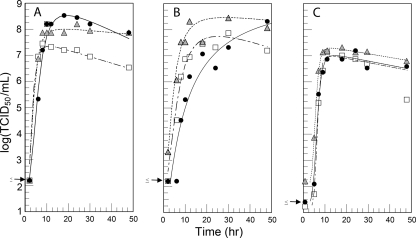
Single-step growth curves (Fig. 7) showed that CVB3/28, CVB3/RD, and CVB3/RDVa all proliferated in both HeLa and RD cells. CVB3/28 proliferation in RD cells was slower than in HeLa cells, but CVB3/28 proliferation was equivalent to that of CVB3/RD and CVB3/RDVa when CAR was expressed in RD cells (Fig. 7C), as previously noted (7). CVB3/28 proliferated more slowly than either CVB3/RD or CVB3/RDVa in both HeLa cells (Fig. 7A) and RD cells (Fig. 7B). Although eventually achieving similar high titers in both cell lines, CVB3/28 required 48 h in RD cells to do so. CVB3/RDVa and CVB3/RD proliferated at comparable rates in HeLa cells, but CVB3/RDVa proliferated more efficiently in the RD and RDt3 cells. The latter result may be due to the measurable infectious CVB3/RDVa associated with these cells at the earliest time points. The results are consistent with increased retention on the cell surface after the inoculation and wash and the notion that the carrier culture environment had selected for a CVB3 strain that was more adapted to the RD environment.
FIG. 7.
Virus growth compared in HeLa, RD, and RDt3 cells. (A to C) Single-step growth curves show that CVB3/28 (black circles), CVB3/RD (white squares), and CVB3/RDVa (gray triangles) all replicate to high titers in HeLa (A), RD (B), and RDt3 (C) cells. The arrows on the y axes indicate the maximum possible log (TCID50/ml) for the corresponding points.
Virus stability at 37°C.
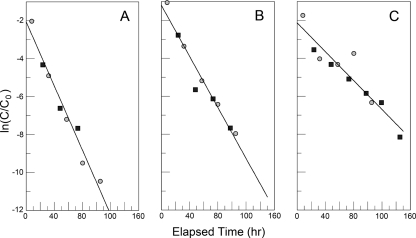
The virus titers decreased at later time points in the growth curves (Fig. 7A), indicating the possibility that the three strains may not be equally stable. Therefore, aliquots of the virus strains were placed at 37°C and sampled over time, and the titers of the viruses were determined again. CVB3/28, CVB3/RD, and CVB3/RDVa all lose activity with first-order kinetics but at different rates (Fig. 8). CVB3/RDVa was the most stable, with a half-life of about 14 h. CVB3/28 was the least stable (half-life of about 7 h). The stability of CVB3/RD was intermediate (half-life of about 9 h).
FIG. 8.
The three CVB3 strains decay in DMEM-10 at 37°C with first-order kinetics but at different rates. (A and B) CVB3/28 (A) and CVB3/RD (B) were assayed on HeLa cells. (C) CVB3/RDVa was assayed on RD cells. Two independent experiments are shown, in which medium was equilibrated in the CO2 incubator before the addition of virus (gray circles) or warmed and used without preequilibration (black squares). Experiments performed so far, including others done as controls in separate studies, provide first-order rate constants of 0.097 ± 0.009/h, 0.076 ± 0.003/h, and 0.048 ± 0.005/h for CVB3/28 (n = 6), CVB3/RD (n = 3), and CVB3/RDVa (n = 4), respectively. All rate constants were statistically different at a P of <0.05 after Bonferroni's correction. C0, concentration of infectious virus determined at the zero time point; C, concentration of infectious virus determined at subsequent time points.
Capsid sequences are largely conserved, with a few key differences.
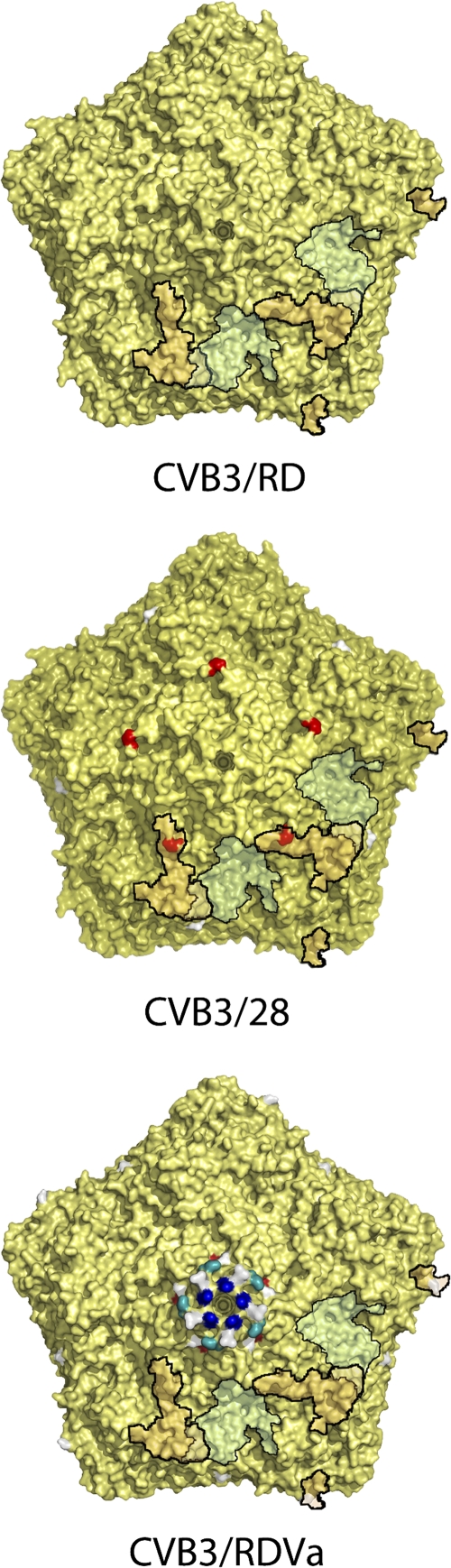
CVB3/28 is propagated from a cDNA for which the sequence is known (22) and confirmed here, with one correction (VP1 Q264). The capsids for the CVB3/RD and CVB3/RDVa populations used in these experiments were also characterized by sequencing the P1 (capsid protein-coding) region of the RNA prepared from the viruses. Results summarized in Table 2 show a total of 11 amino acid differences among these virus strains, and two degenerate positions in the CVB3/RD population sequence. Compared to CVB3/RD, four residues were unique to CVB3/28, one of which maps in the DAF-binding footprint (VP3 Glu234; Fig. 9) (10). The uncharged amino acid, glutamine, occurs at this position in the DAF-binding strains. VP3 Glu76, which is located in the minor DAF footprint near the 3-fold axes of symmetry (10), is present in CVB3/28 and CVB3/RD, but is replaced by glycine in CVB3/RDVa, a pattern that is not consistent with a major role in DAF binding. Residues in both the CAR and DAF-binding regions were conserved in the capsid of CVB3/RDVa. The replacement of proline by histidine at position 126 and glutamine by lysine at position 130 in VP1 result in a dense cluster of positively charged side chains near the prominences associated with the 5-fold axes of symmetry (Fig. 9). As these clustered residues are the only surface-displayed differences compared to CVB3/RD, they are likely responsible for the charge-mediated promiscuous binding observed with CVB3/RDVa.
TABLE 2.
Capsid differences among the three CVB3 strains
Amino acids unique to one of the three virus strains are indicated by boldface type. The two degenerate positions in the CVB3/RD population sequence are indicated by the two shaded rows. Multiple amino acids detected at the same position are separated by a slash (/), and the more prevalent amino acid is listed first. Parentheses indicate that K, a key residue otherwise unique to CVB3/RDVa, was present in a small but detectable amount. The boldface # indicates the glutamate uniquely present in CVB3/28 in the region that has been associated with DAF binding by CVB3/RD. The footprint column indicates amino acid differences that fall within the DAF-binding footprint identified on CVB3/RD (10).
FIG. 9.
Capsid amino acid differences mapped onto the CVB3 structure. Five mature protomers surrounding a 5-fold axis of symmetry (one-twelfth of the capsid) are shown. Black outlines indicate two of the five CAR footprints (pale green) (11) and DAF footprints (dark yellow) (10) which partially overlap at the puff feature. Surface mutations (Table 2) are shown relative to CVB3/RD with red (Asp and Glu), royal blue (Arg and Lys), and blue-green (His) indicating key substitutions with respect to binding characteristics. Other substitutions are shown in light gray. CVB3/28 does not agglutinate red blood cells. All three strains bind CAR. The images were rendered in PyMol based on the structure 1COV.pdb (15).
DISCUSSION
Experiments seeking to explain how CVB3 can require CAR for infection and yet infect RD cells which were reported to lack CAR (7) ultimately demonstrated that RD cells do in fact express a small amount of CAR (5, 14, 20, 21). Under typical conditions of cell culture, small amounts of CAR are transiently present on a subpopulation of cells in the culture (5), the extent of which can vary with culture conditions (21). Subsequent experiments revealed that DAF binding can mediate delivery of CVB3 to CAR sequestered in tight junctions of polarized epithelial cells (6), but this advantageous mechanism was not demonstrable on HeLa cells (16), and it is unlikely to participate during infection of RD cells which are also nonpolar. Consequently, why the DAF-binding phenotype emerged from the CVB3 population passed in RD cells remained unexplained. The current studies support a model wherein virus that is able to bind nonreceptor ligands and that can best survive at 37°C can persist on the cell surface until CAR is expressed by the RD cells. This is especially important in experimental circumstances in which cultures are washed after inoculation and where conditions in a carrier culture maximize competition for cell surface binding sites.
The capsid proteins of CVB3/28, CVB3/RD, and CVB3/RDVa differ by only a few amino acids (Table 2), yet these strains possess binding characteristics that range from exclusive binding to CAR (CVB3/28) to binding CAR, DAF, and numerous other molecules (CVB3/RDVa). The glutamate at position 234 in VP3 of CVB3/28 is singularly located in the landscape associated with DAF binding (10), with glutamine located at this position in both of the DAF-binding strains (Fig. 9). The limited differences between CVB3/28 and CVB3/RD lead to the conclusion that the VP3 Gln234Glu dimorphism is a key determinant of the DAF-binding phenotype. A similar comparison (Fig. 9) of the differences between CVB3/RDVa and the strains that bind CAR or CAR and DAF reveals the obvious cluster of lysine residues flanked by isoleucine and histidine that surround the icosahedral 5-fold axes of symmetry of the CVB3/RDVa capsid. Such a cluster of positive charges, as are present on CVB3/RDVa but not CVB3/28 or CVB3/RD, may mediate the promiscuous binding observed on overlay blots that can be blocked by histones or heparin. Consistent with this finding, CVB3/Nancy-PD, which binds heparan sulfate, also has a positively charged cluster of amino acids near the 5-fold axes of symmetry that results from the replacement of two glutamates by a lysine and an alanine (19).
The ability to bind multiple cell surface ligands confers replicative advantages to these CVB3 strains, as is readily apparent from single-step growth analysis (Fig. 7). All three virus strains replicated with comparable kinetics in CAR-rich HeLa cells and RDt3 cells; CVB3/28 lagged behind the other strains only slightly. All three CVB3 strains have access to CAR on the HeLa cells until the cells are washed at the end of the 1-h inoculation. Most of the HeLa cells are infected and killed in a single cycle of virus replication. CVB3/RD and CVB3/RDVa benefit from a reservoir of virions bound to other molecules on the cell surface, and virus from this reservoir continues to infect cells that escaped infection during the hour of intended inoculation. This effect is greatly amplified on RD cells wherein only a fraction of the cells express CAR during the time they are exposed to virus. Only the RD cells expressing CAR are infected during the planned inoculation, but any reservoir of viruses that remain bound to the cell surface provides for continued infection of cells as they express new CAR. As a result, CVB3/RDVa and CVB3/RD kill most of the RD cells in a single extended round of replication.
CVB3/RDVa emerged from the carrier culture in which both cells and CAR were sparse. The ability to bind CAR, DAF, and multiple ligands present in greater abundance than CAR or DAF may provide an expanded reservoir of viruses competing for scarce and transiently expressed CAR. This CVB3 strain thus competes most effectively for access to new CAR presented on the cell surface. These conditions that necessitate virus survival until CAR becomes available also selected for more stable virions in both the CVB3/RD and CVB3/RDVa populations.
In this study, only the P1 regions of the CVB3 genomes have been sequenced, and mutations in other parts of the genome may contribute to some of the phenotypic variation (e.g., small differences in growth rates and stabilities), but capsid residues are expected to determine whether the virus strains bind DAF or other ligands as well as contribute to capsid stability. Moreover, when all of the strains can use CAR for infection, it is difficult to prove that they cannot utilize some alternative mechanism for infection. Even with these considerations, the model presented is consistent with the data and remains the most parsimonious construct.
The ability of the virus strains to bind new ligands was accompanied by altered CPE on HeLa and/or RD cells, as was found in the original report describing the CVB3/RD strain (18). Even though there is an apparent shift in tropism from cells that express abundant CAR to cells that express little CAR, amino acids in the CAR-binding region (11) are fully conserved in all three CVB3 strains, and both HeLa and RD cells are protected from infection by receptor blockade with monoclonal antibody Rmcb or preincubation of the virus strains with soluble CAR. Thus, the acquisition of the ability to bind new ligands corresponds to the abilities of the viral strains to cause complete CPE on HeLa and/or RD cells, but the strains remain dependent on CAR to infect the cells. Data available at this time provide no compelling reason to invoke use of receptors other than CAR for these viruses to infect cells. Indeed, consideration of these new results in context with historic data provides a model of virus evolution and adaptation in which fitness is defined not so much by the ability to strike and replicate quickly, but by the ability to persist on the cell surface and remain viable, patiently awaiting the appearance of CAR.
Acknowledgments
This work was partially supported by the following grants from the National Institutes of Health: grant R01 AI054551 (S.D.C.), grant K22 AI 079271 (S.H.), and grant R01 AI081830 (N.M.C. and S.T.). S.D.C. was also supported by funds from the Edna Ittner Pediatric Research Support Fund.
Aaron Schumacher purified the sCAR used in these experiments.
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1.Bergelson, J. M., et al. 1995. Coxsackievirus B3 adapted to growth in RD cells binds to decay accelerating factor (CD55). J. Virol. 69:1903-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carson, S. D. 2000. Limited proteolysis of the coxsackievirus and adenovirus receptor (CAR) on HeLa cells exposed to trypsin. FEBS Lett. 484:149-152. [DOI] [PubMed] [Google Scholar]
- 3.Carson, S. D., N. M. Chapman, and S. M. Tracy. 1997. Purification of the putative coxsackievirus B receptor from HeLa cells. Biochem. Biophys. Res. Commun. 233:325-328. [DOI] [PubMed] [Google Scholar]
- 4.Carson, S. D., J. T. Hobbs, S. M. Tracy, and N. M. Chapman. 1999. Expression of the coxsackievirus and adenovirus receptor in cultured human umbilical vein endothelial cells: regulation in response to cell density. J. Virol. 73:7077-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carson, S. D., K.-S. Kim, S. J. Pirruccello, S. Tracy, and N. M. Chapman. 2007. Endogenous low-level expression of the coxsackievirus and adenovirus receptor enables coxsackievirus B3 infection of RD cells. J. Gen. Virol. 88:3031-3038. [DOI] [PubMed] [Google Scholar]
- 6.Coyne, C. B., and J. M. Bergelson. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119-131. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, K. A., N. M. Chapman, and S. D. Carson. 2003. Caspase-3 activation and ERK phosphorylation during CVB3 infection of cells: influence of the coxsackievirus and adenovirus receptor and engineered variants. Virus Res. 92:179-186. [DOI] [PubMed] [Google Scholar]
- 8.de Verdugo, U. R., et al. 1995. Characterization of a 100-kilodalton binding protein for the six serotypes of coxsackie B viruses. J. Virol. 69:6751-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodfellow, I. G., et al. 2005. Inhibition of coxsackie B virus infection by soluble forms of its receptors: binding affinities, altered particle formation, and competition with cellular receptors. J. Virol. 79:12016-12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafenstein, S., et al. 2007. Interaction of decay-accelerating factor with coxsackievirus B3. J. Virol. 81:12927-12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Y., et al. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu, K. H., K. Lonberg-Holm, B. Alstein, and R. L. Crowell. 1988. A monoclonal antibody specific for the cellular receptor for the group B coxsackieviruses. J. Virol. 62:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Milstone, A. M., et al. 2005. Interaction with coxsackievirus and adenovirus receptor, but not with decay-accelerating factor (DAF), induces A-particle formation in a DAF-binding coxsackievirus B3 isolate. J. Virol. 79:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muckelbauer, J. K., et al. 1995. Structure determination of coxsackievirus B3 to 3.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 51:871-877. [DOI] [PubMed] [Google Scholar]
- 16.Patel, K., C. B. Coyne, and J. M. Bergelson. 2009. Dynamin- and lipid raft-dependent entry of decay accelerating factor (DAF)-binding and non-DAF-binding coxsackieviruses into nonpolarized cells. J. Virol. 83:11064-11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell, R. M., T. Ward, I. Goodfellow, J. W. Almond, and D. J. Evans. 1999. Mapping the binding domains on decay accelerating factor (DAF) for hemagglutinating enteroviruses: implications for the evolution of a DAF-binding phenotype. J. Gen. Virol. 80:3145-3152. [DOI] [PubMed] [Google Scholar]
- 18.Reagan, K. J., B. Goldberg, and R. L. Crowell. 1984. Altered receptor specificity of coxsackievirus B3 after growth in rhabdomyosarcoma cells. J. Virol. 49:635-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidtke, M., et al. 2000. Attachment of coxsackievirus B3 variants to various cell lines: mapping of phenotypic differences in capsid protein VP1. Virology 275:77-88. [DOI] [PubMed] [Google Scholar]
- 20.Shafren, D. R., et al. 1995. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J. Virol. 69:3873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafren, D. R., D. T. Williams, and R. D. Barry. 1997. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J. Virol. 71:9844-9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu, Z., et al. 1995. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the 5′ nontranslated region. J. Virol. 69:4607-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zautner, A. E., U. Korner, A. Henke, C. Badorff, and M. Schmidtke. 2003. Heparan sulfates and coxsackievirus-adenovirus receptor: each one mediates coxsackievirus B3 PD infection J. Virol. 77:10071-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]