Abstract
Late domains are short peptide sequences encoded by enveloped viruses to promote the final separation of the nascent virus from the infected cell. These amino acid motifs facilitate viral egress by interacting with components of the ESCRT (endosomal sorting complex required for transport) machinery, ultimately leading to membrane scission by recruiting ESCRT-III to the site of viral budding. PPXY late (L) domains present in viruses such as murine leukemia virus (MLV) or human T-cell leukemia virus type 1 (HTLV-1) access the ESCRT pathway via interaction with HECT ubiquitin ligases (WWP1, WWP2, and Itch). However, the mechanism of ESCRT-III recruitment in this context remains elusive. In this study, we tested the arrestin-related trafficking (ART) proteins, namely, ARRDC1 (arrestin domain-containing protein 1) to ARRDC4 and TXNIP (thioredoxin-interacting protein), for their ability to function as adaptors between HECT ubiquitin ligases and the core ESCRT machinery in PPXY-dependent budding. We present several lines of evidence in support of such a role: ARTs interact with HECT ubiquitin ligases, and they also exhibit multiple interactions with components of the ESCRT pathway, namely, ALIX and Tsg101, and perhaps with an as yet unidentified factor. Additionally, the ARTs can be recruited to the site of viral budding, and their overexpression results in a PPXY-specific inhibition of MLV budding. Lastly, we show that WWP1 changes the ubiquitination status of ARRDC1, suggesting that the ARTs may provide a platform for ubiquitination in PPXY-dependent budding. Taken together, our results support a model whereby ARTs are involved in PPXY-mediated budding by interacting with HECT ubiquitin ligases and providing several alternative routes for ESCRT-III recruitment.
Budding of a wide variety of enveloped viruses is entirely dependent on the presence of short peptide sequences in their Gag or matrix (MA) proteins termed late (L) domains (4, 12, 14, 40). Crucially, the final separation of viral particles from the host cell mediated by every known L domain requires the recruitment of the ESCRT machinery (36, 55, 59), a highly conserved set of protein complexes (ESCRT-0, -I, -II, and -III) that is primarily involved in cellular processes that require the scission of topologically equivalent membrane tethers, namely, multivesicular body (MVB) formation and abscission of the midbody during cytokinesis (7, 46, 48). In the context of MVB biogenesis, the ESCRT machinery selectively recognizes membrane proteins marked for degradation via monoubiquitin tags and recruits them into the lumen of MVBs, subsequently inducing the formation of intralumenal vesicles (ILVs) (22, 49). The different ESCRT complexes are responsible for specific functions in this process: ESCRT-0, -I, and -II are soluble complexes containing ubiquitin binding domains that recognize the endosomal cargo (24, 61), whereas downstream membrane scission events are mediated by activated ESCRT-III subunits that assemble on endosomal membranes (23, 63).
All ESCRT-dependent viruses share a requirement for the membrane scission activity provided by ESCRT-III, but different types of L domains bind different adaptor proteins within the ESCRT machinery: viruses relying on PTAP motifs recruit Tsg101 (9, 13, 37, 58), a subunit of ESCRT-I, while LYPXL motifs bind AIP1/ALIX (11, 36, 55, 59), an ESCRT-associated protein. An L domain more recently identified in the paramyxovirus simian virus 5 (SV5), the FPIV motif (51), interacts with the ESCRT machinery via a currently unknown mechanism. Lastly, viruses such as murine leukemia virus (MLV), human T-cell leukemia virus type 1 (HTLV-1), and Ebola virus encode PPXY motifs that recruit a subset of HECT ubiquitin ligases belonging to the Nedd4 family (5, 16, 17, 28, 34, 62), namely, WWP1, WWP2, and Itch, that are required for tagging membrane proteins for lysosomal degradation (47).
In addition to the role of HECT ubiquitin ligases in PPXY-dependent budding, recent work has highlighted their contributions to retroviral budding through L-domain activities that do not involve the above-mentioned amino acid motifs. For example, it is now established that Nedd4L can stimulate the release and infectivity of HIV-1 viruses that lack the PTAP and LYPXL late domains, and this activity requires the enzymatic activity encoded by Nedd4L (8, 57, 60). Additionally, Itch is recruited by MLV Gag in a PPXY-independent manner to promote viral egress, and, as shown for PPXY-dependent budding, this activity also requires the core ESCRT machinery (25).
Importantly, the mechanism of ESCRT-III recruitment by HECT ubiquitin ligases remains elusive since direct interactions between these proteins and the core ESCRT machinery have not been found (33). The fact that the enzymatic activity of HECT ubiquitin ligases is critical for their function in viral budding (34, 50, 60) suggests two possible scenarios for ESCRT-III recruitment in PPXY-mediated budding: the target for ubiquitination by HECT ubiquitin ligases could be a viral protein that would subsequently recruit ESCRT-III via interactions of ESCRT proteins with ubiquitin, or, alternatively, an unidentified ubiquitin acceptor could serve as a bridge between HECT ubiquitin ligases and the core ESCRT machinery. Accordingly, the observation that cumulative mutation of lysine residues in Gag can lead to a late budding arrest in HIV-1 (15) and Rous sarcoma virus (RSV) (52) suggested that Gag might be the relevant substrate for L-domain activity, a hypothesis that is consistent with the functional rescue of an L-domain-deficient Gag upon fusion to ubiquitin (26). However, several lines of evidence clearly indicate that ubiquitination of viral proteins is not exclusively responsible for ESCRT recruitment in PPXY-mediated budding. First, only a small fraction of retroviral Gag is ubiquitinated (43, 44, 54) and the PPXY motif is unable to induce viral budding in the context of HIV-1; yet its presence leads to increased levels of Gag ubiquitination (35). Crucially, Zhadina et al. also have been able to demonstrate that PPXY-dependent egress in prototypic foamy virus (PFV) remains dependent on the ubiquitin ligase activity of WWP1 in the complete absence of ubiquitin acceptor sites in Gag (65). Similarly, HTLV Gag featuring a mutation in the exclusive ubiquitin acceptor in MA exhibits only a modest defect in particle release, and this mutant is also hypersensitive to the inhibitory effect of catalytically inactive WWP1 (18). Intriguingly, a cellular scenario that parallels findings in PPXY-mediated budding has been described; that is, sorting of the endosomal cargo Sna3 into MVBs is dependent on the ubiquitin ligase activity of Rsp5, the yeast homologue of WWP1, but ubiquitination of the cargo itself is dispensable (39). Altogether, these findings clearly argue for the existence of one or several cellular factors as the target for HECT ubiquitin ligases, but the identity of these adaptor proteins is unknown and has remained a sought-after factor in the field.
In order to identify potential adaptor proteins required for PPXY-dependent budding, we decided to study the human homologues of the arrestin-related trafficking adaptors (ARTs), a family of proteins that is involved in regulation of protein turnover at the plasma membrane (32, 41, 42). Importantly, the interaction between the Saccharomyces cerevisiae ARTs and the Rsp5/Nedd4-like ubiquitin ligase results in the ubiquitination of both the cargoes and the ARTs (27, 32, 45), and, as a consequence of this interaction, plasma membrane proteins are internalized and targeted to the MVBs for degradation, thus suggesting potential interactions with the ESCRT machinery. Crucially, we show in this study that members of the ART family interact with the HECT ubiquitin ligases involved in retroviral budding (WWP1, WWP2, and Itch), and we also find interactions with Tsg101 and ALIX, supporting their role as functional adaptors between HECT ubiquitin ligases and the ESCRT machinery. The role of the ARTs in retroviral budding is also supported by their recruitment to the sites of viral budding and by the dominant negative inhibition of PPXY-dependent budding.
MATERIALS AND METHODS
Expression constructs.
Expression constructs for ARRDC1 (arrestin domain-containing protein 1), -2, -3, and -4 and for TXNIP (thioredoxin-interacting protein) and Arrestin-2 were derived from image clones 5211669 (ARRDC1), 4817429 (ARRDC2), 3908916 (ARRDC3), 30345612 (ARRDC4), 7939549 (TXNIP), and 3604829 (Arrestin-2). Complete coding sequences were amplified by PCR that introduced EcoRI/XhoI (ARRDC1, ARRDC3, and TXNIP), EcoRI/NotI (ARRDC2), and NotI/NotI (ARRDC4 and Arrestin-2) restriction sites on the 5′ and 3′ ends. Coding sequences were cloned into pVP16 and pGBKT7 for yeast two-hybrid assays, and pCR3.1-yellow fluorescent protein (YFP), pCR3.1-cyan fluorescent protein (CFP), pCR3.1-mCherry, pCR3.1-hemagglutinin (HA) and pCAG-glutathione S-transferase (GST) were used for protein expression experiments in mammalian cells. Deletion constructs for ARRDC1 were also generated by PCR; all additional expression constructs used in this study have been described elsewhere (1, 6, 34, 38).
Cells and reagents.
293T and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM)-GlutaMAX high glucose supplemented with 10% fetal calf serum (FCS) and Gentamicin (20 μg/ml) (all from Invitrogen). S. cerevisiae (strain Y190) was grown on YPDA medium (yeast peptone, dextrose, and adenine) (Sigma-Aldrich); transformed yeast was selected in synthetic defined (SD) medium lacking tryptophan and leucine (Sigma-Aldrich). The following antibodies were used: rabbit polyclonal anti-ARRDC1 (Abcam), rabbit polyclonal anti-ARRDC3 (Abcam), mouse anti-green fluorescent protein (GFP), mixture of two monoclonal antibodies (7.1 and 13.1; Roche Applied Science), horse polyclonal anti-equine infectious anemia virus (EIAV) Gag (Lady serum; kindly provided by R. Montelaro, University of Pittsburgh, Pittsburgh, PA), rabbit polyclonal anti-HA epitope tag (Rockland), mouse monoclonal anti-HA epitope (H.11; Covance), mouse monoclonal anti-HIV-1 p24 (hybridoma cell line, clone 183-H12-5C; AIDS Repository Reagent Program), rabbit polyclonal anti-Hsp90 (Santa Cruz Biotechnology), mouse monoclonal anti-tubulin (DM1A; Sigma), IRDye 680 goat anti-mouse IgG (Li-Cor), IRDye 800CW goat anti-rabbit IgG (Li-Cor), and horseradish peroxidase-conjugated goat anti-horse IgG(H+L) (Jackson ImmunoResearch Europe Ltd). Small interfering RNAs (siRNAs) targeting Tsg101 (CCUCCAGUCUUCUCUCGUC), ALIX (GAAGGAUGCUUUCGAUAAAUU), ARRDC2 (GACAACGGCUCCACACGUC), ARRDC3 (CCACAGACACCACUCGCUA), ARRDC4 (GAGAAGCUAUUCCAAUCUA), and TXNIP (CAACUCCUCUGCUAGAUGA) were obtained from Dharmacon RNA Technologies. siRNA constructs targeting ARRDC1 (CAGCCUCGUGUUCUAUAUCUU) were purchased from Qiagen.
Yeast two-hybrid screening.
Yeast cells were transformed using 1 μg each of pGBKT7 and pVp16 expression constructs and grown on selection medium for 3 days at 30°C. Protein interactions were determined by the levels of β-galactosidase activity expressed as previously described (37).
Coprecipitation assays.
293T cells previously seeded in six-well plates were transiently transfected with a total of 4 μg of GST, HA, or YFP expression constructs using polyethylenimine (PEI; Polysciences, Inc.) (10). At 48 h posttransfection, cells were lysed in 1 ml of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5% glycerol, 1% Triton X-100, and a protease inhibitor mixture (complete mini-EDTA-free; Roche Applied Science). Clarified lysates were incubated with glutathione-Sepharose beads (Amersham Biosciences) for 3 h at 4°C on a rotating wheel and subsequently washed three times in a buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5% glycerol, and 0.1% Triton X-100. Proteins bound to Sepharose beads were eluted by addition of 100 μl of sodium dodecyl sulfate (SDS) sample buffer and analyzed by SDS-PAGE, followed by Western blotting or Coomassie staining.
Ubiquitination assays.
293T cells grown in six-well plates were transiently transfected with pMT123 HA-ubiquitin (1 μg), pCAG-GST-ARRDC1 (1.5 μg), and pCR3.1-YFP constructs as indicated on figures (1.5 μg). Cells were lysed 48 h posttransfection in 1 ml of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5% glycerol, 1% Triton X-100, and a protease inhibitor mixture (complete mini-EDTA-free; Roche Applied Science), and GST-ARRDC1 was isolated using glutathione-Sepharose beads as described above. Proteins were eluted and analyzed by SDS-PAGE followed by Western blotting.
Infectious particle production.
For analysis of MLV infectious particle production, 293T cells in a 24-well format were cotransfected with expression constructs encoding MLV provirus, vesicular stomatitis virus G protein (VSV-G) envelope, and HIV Tat. The following constructs were used: pMLV NCS (MLV) or pMLV p6pY (MLV human PTAP [hPTAP]) (64) (200 ng), pHIT VSV-G (100 ng), pMSCV Tat (200 ng; where MSCV is murine stem cell virus), and constructs encoding YFP-ARTs as indicated on figures. For HIV-1 infectious particle release assays, the proviral construct pNL/HXB STOP (300 ng) lacking a late domain was cotransfected with the trans-complementing construct pNL/HxB p6 or pNL/HxB p9 (200 ng). Supernatants were harvested 48 h posttransfection, and 100 μl of filtered (22-μm pore size) supernatants was used for infection of TZM-bl indicator cells (CD4+, CXCR4+, CCR5+, HIV-1 LTR-lacZ [where LTR is long terminal repeat]). Infectious virus release was determined by β-galactosidase activity in TZM lysates using Galacto-Star detection reagent (Applied Biosystems).
For analyzing the effect of siRNA-mediated knockdown on infectious particle production, 293T cells were transfected with 50 pmol of siRNAs using Dharmafect 1 (Dharmacon RNA Technologies) and transfected a second time, together with proviral plasmids as described above, 48 h after the initial transfection using Lipofectamine 2000 (Invitrogen).
VLP production.
At 36 h posttransfection, supernatants of 293T cells transiently coexpressing HA-tagged MLV Gag and YFP fusion proteins or EIAV Gag constructs (kindly provided by E. Freed, National Cancer Institute, Frederick, MD) in cells previously treated with siRNAs as described above were harvested, filtered (22-μm pore size), and centrifuged through 20% sucrose (at 20,000 × g for 120 min). Isolated virus-like particles (VLPs) as well as cellular lysates were analyzed by SDS-PAGE, followed by Western blotting.
Microscopy.
293T or HeLa cells grown on 13-mm glass coverslips were transiently transfected with a total of 1 μg of expression construct encoding YFP, CFP, or mCherry fusion protein, using PEI (293T cells) or Lipofectamine 2000 (Invitrogen) (HeLa cells) as transfectants. For colocalization studies of ARRDC1 with Ebola VP40, cells were additionally transfected with pCR3.1 myc-VP40 constructs (35). Cells were harvested at 48 h posttransfection, fixed in 4% paraformaldehyde (PFA), and mounted in Mowiol (Polysciences, Inc.). Samples were analyzed using a TCS SP2 AOBS confocal microscope (equipped with an HCX PL APO CS [confocal scanning], 63.0×/1.4 oil objective; Leica).
Western blotting.
Samples were separated on 8% (ubiquitination assays), 14% (HIV Gag), or 10% (all other assays) polyacrylamide gels and transferred to nitrocellulose membranes. Proteins were detected using a Li-Cor Odyssey Infrared Imaging system, and signal intensities were determined using the Odyssey, version 3.0, software.
RESULTS
ART proteins interact both with HECT ubiquitin ligases and the ESCRT machinery.
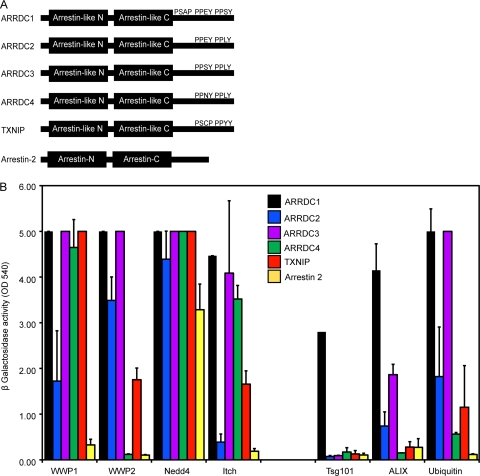
The group of ART proteins is characterized by two domains with a predicted arrestin fold that are connected by a flexible linker as well as two PPXY motifs at the C terminus (32). The human genome encodes five proteins with these characteristics that were selected for our study: ARRDC1 (arrestin domain-containing protein 1), ARRDC2, ARRDC3, ARRDC4, and TXNIP (thioredoxin-interacting protein) (Fig. 1 A). We also chose Arrestin-2, a member of the arrestin family that lacks PPXY motifs in its C terminus, as a control for our studies (3). A yeast two-hybrid-based approach was followed to determine the interaction of the ARTs with WWP1, WWP2, Nedd4, and Itch, showing that each of the ARTs studied interacted with all tested HECT ubiquitin ligases, with the exception of ARRDC2 and ARRDC4 that lacked detectable interactions with Itch and WWP2, respectively (Fig. 1B). In contrast, Arrestin-2 exhibited binding only to Nedd4 while the previously reported interaction with Itch (3) was not detectable in our system.
FIG. 1.
(A) Schematic drawing of human ARTs featuring N- and C-terminal arrestin-like domains as well as PPXY and PSAP motifs in the C terminus as indicated. (B) Yeast two-hybrid screen for interactions of ART proteins with HECT ubiquitin ligases, components of the ESCRT machinery, and ubiquitin. ARTs were expressed as fusion constructs to the Vp16 activation domain for screening against WWP1, Nedd4, and Itch or fused to the GAL4 DNA binding domain for testing binding to WWP2, Tsg101, ALIX, and ubiquitin. Protein interactions were determined by measuring β-galactosidase activity levels and are shown as absorbance units (optical density at 540 nm [OD 540]). Values above saturation levels were arbitrarily set to 5. Error bars indicate the standard deviations from the mean of two independent experiments measured in triplicates.
In a parallel approach, the ability of the ARTs to interact with components of the ESCRT machinery was tested using a panel of all known components of the ESCRT complexes as well as ESCRT-associated proteins (see supplemental Table 1 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf). These experiments revealed a surprising interaction of ARRDC1, ARRDC2, and ARRDC3 with ALIX, an ESCRT-III associated protein, and ARRDC1, the only ART that features a PSAP motif in its C terminus, was also found to interact with Tsg101, confirming previous results in yeast (19). No binding partners of ARRDC4, TXNIP, and Arrestin-2 could be detected using this approach (Fig. 1B). In addition to these interactions with the ESCRT machinery, all tested ARTs seemed to bind ubiquitin, reinforcing the notion that this family of proteins is closely linked to the endolysosomal sorting machinery (Fig. 1B).
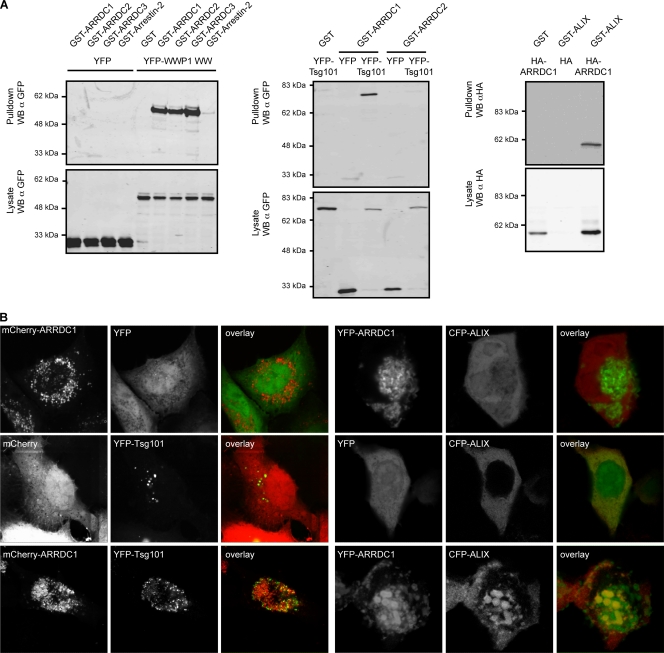
We next performed glutathione S-transferase (GST) coprecipitation assays to confirm the interactions identified above. For this, the region of WWP1 containing the four WW domains, which mediate binding to PPXY motifs (53), was fused to YFP and tested for interactions with GST fusions to ARRDC1, -2, and -3 or to Arrestin-2. These experiments showed that the WW domains coprecipitated with ARRDC1, -2, and -3 but not with Arrestin-2 (Fig. 2 A, left panel). Parallel experiments expressing the WW domains of WWP2, Nedd4, and Itch yielded comparable results (see supplemental Fig. 1 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf). In agreement with the interactions identified by yeast two-hybrid assay, coprecipitation of ARRDC1 with both Tsg101 and ALIX could also be confirmed in GST pulldown assays (Fig. 2A, middle and right panels).
FIG. 2.
(A) Coprecipitation assays showing interactions of ARRDC1, -2, and -3 with WWP1 (WW domains) (left), of ARRDC1 and -2 with Tsg101 (middle), and of ARRDC1 with ALIX (right). Proteins fused to GST, HA, or YFP were transiently expressed in 293T cells as indicated, and GST-tagged proteins were precipitated using glutathione-Sepharose beads. Samples were analyzed for coprecipitated binding partners by SDS-PAGE followed by Western blotting (WB). Figures shown are representative of at least two independent experiments. α, anti. (B) Colocalization of ALIX and Tsg101 with ARRD1. Proteins fused to mCherry, YFP, or CFP as indicated were transiently expressed in 293T cells, and fixed cells were analyzed for protein colocalization by confocal microscopy. Pictures shown are representative of at least two independent experiments. Colocalization of coexpressed proteins was quantified by calculating the Pearson's correlation coefficient using Image J public domain image processing software (plug-in). This analysis yielded the following results: mCherry-ARRDC1/YFP, −0.07 ± 0.18 (n = 7); mCherry/YFP-Tsg101, 0.10 ± 0.07 (n = 8); mCherry-ARRDC1/YFP-Tsg101, 0.7 ± 0.18 (n = 11); YFP-ARRDC1/CFP, −0.26 ± 0.16(n = 12); YFP/CFP-ALIX, 0.55 ± 0.11 (n = 7); YFP-ARRDC1/CFP-ALIX, 0.69 ± 0.11 (n = 9). Data are represented as average value ± standard deviation. n, number of analyzed cells.
As a second approach to confirm the ARRDC1 interactions with the ESCRT machinery, the cellular localization of exogenous ARRDC1 was next analyzed upon coexpression with WWP1, Tsg101, and ALIX. In these experiments mCherry-ARRDC1 showed a punctate distribution that was partly relocalized to the plasma membrane upon cotransfection with YFP-WWP1 (see supplemental Fig. 2 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf). Moreover, coexpression of mCherry-ARRDC1 with YFP-Tsg101 resulted in a marked relocalization of YFP-Tsg101 to ARRDC1-positive structures (Fig. 2B, left panel). While this result provides indirect evidence of an interaction between these two proteins, it has to be noted that the observed colocalization could also be the consequence of aberrant endosome induction in cells overexpressing Tsg101 and possibly ARRDC1, a phenotype that would likewise lead to an overlay of both signals. Overexpression of YFP-ARRDC1 also changed the diffuse cytoplasmic localization of CFP-ALIX to a punctate distribution that overlaid with YFP-ARRDC1 (Fig. 2B, right panel). In summary, these results showed that the mammalian ARTs were indeed able to interact with both HECT ubiquitin ligases and components of the ESCRT machinery, suggesting that proteins of this family are likely candidates to serve as adaptor proteins in PPXY-mediated viral budding.
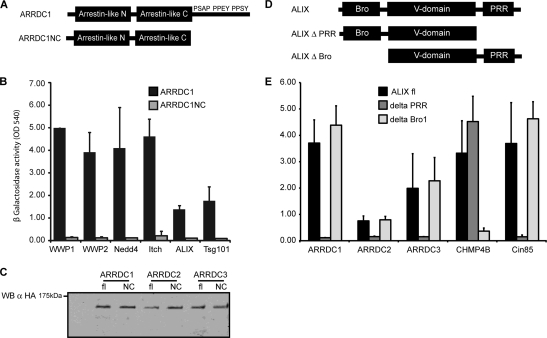
We next aimed to gain insights into the functional domains responsible for interactions between the ARTs, the HECT ubiquitin ligases, and the ESCRT machinery. The localization of the PPXY and PSAP motifs in ARRDC1 suggested that interactions with HECT ubiquitin ligases and Tsg101, respectively, could take place via the C-terminal region of the protein. We therefore generated a deletion mutant of ARRDC1 that was deficient for all residues C-terminal of the arrestin-like C domain (ARRDC1NC, ARRDC1 containing the N- and C-terminal arrestin-like domains [NC construct]) (Fig. 3 A). As expected, ARRDC1NC lost its ability to interact with WWP1, WWP2, Nedd4, and Itch (Fig. 3B), and this construct also failed to interact with Tsg101, likely due to the deletion of the PSAP motif. Surprisingly, ARRDC1NC was also unable to interact with ALIX despite the lack of known interaction motifs at the C terminus (Fig. 3B). Analysis of ARRDC2NC and ARRDC3NC interactions yielded equivalent results (data not shown), showing that deletion of the C-terminal region in the ARTs impaired their binding to all identified interaction partners. Importantly, the lack of detectable interactions by yeast two-hybrid was not due to reduced protein expression in yeast since the expression levels of the full-length and deleted versions were entirely comparable (Fig. 3C). In a parallel approach, the interaction domains in ALIX were also analyzed, showing that the proline-rich region was required for the interaction with the ARTs (Fig. 3D and E).
FIG. 3.
(A) Schematic drawing of ARRDC1 wild type (ARRDC1) and truncation mutant (ARRDC1NC) lacking the C terminus. (B) Binding of ARRDC1 to HECT ubiquitin ligases, ALIX, and Tsg101 is dependent on binding motifs in the C terminus. C-terminally truncated ARRDC1 fused to the Vp16 activation domain or GAL4 DNA binding domain was tested for its ability to interact with WWP1, WWP2, Nedd4, Itch, ALIX, and Tsg101 by yeast two-hybrid screening. Protein interactions were detected by measuring β-galactosidase activity in yeast lysates at an optical density at 540 nm (OD 540). Error bars indicate the standard deviations from the mean of two independent experiments measured in triplicates. (C) Expression of full-length (fl) and C-terminally truncated (NC) ARRDC1, ARRDC2, and ARRDC3 in yeast lysates. ARTs were expressed in the context of the HB18 vector (Vp16 activation domain), thereby generating an HA fusion protein. Protein expression was determined by lysis of yeast in protein sample buffer, followed by SDS-PAGE and Western blotting (WB) against HA. α, anti. (D) Schematic drawing of ALIX featuring the Bro1 and the V domain as well as the proline-rich region (PRR). (E) Binding of ARRDC1, ARRDC2, and ARRDC3 to ALIX is dependent on the proline-rich region in Alix. ARTs, CHMP4B, and Cin85 expressed as fusion proteins to the GAL4 DNA binding domain were tested for binding to ALIX by yeast two-hybrid screening as described for panel A.
HECT-dependent ubiquitination of ARTs.
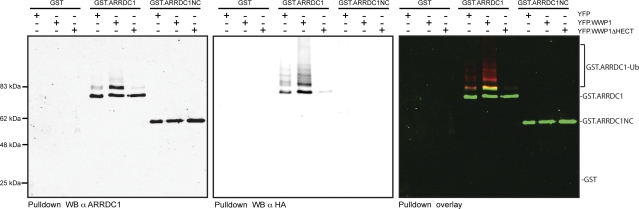
As discussed above, the enzymatic activity of WWP1 is required for PPXY-dependent budding, but the target for this ubiquitination remains unclear. Previous studies of yeast and Aspergillus nidulans have shown that the ARTs are monoubiquitinated by HECT ubiquitin ligases as a regulatory mechanism (19, 20, 32). Thus, it seemed likely that human ARTs could also be modified by the subset of HECT ubiquitin ligases that promote retroviral budding. In order to test this hypothesis, we determined the impact of WWP1 on the ubiquitination status of exogenous ARRDC1. These experiments revealed the presence of two immunoreactive bands of different mobilities upon pulldown of transfected GST-ARRDC1 (Fig. 4), consistent with ARRDC1 monoubiquitination in cell lysates. The identity of the higher band as a monoubiquitinated form of ARRDC1 was confirmed by cotransfection with HA-ubiquitin, which allowed the simultaneous detection of ARRDC1 and ubiquitin in the purified GST-ARRDC1 fraction. Importantly, the overexpression of YFP-WWP1 resulted in an approximately 3- to 4-fold increase in the monoubiquitination status of ARRDC1, and a higher band that is likely to represent ARRDC1 attached to a second ubiquitin moiety was also increased, providing further proof for a functional interaction between ARRDC1 and WWP1. Accordingly, the expression of WWP1 ΔHECT, a WWP1 version that lacks the catalytic domain, dramatically reduced ubiquitination of ARRDC1. It is important to note that WWP1 ΔHECT exhibits a very potent dominant negative activity in PPXY-dependent budding (34), suggesting that at least part of this activity might be due to changes in the ubiquitination status of the endogenous ARTs. In order to ensure specificity of the observed effects, we next analyzed potential ubiquitination of GST-tagged ARRDC1NC, which was found to be devoid of binding to HECT ubiquitin ligases (Fig. 3). This analysis showed that despite efficient isolation of GST-ARRDC1NC (Fig. 4, left panel), no ubiquitination of this mutant could be detected (Fig. 4, middle panel). This result, together with the lack of ubiquitination of GST alone used as a control, clearly argues for the ability of WWP1 to specifically modify ARRDC1 via ubiquitination and excludes a mere global increase of protein ubiquitination by overexpression of this HECT ubiquitin ligase.
FIG. 4.
ARRDC1 can be ubiquitinated by WWP1. 293T cells were transiently transfected with expression constructs for HA-ubiquitin, GST, GST-ARRDC1, or GST-ARRDC1NC together with YFP constructs as indicated. GST-tagged proteins were subsequently isolated using glutathione-Sepharose beads, and samples were analyzed by Western blotting (WB). Western blots were performed using an infrared imaging system that allowed detection of total amounts of ARRDC1 (left panel) as well as its ubiquitinated forms using anti-HA antibodies (middle panel) on the same membrane. An overlay picture of signals acquired in both channels is shown in the right panel. Results presented are representative of three independent experiments. α, anti.
Recruitment of ARRDC1 to the site of viral budding.
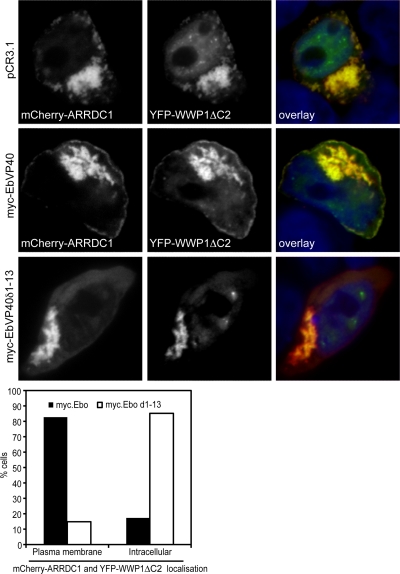
As a first indication of a role of the ARTs in PPXY-mediated budding, we tested if members of this family were recruited to the site of viral budding. We therefore coexpressed mCherry-ARRDC1 with GFP, GFP-tagged wild-type Gag, or a late-domain-deficient Gag mutant. In these experiments we observed that mCherry-ARRDC1 could be recruited to the plasma membrane by the wild type but not by Gag lacking its PPXY motif (see supplemental Fig. 3 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf). However, recruitment was inefficient and occurred in only a small percentage of cells, maybe due to the indirect recruitment of ARRDC1. We therefore addressed this question in an additional system. For this purpose, mCherry-ARRDC1 was coexpressed with the Ebola virus matrix protein VP40 and YFP-WWP1 ΔC2, a truncated version of WWP1 that lacks the membrane recruitment domain and whose localization at the plasma membrane is therefore dependent on its interaction with VP40 (34). In fact, confocal microscopy analysis showed the pronounced relocalization of mCherry-ARRDC1 and YFP-WWP1 ΔC2 from a punctate intracellular distribution to the plasma membrane upon coexpression with Ebola virus VP40 (Fig. 5). Importantly, this relocalization of mCherry-ARRDC1 was not observed in the absence of the PTAPPEY motif in VP40, supporting a model in which ARRDC1 can be recruited to the plasma membrane during PPXY-mediated viral budding via interaction with WWP1.
FIG. 5.
Ebola virus VP40 can recruit ARRDC1 to the plasma membrane, depending on its PTAPPEY motif. 293T cells were transiently transfected with expression constructs for mCherry-ARRDC1 and YFP-WWP1 featuring a deletion of its membrane-targeting domain (YFP-WWP1ΔC2). Localization of ARRDC1 and WWP1ΔC2 upon expression of either empty vector (pCR3.1), wild-type Ebola virus matrix protein VP40 (myc-EbVP40), or a VP40 deletion mutant lacking the late domain (myc-EbVP40δ1-13) was analyzed by confocal microscopy after cells were fixed and nuclei were stained with Hoechst. Localization of mCherry-ARRDC1 and CFP-WWP1ΔC2, either intracellular or at the plasma membrane, was determined by counting at least 50 cells for samples expressing wild-type or late-domain-deficient VP40 (graph). Pictures and graph shown are representative of two independent experiments.
Functional characterization of the ARTs in PPXY-dependent budding.
In order to assess the role of the ARTs in retroviral budding, we first followed an siRNA-based approach to deplete each of them individually or in several combinations. The experiments failed to provide conclusive evidence supporting an absolute requirement for these proteins in PPXY-dependent budding (see supplemental Fig. 4 and 5 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf; also data not shown), but the interpretation of these experiments is unclear since we cannot exclude an incomplete depletion of the ARTs due to the lack of available antibodies that can detect all the targeted endogenous proteins (see supplemental Fig. 4 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf). A second possibility is that each member of the family may play redundant roles in viral budding that would not be revealed with single knockdowns.
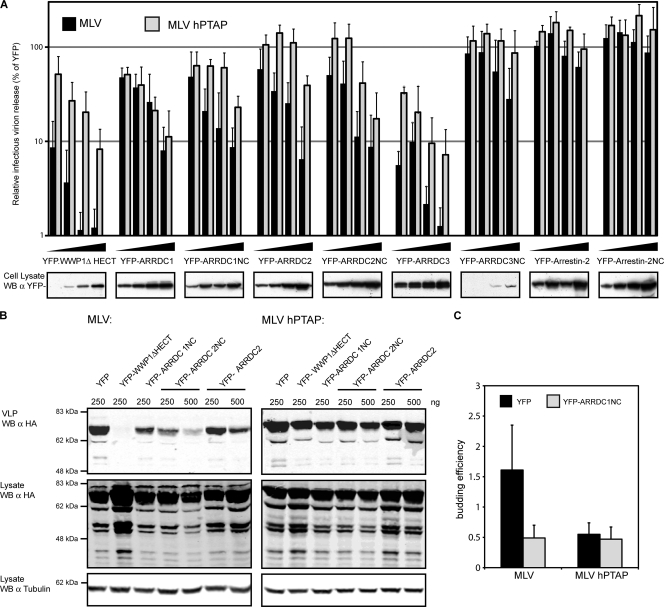
In order to test the latter possibility, a dominant negative approach was followed by determining the effect of overexpressing each of the ARTs on murine leukemia virus (MLV) infectious virus production, using an MLV version that harbors a PTAP motif (MLV hPTAP) to replace the PPXY motif (64) as the control for L-domain-specific effects. These experiments showed a dose-dependent reduction in MLV infectious particle production upon expression of full-length ARRDC1, ARRDC2, and ARRDC3 (Fig. 6 A) while the expression of Arrestin-2 did not exhibit this inhibitory phenotype. Importantly, the observed effects, with the exception of ARRDC1, were specific for PPXY-dependent budding although a comparison with a WWP1 ΔHECT showed less potent effects in cells overexpressing the ARTs. In agreement with this specificity of ART protein overexpression, no effect was observed on HIV-1 infectious virus production in parallel experiments using either HIV relying on its wild-type PTAP or a YPDL L domain from EIAV p9 (see supplemental Fig. 6 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf). One interpretation of these results is that the presence of PPXY motifs in the ARTs might simply compete with the MLV L domain for binding the endogenous HECTs, a possibility that would be in agreement with the lack of specificity of ARRDC1, which contains both PPXY and PTAP motifs. To rule out this possibility, we did similar experiments using versions of the ARTs lacking the PPXY motifs through the deletion of the entire C-terminal region (NC constructs). Importantly, these deletions retain the PPXY-specific inhibition of infectious virus release, whereas a similar deletion in Arrestin-2 does not exhibit this phenotype (Fig. 6A).
FIG. 6.
PPXY-mediated viral particle production is specifically reduced upon transient expression of ART proteins. (A) Analysis of infectious particle production. 293T cells were transfected with MLV provirus expression vectors containing its natural PPXY L domain (MLV) or a mutant depending on a PTAP motif (MLV hPTAP) as well as expression constructs encoding YFP fusion proteins as indicated. Infectious virus release was analyzed using a chemiluminescent assay after infection of TZM-bl reporter cells with supernatants harvested from the transfected 293T cells. In these experiments, absolute infectivity values of 2.4 × 106 for MLV and 1.0 × 106 for MLV hPTAP were obtained on average for cells expressing unfused YFP, and these values were set to 100% to calculate relative infectious virion release for either MLV or MLV hPTAP. Error bars indicate the standard deviation from the mean of at least three independent experiments. (B) Analysis of virus-like particle release. Virus-like particles (VLPs) were isolated from 293T cells transiently expressing HA-tagged MLV Gag featuring either a PPXY (MLV) or a mutant PTAP L domain (MLV hPTAP) together with the indicated YFP fusion proteins. VLPs and cellular lysates were analyzed by Western blotting (WB) against the MLV HA tag; signal intensities were measured using Odyssey, version 3.0, software, and budding efficiency was determined. The depicted Western blots are representative of three independent experiments; the graph illustrates the budding efficiency gained upon expression of YFP-ARRDC1NC. Error bars indicate standard deviations from three independent experiments. α, anti.
In order to confirm the inhibition of the L-domain activity in these experiments, the effect of the NC constructs was also determined for virus-like particle (VLP) production in cells transfected with MLV Gag (Fig. 6B and C). In agreement with data gained for viral infectivity, these experiments confirmed a PPXY-specific inhibition of VLP production and budding efficiency in cells expressing ARRDC1NC and ARRDC2NC (Fig. 6B). Interestingly, the NC construct by itself did not bind HECT ubiquitin ligases, ALIX, Tsg101 (Fig. 3), or ubiquitin (data not shown), thus suggesting the interaction of this region with unknown cellular cofactors required for PPXY budding.
Multiple interactions between ARTs and the ESCRT pathway.
Our results so far suggest that the ARTs may provide a functional bridge between HECT ubiquitin ligases and the ESCRT machinery. One obvious possibility derived from our data is that this bridging activity might occur at least in part through Tsg101 and ALIX, but the codepletion of these proteins has minor effects on MLV budding (see supplemental Fig. 7 at http://www.kcl.ac.uk/content/1/c6/08/27/36/SupplJVRauch.pdf), a result that is also consistent with the modest contribution to viral release of the Tsg101 and ALIX binding sites in MLV Gag (25). This negative result can be rationalized by proposing a potential interaction between the NC region and the ESCRT machinery as suggested above. However, no interaction partners were detectable in a yeast two-hybrid screen using this region as a bait against known components of the ESCRT machinery (data not shown).
FIG. 7.
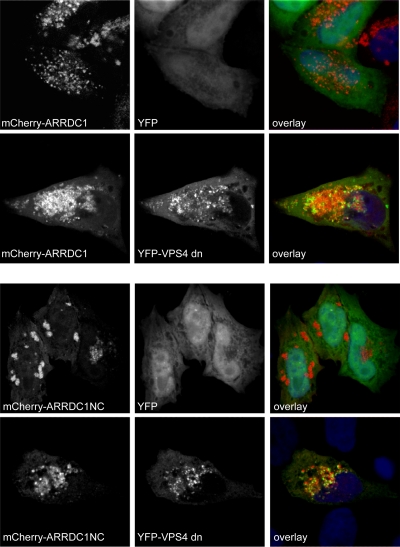
Full-length as well as dominant negative ARRDC1 colocalizes with aberrant endosomes induced by dominant negative VPS4. HeLa cells were transiently transfected with expression constructs encoding mCherry-ARRDC1 or mCherry-ARRDC1NC and dominant negative YFP-VPS4 (YFP-VPS4dn). Cells were fixed, and nuclei were stained using Hoechst stain and analyzed by confocal microscopy for colocalization of ARRDC1 proteins with aberrant endosomes induced by the expression of dominant negative VPS4. Pictures shown are representative of three independent experiments. Colocalization of ARRDC1 constructs with dominant negative VPS4 was quantified by calculating the Pearson's correlation coefficient using Image J public domain image processing software (plug-in). The following values were obtained: mCherry-ARRDC1/YFP, 0.16 ± 0.04 (n = 3); mCherry-ARRDC1/YFP-VPS4dn, 0.66 ± 0.10 (n = 7); mCherry-ARRDC1NC/YFP, −0.24 ± 0.15 (n = 7); mCherry-ARRDC1NC/YFP-VPS4dn, 0.76 ± 0.15 (n = 5). Data are represented as average value ± standard deviation. n, number of analyzed cells.
In order to determine if the NC region of ART proteins might still be able to interact with the ESCRT machinery, via direct or indirect mechanisms, we next determined the relocalization to VPS4-induced aberrant endosomes. Localization to these compartments represents a well-known feature of proteins associated with the ESCRT pathway (2) although it cannot be excluded that colocalization with dominant negative VPS4 (VPS4dn) is induced by interactions with elements of the endocytic pathway other than ESCRT subunits. These experiments showed that catalytically inactive VPS4 seems to exacerbate the punctate distribution of mCherry-ARRDC1, and a clear localization of this construct with YFP-VPS4dn was also observed (Fig. 7, top panels), representing a further indication for the interaction of ARRDC1 with the ESCRT machinery. More interestingly, ARRDC1NC also colocalized with VPS4-induced aberrant endosomes, and, in fact, this construct colocalized more evidently with YFP-VPS4dn than the full-length protein (Fig. 7, bottom panels). Consequently, these results allow the hypothesis that ARRDC1 retains as yet uncharacterized interactions with the ESCRT machinery in the absence of its C terminus.
DISCUSSION
Several lines of evidence presented in this work support a model whereby the ARTs can act as adaptor proteins between HECT ubiquitin ligases and the ESCRT machinery in PPXY-dependent viral budding. First, the ARTs are able to bind both the HECT ubiquitin ligases required for PPXY-mediated budding and components of the ESCRT machinery, namely, ALIX and Tsg101, through their C-terminal regions as well as possible additional interactions of the NC region with some as yet unidentified component of the ESCRT pathway. Second, the ARTs can localize to the site of viral assembly, and their overexpression results in a PPXY-specific inhibition of MLV budding. Importantly, this phenotype is also induced by ART constructs lacking the PPXY motifs, thus excluding a mere competition with MLV Gag for binding to endogenous HECT ubiquitin ligases. Third, the ARTs are themselves substrates for WWP1-dependent ubiquitination, perhaps explaining, at least in part, the requirement for the enzymatic activity of the HECT ubiquitin ligases in retroviral budding.
It remains to be addressed whether ART ubiquitination is required for PPXY-dependent budding, and it is also unclear which residues are the target for ubiquitination. Studies of the yeast homologues Art1/Cvs7 and Art9/Rim8 identified a conserved lysine residue in the C terminus as the target for ubiquitination in these proteins (19, 21, 32). However, this residue is conserved only in ARRDC1 and TXNIP while ARRDC2, ARRDC3, and ARRDC4 lack lysine residues in their C termini, suggesting that lysines in the NC region may be modified by ubiquitination in humans. It also remains to be addressed whether ART ubiquitination by WWP1 is K63 linked, a posttranslational modification that has been involved in MVB sorting (30) and retroviral budding (56, 60). In fact, WWP1 is likely to synthesize K63-linked ubiquitin chains (29), suggesting that the ARTs might simply provide a platform for this type of ubiquitination by virtue of being at the sites of viral assembly.
One important piece of evidence to support our model remains elusive; namely, individual depletion of the ARTs has minor effects in PPXY-dependent budding. However, it is entirely possible that redundancy among the ARTs, as already shown for their endocytic functions in yeast (42), could hamper the siRNA approach. In fact, the emerging picture for PPXY motifs suggests that this L domain exploits a highly redundant system, which can be very advantageous for viral replication. In this scenario, the alternative use of WWP1, WWP2, and Itch already introduces a first level of redundancy in PPXY-mediated budding (33), and, based on our results, it seems likely that downstream adaptors may add a second layer of redundancy in the system. It must also be noted that the L-domain inhibition by ART overexpression is less pronounced than the phenotype induced by WWP1 ΔHECT. It could be speculated that WWP1 ΔHECT inhibits viral budding by preventing the interaction of the L domain with the endogenous HECT ubiquitin ligases as well as by inducing downstream inhibitory effects while the ARTs do not bind the L domain directly and might therefore inhibit viral budding less effectively. It is also possible that the ARTs are not the only downstream factors targeted by HECT ubiquitin ligases during PPXY-mediated viral budding. Accordingly, functional redundancy between the ARTs and the related proteins Bul1 and Bul2 (41) has been shown in yeast, and it is also evident that the endocytic function of Rsp5 in yeast requires additional PPXY-containing adaptors such as Ear1p and Ssh4p (31). Additionally, our previous results suggesting an interaction of the HECT ubiquitin ligases with as yet unknown components of the ESCRT machinery through the isolated HECT domains (34) could also be indicative of redundant interactions between these ubiquitin ligases and the ESCRT complexes.
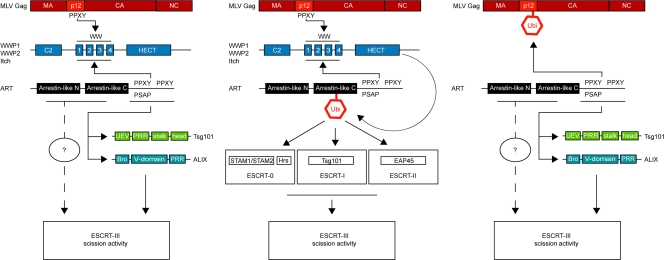
At a more mechanistic level, our findings suggest that the ARTs could exert their role in PPXY-mediated budding via three possible mechanisms that are not mutually exclusive. First, the ARTs could provide a mere physical link between HECT ubiquitin ligases and components of the core ESCRT machinery via interactions with ALIX and Tsg101 through the C terminus as well as with potential interaction partners through the NC region (Fig. 8). Second, ubiquitination of the ARTs by HECT ubiquitin ligases suggests that recruitment of ESCRT components could also take place via recognition of the ubiquitin moiety attached to the ART by ubiquitin binding domains in the ESCRT-0, -I, and -II complexes. Lastly, our preliminary finding that ARTs can bind to ubiquitin moieties themselves supports a possibility in which ARTs could interact with ubiquitinated viral proteins, followed by ESCRT recruitment via the mechanisms described above.
FIG. 8.
Schematic drawing of three possible mechanisms of ESCRT-III recruitment by ART proteins during PPXY-mediated viral budding. ARTs might function by binding to both HECT ubiquitin ligases and components of the ESCRT machinery, i.e., ALIX and Tsg101, as well as an uncharacterized factor, thereby recruiting the ESCRT-III scission activity (left panel). Second, ubiquitination of ARTs induced by HECT ubiquitin ligases might recruit ESCRT-III via interactions of ESCRT-0, -I, and -II with ubiquitin moieties (middle panel). Third, binding of ARTs to ubiquitinated viral proteins might induce ESCRT-III recruitment via mechanisms described above (right panel). CA, capsid; NC, nucleocapsid.
Overall, these results suggest that the recruitment of downstream factors by HECT ubiquitin ligases during PPXY-mediated viral budding might not be restricted to one protein acting as the exclusive adaptor with the ESCRT machinery. We rather speculate that a family of proteins, including the ARTs, ubiquitin, and perhaps additional factors, could act in parallel to exert this function via different mechanisms.
Acknowledgments
We thank Eric Freed for kindly providing us with expression constructs for EIAV Gag as well as Ron Montelaro for the anti-EIAV Gag serum.
This work was supported by the Medical Research Council UK, the Lister Institute of Preventive Medicine, and the EMBO Young Investigator Programme. S.R. is a Deutsche Forschungsgemeinschaft Fellow.
Footnotes
Published ahead of print on 29 December 2010.
REFERENCES
- 1.Agromayor, M., and J. Martin-Serrano. 2006. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 281:23083-23091. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhandari, D., J. Trejo, J. L. Benovic, and A. Marchese. 2007. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J. Biol. Chem. 282:36971-36979. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 5.Bouamr, F., et al. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. (Author's correction, 78:4383, 2004.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlton, J. G., M. Agromayor, and J. Martin-Serrano. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U. S. A. 105:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlton, J. G., and J. Martin-Serrano. 2009. The ESCRT machinery: new functions in viral and cellular biology. Biochem. Soc Trans. 37:195-199. [DOI] [PubMed] [Google Scholar]
- 8.Chung, H. Y., et al. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82:4884-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, R. D., et al. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841-852. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Gottlinger, H. G. 2001. The HIV-1 assembly machine. AIDS 15(Suppl. 5):S13-S20. [DOI] [PubMed] [Google Scholar]
- 15.Gottwein, E., S. Jager, A. Habermann, and H. G. Krausslich. 2006. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 Gag cause a late budding defect. J. Virol. 80:6267-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty, R. N., M. E. Brown, G. L. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U. S. A. 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidecker, G., P. A. Lloyd, F. Soheilian, K. Nagashima, and D. Derse. 2007. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J. Virol. 81:9769-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrador, A., S. Herranz, D. Lara, and O. Vincent. 2010. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 30:897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herranz, S., et al. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U. S. A. 102:12141-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hitchcock, A. L., K. Auld, S. P. Gygi, and P. A. Silver. 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. U. S. A. 100:12735-12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurley, J. H., and S. D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurley, J. H., and P. I. Hanson. 2010. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurley, J. H., et al. 2009. Piecing together the ESCRTs. Biochem. Soc Trans. 37:161-166. [DOI] [PubMed] [Google Scholar]
- 25.Jadwin, J. A., V. Rudd, P. Sette, S. Challa, and F. Bouamr. 2010. Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch. J. Virol. 84:704-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi, A., U. Munshi, S. D. Ablan, K. Nagashima, and E. O. Freed. 2008. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic 9:1972-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kee, Y., W. Munoz, N. Lyon, and J. M. Huibregtse. 2006. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J. Biol. Chem. 281:36724-36731. [DOI] [PubMed] [Google Scholar]
- 28.Kikonyogo, A., et al. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. U. S. A. 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, H. C., and J. M. Huibregtse. 2009. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29:3307-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauwers, E., C. Jacob, and B. Andre. 2009. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185:493-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leon, S., Z. Erpapazoglou, and R. Haguenauer-Tsapis. 2008. Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo and sorting at multivesicular bodies. Mol. Biol. Cell 19:2379-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, C. H., J. A. MacGurn, T. Chu, C. J. Stefan, and S. D. Emr. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135:714-725. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Serrano, J. 2007. The role of ubiquitin in retroviral egress. Traffic 8:1297-1303. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 38.McDonald, B., and J. Martin-Serrano. 2008. Regulation of Tsg101 expression by the steadiness box: a role of Tsg101-associated ligase. Mol. Biol. Cell 19:754-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNatt, M. W., I. McKittrick, M. West, and G. Odorizzi. 2007. Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol. Biol. Cell 18:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 41.Nikko, E., and H. R. B. Pelham. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikko, E., J. A. Sullivan, and H. R. Pelham. 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9:1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 44.Ott, D. E., et al. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng, J., et al. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921-926. [DOI] [PubMed] [Google Scholar]
- 46.Raiborg, C., and H. Stenmark. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445-452. [DOI] [PubMed] [Google Scholar]
- 47.Rotin, D., and S. Kumar. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10:398-409. [DOI] [PubMed] [Google Scholar]
- 48.Saksena, S., and S. D. Emr. 2009. ESCRTs and human disease. Biochem. Soc. Trans. 37:167-172. [DOI] [PubMed] [Google Scholar]
- 49.Saksena, S., J. Sun, T. Chu, and S. D. Emr. 2007. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32:561-573. [DOI] [PubMed] [Google Scholar]
- 50.Sakurai, A., et al. 2004. Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect. 6:150-156. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt, A. P., G. P. Leser, E. Morita, W. I. Sundquist, and R. A. Lamb. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spidel, J. L., et al. 2004. Lysines close to the Rous sarcoma virus late domain critical for budding. J. Virol. 78:10606-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staub, O., et al. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 54.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. U. S. A. 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 56.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Usami, Y., S. Popov, E. Popova, and H. G. Gottlinger. 2008. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 82:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VerPlank, L., et al. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. U. S. A. 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Schwedler, U. K., et al. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 60.Weiss, E. R., et al. 2010. Rescue of HIV-1 release by targeting widely divergent NEDD4-Type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 6:e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, R. L., and S. Urbe. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355-368. [DOI] [PubMed] [Google Scholar]
- 62.Wills, J. W., et al. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wollert, T., C. Wunder, J. Lippincott-Schwartz, and J. H. Hurley. 2009. Membrane scission by the ESCRT-III complex. Nature 458:172-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhadina, M., M. O. McClure, M. C. Johnson, and P. D. Bieniasz. 2007. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 104:20031-20036. [DOI] [PMC free article] [PubMed] [Google Scholar]