Abstract
Arenaviruses are negative-strand RNA viruses that cause human diseases such as lymphocytic choriomeningitis, Bolivian hemorrhagic fever, and Lassa hemorrhagic fever. No licensed vaccines exist, and current treatment is limited to ribavirin. The prototypic arenavirus, lymphocytic choriomeningitis virus (LCMV), is a model for dissecting virus-host interactions in persistent and acute disease. The RING finger protein Z has been identified as the driving force of arenaviral budding and acts as the viral matrix protein. While residues in Z required for viral budding have been described, residues that govern the Z matrix function(s) have yet to be fully elucidated. Because this matrix function is integral to viral assembly, we reasoned that this would be reflected in sequence conservation. Using sequence alignment, we identified several conserved residues in Z outside the RING and late domains. Nine residues were each mutated to alanine in Lassa fever virus Z. All of the mutations affected the expression of an LCMV minigenome and the infectivity of virus-like particles, but to greatly varying degrees. Interestingly, no mutations appeared to affect Z-mediated budding or association with viral GP. Our findings provide direct experimental evidence supporting a role for Z in the modulation of the activity of the viral ribonucleoprotein (RNP) complex and its packaging into mature infectious viral particles.
Arenaviruses are the causative agents of a number of human diseases, including lymphocytic choriomeningitis, Lassa fever, and Bolivian hemorrhagic fever. These viruses are carried asymptomatically in wild rodents and are transmitted through contact with contaminated excretions. Endemic infection in these rodents maintains a reservoir of virus that can reemerge in human populations as outbreaks (1, 18, 30) or as novel pathogenic species (6). No FDA-licensed vaccines exist for these viruses, and current antiarenaviral therapy is limited to the off-label use of the nucleoside analogue ribavirin, which has had mixed success in treating cases of arenaviral hemorrhagic fever (HF) disease, while being associated with significant side effects (5).
The prototypic arenavirus, lymphocytic choriomeningitis virus (LCMV), is ubiquitous in wild rodents and can infect domesticated mice and hamsters. These infected hamsters have most recently been responsible for reported disease in humans (2, 34). Additionally, reports of transplant-associated LCMV disease and fatality highlight the clinical relevance of arenaviral infection in immunocompromised individuals (15). In addition, congenital transmission of LCMV has been implicated in a variety of clinical conditions that affect mainly the retina and brain (3, 4).
Arenaviruses are enveloped viruses bearing a bi-segmented, negative-sense RNA genome. Each segment contains two open reading frames (ORFs) in the ambisense orientation that are separated by a noncoding intergenic region (IGR) and bounded at their 5′ and 3′ ends by untranslated regions (UTRs) that are critical for the control of viral RNA replication and gene expression (7). The S segment (3.5 kb) encodes the glycoprotein precursor (GPC) and nucleoprotein (NP). The L segment (7.3 kb) encodes the viral-RNA-dependent RNA polymerase L and the Z protein. The GPC is posttranslationally cleaved into three subunits consisting of a stable signal peptide (SSP), glycoprotein subunit 1 (GP1), and GP2, which are stably associated in a GP complex that decorates the surfaces of viral particles. GP1 mediates cell entry via receptor-mediated endocytosis, GP2 provides a pH-dependent fusogenic activity required for the delivery of the viral ribonucleoprotein (RNP) complex into the cell cytoplasm, and the SSP has been shown to be necessary for proper GP cleavage. NP is required for forming the viral RNP responsible for directing both viral RNA replication and gene transcription.
The Z protein (90 to 99 amino acids [aa], depending on the species of arenavirus) has been shown to be the driving force of arenaviral budding (25, 32), and cryo-electron microscopy (cryo-EM) studies suggest that Z is located between GP and NP in arenaviral particles (22). These observations, together with Z's ability to negatively regulate RNA synthesis by the viral polymerase (13), have led us to consider Z as the arenaviral counterpart of the matrix protein found in most enveloped, negative-sense (NS) RNA viruses (21). Mutation-function studies have identified several functional domains within Z. The conserved central RING domain has been shown to be required for downregulation of viral RNA synthesis (12) and budding mediated by Z. Bona fide, conserved, late domain motifs are present at the C termini of all known arenaviral Z proteins. As with many other viruses, budding mediated by Z late domain motifs involves the interaction of Z with members of the cellular multivesicular body (MVB) pathway (25, 32, 35). Further, the glycine residue at position 2 (G2) in Z was found to be a myristoylation site, and a G2A mutation abrogated budding, indicating a requirement for Z insertion into membranes to promote viral budding (27, 33). G2 was also found to be required for Z-GP interaction (10). Most recently, budding by Z has been shown to be enhanced in the presence of viral NP (16). Z has also been shown to interact with NP and L (11, 14, 17, 31, 38), but the specific determinants in Z that mediate these viral protein interactions have yet to be fully elucidated.
Given that Z functions through highly conserved residues for Z-mediated budding and downregulation of viral RNA synthesis, we reasoned that residues that mediate other Z-dependent functions and interactions would also be conserved. A deeper understanding of these interactions would be complemented further by the published structure of Lassa fever virus (LASV) Z (36, 37). Here we present evidence that several conserved residues in Z located outside the RING and late domain motifs are critical for viral infectivity by their involvement in the control of viral RNA replication and gene transcription, representing potential contact points between Z and the biologically active RNP.
MATERIALS AND METHODS
Cells and virus.
HEK 293T (293T) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 2 mM glutamine, and 100 U/ml penicillin-streptomycin. BHK21 cells were purchased from the American Type Culture Collection and were cultured using the same medium conditions as for 293T cells. LCMV strain Armstrong was used for viral infections.
Plasmid constructs.
All constructs were made in pCAGGS (23), herein referred to as pC, unless otherwise indicated. Several plasmids have been described previously. Plasmids pC-NP, pC-L, and pC-T7 were described by Lee and colleagues (20), where NP and L were subcloned from pCITE-2a and pGEM-4Z, respectively (19), and pC-T7 was designed to promote T7 expression in the cytoplasm of cells. Plasmids pC-LASV-Z-HA and pC-LASV-GP-Flag were originally named pC-LFV-Z-HA and pC-LFV-GP-Flag, respectively (10, 25), and were renamed to be consistent with arenavirus nomenclature. Plasmid pMG#7Δ2G contains an LCMV minigenome (MG) utilizing a chloramphenicol acetyltransferase (CAT) reporter, which is further bounded by T7 RNA polymerase promoter and terminator sequences to mediate transcription by T7 polymerase (25, 26).
Alanine point mutagenesis.
The LASV Z ORF was PCR amplified such that a hemagglutinin (HA) tag was added and then cloned into the EcoRI and BamHI sites of pGEM3Z to generate pGEM-LASV-Z-HA. Primer pairs to generate targeted alanine mutations were designed using the QuikChange primer design program (Stratagene, La Jolla, CA), and pGEM-LASV-Z-HA was used as a template for mutagenic PCR with the QuikChange Lightning site-directed mutagenesis kit (Stratagene) by following the manufacturer's instructions. Sequences for each oligonucleotide are available upon request. Constructs were sequenced for successful mutagenesis, and then ORFs were liberated using EcoRI and BamHI and cloned into the EcoRI and BglII sites of pC. Plasmids that were isolated after being subcloned into pC were sequenced again before use in experiments.
Cell transfection.
All DNAs were prepared for transfection using Qiagen (Valencia, CA) reagents. 293T cells were transfected in 12-well or 6-well plates using Lipofectamine 2000 (LF2000; Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Immunoblotting.
SDS-PAGE was done using precast Novex gels (Invitrogen) containing 12% or 16% acrylamide. Following electrophoresis, gels were transferred to nitrocellulose using a semidry apparatus (Bio-Rad, Hercules, CA). Blocking was done using 5% nonfat dry milk in phosphate-buffered saline (PBS) containing 0.2% Tween 20 (PBS-Tween 20). All incubations with antibody were done in 5% milk, and washes were done using PBS-Tween 20. Anti-HA monoclonal antibody was from Covance (Princeton, NJ), anti-Flag polyclonal antibody was from Cayman Chemical (Ann Arbor, MI), anti-NP antibody was as previously described (8), and secondary alkaline phosphatase (AP)-conjugated antibodies were from Bio-Rad. Chemiluminescence reactions were done using Immun-Star AP substrate (Bio-Rad). Blots were visualized using the Chemi-Doc XRS gel documentation system and quantified using the Quantity One program (Bio-Rad).
VLP isolation.
Virus-like particle (VLP) assays were done as previously described (25). Briefly, 293T cells were transfected with the individual plasmids for Z using LF2000 (Invitrogen). At 48 to 72 h posttransfection, cell extracts (CE) were collected in lysis buffer (50 mM Tris-Cl [pH 8.0]-62.5 mM EDTA-1% NP-40-0.4% deoxycholate [DOC]) and clarified culture supernatant was centrifuged through a 20% sucrose cushion at 100,000 × g at 4°C for 30 min. The resulting pellet containing VLPs was dissolved in 2× reducing SDS sample buffer. VLPs and CE were analyzed by immunoblotting using HA-specific antibody to detect Z-HA.
IP.
Immunoprecipitation (IP) of Flag-tagged GP was done according to the method described previously, with slight revision (10). Briefly, transfected 293T cells were scraped from wells on ice into 0.5 ml lysis buffer containing 1% beta-octyl glucoside and protease inhibitors (Research Products International, Mt. Prospect, IL) to prepare CE (Complete lysis buffer). After 30 min of end-over-end incubation, the soluble fraction was collected after a brief centrifugation, and 90% of the CE was used for immunoprecipitation with M2 anti-Flag affinity gel (Sigma, St. Louis, MO). The remaining 10% of the CE was used directly for immunoblot analysis. The bead-containing gel was washed three times using Complete lysis buffer and then once in lysis buffer lacking detergent and protease inhibitors. Proteins were eluted using 20 μl of 2× sample buffer and heating at 95°C for 5 min before analysis by immunoblotting.
Immunofluorescence assay.
Transfected 293T cells were grown on glass coverslips and prepared for indirect immunofluorescence as previously described (10). Mouse anti-HA antibody was incubated with cells at a 1:1,000 dilution, and DyLight 594-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) was incubated with cells at a 1:1,000 dilution. Counterstaining for nuclei was done by mounting cells with DAPI (4′,6-diamidino-2-phenylindole) Fluoromount G (Southern Biotech, Birmingham, AL). Cells were visualized using a Nikon Eclipse Ti-U inverted microscope fitted with a Nikon D-Eclipse confocal laser assembly and a D-Eclipse C1 controller (Nikon, Melville, NY). Images were acquired using the Nikon EZ-C1 program, analyzed using the programs Nikon NIS Elements and Image J, and assembled using Adobe Photoshop.
LCMV minigenome reporter assay.
293T cells grown in 12-well plates were transfected to mediate transcription and replication of the LCMV minigenome carrying the CAT reporter gene (25). Transfection mixtures lacking L plasmid were prepared as negative controls. Addition of different amounts of pC-Z was done as indicated in the figures below, and empty pC was used to equalize the amounts of total DNA per transfection. At 48 to 72 h posttransfection, cell extracts were prepared by freeze-thawing in 0.25 M Tris-Cl, pH 7.4, and analyzed for CAT activity with the FAST-CAT Green assay kit (Invitrogen).
Infectious-VLP assay.
293T cells were transfected to mediate the formation of infectious virus-like particles (25). Briefly, cells were transfected in 12-well plates with pC-T7 (0.25 μg), pMG#7Δ2G (0.125 μg), pC-L (0.2 μg), pC-NP (0.2 μg), pC-Z (0.1 μg), and pGP (0.2 μg). At 48 h posttransfection, clarified cell supernatants were collected and incubated with BHK21 cell monolayers, which were subsequently infected with LCMV Armstrong at a multiplicity of infection (MOI) of 3 to provide the trans-acting factors NP and L that mediate the transcription and replication of the CAT minigenome from the infectious VLPs. Cell extracts were analyzed for CAT activity using the FAST-CAT Green assay kit.
RESULTS
Selection of mutations in LASV Z.
In order to examine the relationship between the recently published structure of LASV Z and its function, we used the ClustalW algorithm to compare currently available arenavirus Z protein sequences and identify conserved residues whose functional roles have not been previously investigated. We identified, in addition to G2 and conserved residues within the RING and late domains, nine residues in LASV Z that showed conservation either across all the arenaviruses or in a clade-specific manner (Fig. 1). Specifically, G27, L71, and P72 were conserved across the whole family, P21, D22, P28, and K68 were conserved among Old World arenaviruses, and R16 and T73 were conserved in all arenaviruses except representatives of clade A or B of New World arenaviruses. These residues were individually mutated to alanine, except for P21 and D22, which were mutated together to result in a double-alanine mutant (P21A+D22A).
FIG. 1.
ClustalW alignment of arenavirus Z proteins. The Z proteins are arranged by clade, as indicated to the left of the alignment. The myristoylation site glycine, RING domain, and late domains are shown in gray boxes, while the residues targeted for mutagenesis are shown in black boxes. The position of each residue in LASV Z is indicated above the alignment, and the Z amino acid sequences from the following species (indicated by GenBank accession no. or NCBI reference sequence no.) were used: NP_694871.1 (LASV Josiah), AAO59510.1 (LASV NL), AAO59508.1 (LASV AV), AAO59514.1 (LASV CSF), AAD03395.1 (LCMV WE), ABC96003 (LCMV Armstrong), ABY20731 (Dandenong virus), ABC71138.1 (Mobala virus [MOBV] Acar), AAV54106.1 (Mopeia virus [MOPV] AN20410), ABC71136.1 (MOPV Mozambique), ABC71142.1 (Ippy virus [IPPYV] DakAnB), YP_002929492 (Lujo virus [LUJV]), YP_001649224 (Bear Canyon virus [BCNV]), YP_001911117 (Tamiami virus [TAMV]), YP_001911119 (Whitewater Arroyo virus [WWAV]), YP_001649213 (Allpahuayo virus [ALLV]), YP_001936023 (Flexal virus [FLEV]), YP_001936027 (Parana virus [PARV]), YP_138535 (Pichinde virus [PICV]), YP_025092 (Pirital virus [PIRV]), YP_001649217 (Amapari virus [AMAV]), YP_001649219 (Cupixi virus [CPXV]), NP_899216 (JUNV XJ13), AAV68494 (JUNV Candid), NP_899220 (Guanarito virus [GTOV]), NP_899214 (Machupo virus [MACV]), YP_001816784 (Chapare virus), ABY59837 (Sabia virus [SABV]), Q88470 (Tacaribe virus [TCRV]), YP_001936025 (Latino virus [LATV]), and YP_001649215 (Oliveros virus [OLVV]).
Effects of mutations of selected conserved residues on the production of infectious virus-like particles.
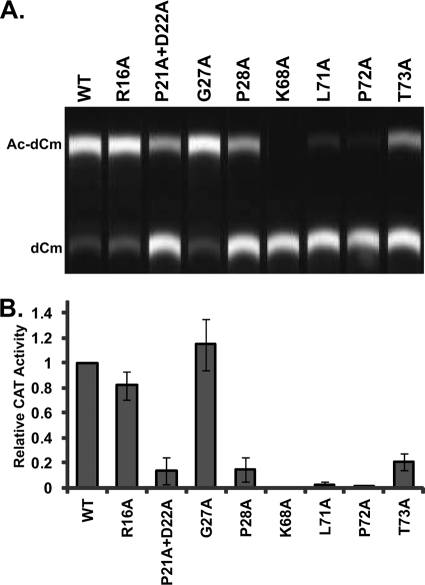
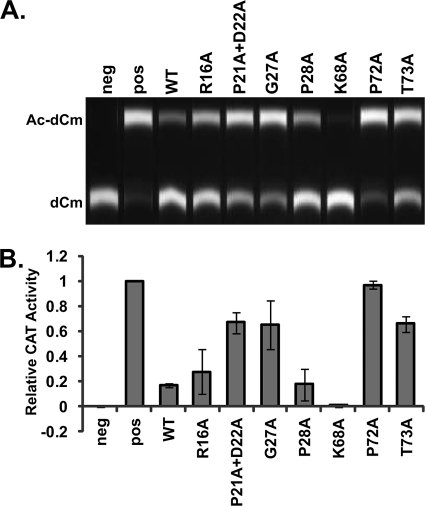
To determine whether these mutations perturbed infectivity, we utilized a well-characterized in vitro assay based on LCMV; it was previously shown that LASV Z is interchangeable with LCMV Z (10, 25). We first assessed the ability of these mutant Z proteins to mediate the production of infectious VLPs (Fig. 2). We observed that an R16A mutant showed slightly reduced infectivity and that a G27A mutant showed slightly greater levels of infectious VLPs than the wild type (WT). The remaining mutations resulted in a decrease in the number of infectious VLPs of more than 4-fold.
FIG. 2.
Point mutations in Z affect VLP infectivity. HEK 293T cells were transfected to mediate the formation of infectious VLPs containing the CAT minigenome. Clarified supernatants were transferred to BHK21 cells, which were subsequently infected with WT LCMV to mediate expression of the CAT reporter gene carried by infectious VLPs. (A) CAT activity from BHK21 cells infected with VLPs. Ac-dCm, acetylated deoxychloramphenicol; dCm, deoxychloramphenicol. (B) Relative CAT activities in cells, where activity in the presence of WT Z was set at 1. These data represent the results of three independent experiments, and standard deviations of the means were calculated.
We also generated an alanine mutation at LASV Z L71, the residue homologous to L79 in Junin virus (JUNV) Z, which has been shown to contribute to infectivity by mediating NP recruitment into VLPs (11). We observed a similar loss of infectivity with Z L71A, as previously described (Fig. 2), and chose not to pursue this mutant further.
Effects of selected mutations on LASV Z-mediated budding and Z-GP interaction.
Differences among the Z mutant proteins in promoting infectious VLP production might have reflected changes in budding or interaction with GP, both of which are required for the generation of infectious VLPs. We therefore examined each Z mutant for budding activity and the ability to biochemically associate with GP.
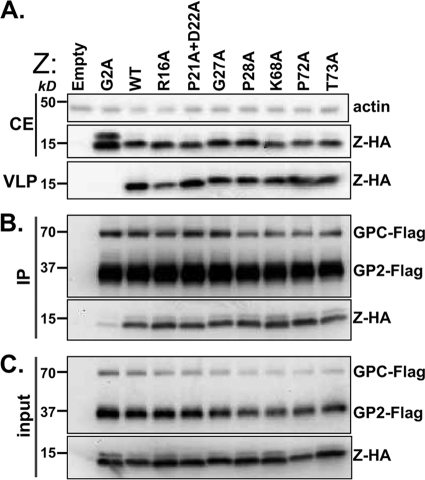
The budding activity of each mutant was determined using a previously described assay (25). As controls for the budding assay, we used Z WT and Z G2A. The G2A mutant has been shown to lack budding activity due to loss of myristoylation (25, 27, 33). The Z WT and Z mutants tested were expressed similarly in transfected cells, with the exception of the G2A mutant, which, consistently with our previous findings (10), was expressed to slightly higher levels (Fig. 3 A, blot CE). Likewise, with the exception of the G2A mutant, all Z mutants tested exhibited WT budding activity (Fig. 3A, blot VLP).
FIG. 3.
LASV Z mutants are competent to form VLP and interact with viral GP. (A) VLP analysis. HEK 293T cells were transfected with single plasmids containing different Z-HA proteins, indicated above the immunoblots. At 48 h posttransfection, cell extracts (CE) were prepared and VLPs collected from clarified tissue culture supernatants using ultracentrifugation. Aliquots of each fraction were submitted to immunoblotting using an anti-HA antibody. (B) Coimmunoprecipitation of LASV GP and Z. HEK 293T cells were cotransfected with pC-LASV-GP-Flag and one of the Z-containing plasmids indicated above the panels. Flag immunoprecipitates were separated on a 16% SDS-PAGE gel and subjected to immunoblotting with either anti-Flag or anti-HA antibody to detect LASV GP-Flag or LASV Z-HA, respectively. (C) Immunoblots of cell extracts prior to Flag immunoprecipitation. These results are representative of two independent experiments.
To address whether these selected mutations in LASV Z affected Z's biochemical association with GP, each LASV Z mutant was coexpressed with GP-Flag, and at 48 h posttransfection, cell lysates were prepared for immunoprecipitation (IP) assays. As controls, again we included Z WT and Z G2A. Cell lysates were subjected to IP using an antibody to Flag, and immunoprecipitated proteins were examined by immunoblotting using antibodies to either Flag or HA to detect GP and Z, respectively. Control conditions were consistent with previous findings (10); Z WT coimmunoprecipitated efficiently with GP, while the G2A mutant was poorly coimmunoprecipitated by GP-Flag (Fig. 3B). Examination of Flag immunoprecipitates revealed that all LASV Z mutants tested coimmunoprecipitated with GP-Flag at levels similar to that of Z WT (Fig. 3B). In cells cotransfected with Z and GP, all LASV Z mutants tested exhibited similar cell expression levels, with the exception of the G2A mutant, which, as previously documented (10, 27), was present at slightly higher levels (Fig. 3C).
We also observed a species corresponding to Z that migrated more slowly than expected for its molecular weight; this species was more pronounced in the G2A mutant. This has been observed in other work with LASV Z (27, 28, 33); however, mutations aside from G2A did not greatly affect the appearance of this extra species.
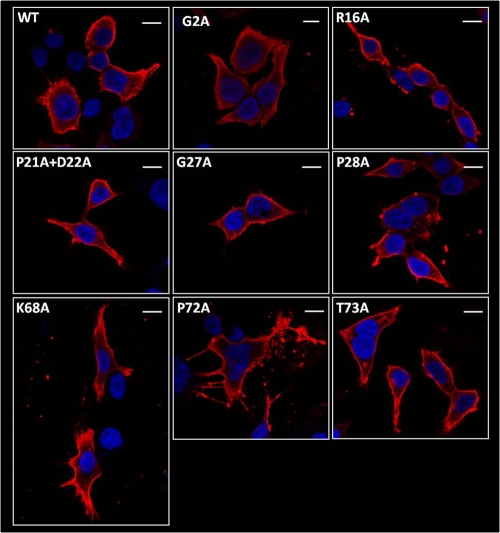
Plasma membrane localization of LASV Z mutants.
We next addressed whether the selected mutations introduced in LASV Z affected its subcellular location. For this, we examined cells transfected with either Z WT or individual mutants by immunofluorescence and confocal microscopy. Consistently with previously published results (10), Z WT showed accumulation on the plasma membrane, while G2A mutant expression was restricted mainly to the cytosol (Fig. 4). The subcellular distribution of each of the Z mutants examined was similar to that of Z WT (Fig. 4).
FIG. 4.
LASV Z mutants localize to the plasma membrane. HEK 293T cells were transfected to express the indicated Z protein, seeded 1 day after transfection to coverslips, and then processed for indirect immunofluorescence confocal microscopy using an anti-HA antibody and DyLight 594-conjugated secondary antibody. DAPI was used to counterstain nuclei. Mutations are indicated in each image, and the white line in each panel represents 10 μm.
Effects of selected mutations in LASV Z on the expression of an LCMV minigenome.
Since the altered production of infectious VLPs associated with Z mutants in this study was not due to defects in budding or an association with viral GP, we reasoned that these mutations might have affected Z functions for controlling viral RNA synthesis such that lower levels of the viral genome would be available for packaging into viral particles. To investigate this possibility, we first examined the effects of these Z mutants on levels of expression of a CAT reporter gene within an LCMV minigenome (MG), where Z WT exerts a dose-dependent inhibitory effect on MG RNA replication and expression (13).
Compared to Z WT, the P28A mutant appeared similar to the WT (Fig. 5 A and B). Interestingly, the R16A, P21A+D22A, G27A, P72A, and T73A mutants were impaired in their ability to inhibit MG expression to various degrees, which we quantified. In contrast, the K68A mutant exhibited greater inhibition of MG expression.
FIG. 5.
Effect of point mutations in LASV Z on LCMV minigenome expression. HEK 293T cells were transfected to mediate the transcription and replication of the CAT minigenome in the presence or absence of various Z proteins. (A) Point mutations in Z differ in their inhibitory activities of model-genome expression. CAT activities from transfected cells in the presence of Z are shown. Plasmid DNA for Z was transfected at 50 ng per transfection. (B) Quantification was done for the CAT thin-layer chromatograph (TLC) shown in panel A, and values are shown as relative to that for the positive control (pos). These results were representative of four independent experiments, and standard deviations of the means were calculated. Negative-control transfections were done in the absence of both pL and pZ (neg), and positive-control transfection was done with pL but in the absence of pZ (pos).
Effect of WT Z expression on phenotypes associated with the G27A and P72A mutants.
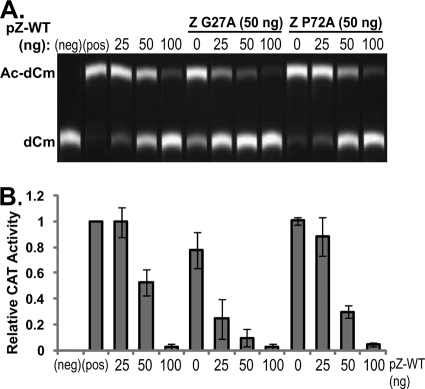
We noted that two mutants, Z G27A and Z P72A, showed a decreased ability to inhibit MG expression yet showed opposite phenotypes in the infectivity assay. We then asked if the MG expression phenotype with the G27A and P72A mutants could be complemented in trans by coexpression of Z WT. For this, we transfected cells with a fixed amount (50 ng) of the pC-Z(G27A) or pC-Z(P72A) expression plasmid together with an increasing amount (0 to 100 ng) of the pC-Z(WT) plasmid and fixed amounts of the plasmids for the other components of the MG system. At 48 h posttransfection, we determined levels of CAT activity in each sample (Fig. 6).
FIG. 6.
Reversible phenotype of LASV Z mutants to inhibit the expression of a minigenome reporter. (A) CAT activity resulting from transcription and replication of an LCMV minigenome was measured in transfected cells, where a Z mutant was cotransfected with increasing amounts of the WT Z plasmid. The transfection scheme is indicated. A representative TLC is shown here. (B) Quantification was done for three independent experiments, and standard deviations of the means were calculated. Negative-control transfections were done in the absence of both pL and pZ (neg), and positive-control transfection was done with pL but in the absence of pZ (pos).
Intriguingly, the coexpression of Z WT with the G27A mutant showed better inhibition of MG expression than did Z WT alone (Fig. 6B). Coexpression of Z WT with Z P72A also showed a higher level of inhibition than the WT, but to a lesser degree than the G27A mutant.
These findings suggested that neither protein exerted a dominant effect in the presence of the WT and that both proteins retained some inhibitory activity, thus resulting in an additive effect when the WT and these mutants were expressed together. Further, the G27A mutant retained a more inhibitory activity than the P72A mutant, which was reflected in the lower levels of CAT activity observed.
DISCUSSION
In this work, we sought to expand our knowledge about structure-function relationships of LASV Z by introducing alanine substitutions at phylogenetically conserved residues within Z and assessing their phenotypes in cell-based assays. We used reverse genetics tools with LCMV proteins that have been shown to accommodate the use of heterologous Z protein (25). The resulting mutants showed a range of phenotypes when tested in a VLP infectivity assay. While mutation at R16 resulted in some impairment in the formation of infectious VLPs, the G27A mutation resulted in slightly increased levels of production of infectious VLPs that correlated with increased levels of MG expression. Each of the other five mutants examined (the P21A+D22A, P28A, K68A, L71A, and T73A mutants) showed >4-fold decreases in infectivity. These results have been summarized in Table 1.
TABLE 1.
Summary of phenotypes observed for the LASV Z point mutants discussed in this work
| LASV Z | Fold reduction of CAT reporter expressiona | Biochemical association with GPb | Plasma membrane localizationb | Budding activityb | % of WT VLP infectivityc |
|---|---|---|---|---|---|
| Z WT | 6 | + | + | + | 100 |
| Z R16A | 3.6 | + | + | + | 82 |
| Z P21A+ D22A | 1.5 | + | + | + | 13.7 |
| Z G27A | 1.5 | + | + | + | 115 |
| Z P28A | 5.6 | + | + | + | 15 |
| Z K68A | 200 | + | + | + | 0 |
| Z L71A | NDe | ND | ND | ND | 2 |
| Z P72A | 1 | + | + | + | 0.7 |
| Z T73A | 1.5 | + | + | + | 21 |
Fold reduction of CAT reporter expression = (1/relative CAT activity). Mean relative CAT activities are reported in Fig. 5.
The mutations discussed did not affect the biochemical association with viral GP, plasma membrane localization, or Z-mediated budding (Fig. 3 and 4).
The percentage of WT VLP infectivity = (100 × relative CAT activity), using the mean relative CAT activities reported in Fig. 2.
ND, not done.
None of the Z mutations examined in this work had significant effects on the plasma membrane localization of Z, biochemical interaction with GP, or Z budding activity. However, the Z mutants examined (with the mutations P21A+D22A, G27A, P28A, K68A, P72A, and T73A) showed different effects on the replication and gene expression of an LCMV MG. Thus, mutation G27A was associated with the loss of Z-mediated inhibition of viral RNA synthesis but slightly increased VLP infectivity. Contrastingly, the P28A mutant retained and the K68A mutant gained Z inhibitory effects on viral RNA but were associated with the loss of VLP infectivity. Finally, the R16A, P21A+D22A, P72A, and T73A mutants caused the loss of both Z-mediated inhibition of viral RNA synthesis and VLP infectivity.
A recent study by Casabona et al. showed that Z L79 was involved in Junin virus (JUNV) Z-mediated incorporation of NP into VLP (11) and that P80 was not involved in this process. Our results have shown that LASV Z's P72, the residue homologous to JUNV Z P80, was indeed also involved in mediating the infectivity of VLPs via mechanisms yet to be determined. A recent study showed that regions in the C terminus of arenavirus NP are necessary for the incorporation of NP into Z-containing VLPs (31). Interestingly, a C-terminal region of NP showed high sequence variability among members of the family (9), suggesting that NP-Z interactions are likely affected by evolutionary changes within the NP region involved in the NP-Z interaction, which may contribute to the restricted generation of reassortants observed among genetically distant arenaviruses.
None of the new mutations examined in this work had significant effects on Z's budding activity or its ability to interact with GP. We consistently observed a Z-related species with slower mobility in cell extract from transfected cells, especially with the G2A mutant. As the other mutations did not greatly affect this species, we do not attribute the phenotypes that we observed here to its presence. It is possible that this species reflects some posttranslational modification; however, to date, there is no published evidence for modifications of Z other than myristoylation.
The major effects found to be associated with the Z mutants were related to changes in MG expression, suggesting that these residues contribute to Z-RNP interactions. Previous studies indicated that the N (residues 1 to 16) and C (residues 79 to 90) termini of Z were not required for the inhibitory activity of Z on RNA synthesis by the arenavirus polymerase (12). Except for R16A, the mutations described herein that resulted in the loss of Z inhibitory activity, P21A D22A, G27A, L71A, P72A, and T73A, were located outside these regions. Interestingly, these regions in Z have also been suggested to be necessary for the biochemical interaction(s) with the viral L RNA-dependent RNA polymerase, and that work argued that these interactions directed the inhibition of transcription and replication by Z (17). While the previous studies did not go further to describe the impact of these regions on the infectivity of VLPs, the results of our experiments that measured viral transcription and replication were quite consistent. Further experiments will be necessary to determine how these residues affect Z-L and Z-NP interactions during viral infection.
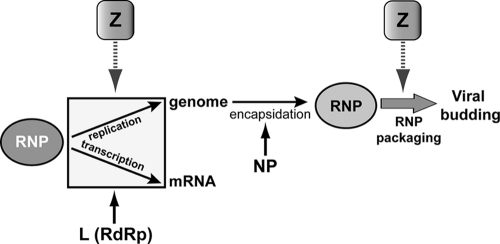
Two mutants of LASV Z, the G27A and P72A mutants, showed opposite phenotypes in the production of infectious VLPs yet exhibited similar losses of function in inhibiting viral RNA synthesis. Our findings have provided strong evidence that functional domains and residues within Z responsible for the inhibition of viral RNA synthesis can be segregated from those involved in the budding of VLPs. The Z WT dose-dependent inhibitory effect on viral RNA synthesis augmented the loss of this effect by the G27A or P72A mutant, suggesting that residues G27 and P72 participate in interactions between Z and viral RNP that specifically regulate viral transcription and replication. The difference in phenotype between these two mutants may also reflect an impact on a downstream event that requires Z function, such as packaging (Fig. 7). Distinguishing between these events is a topic of future research.
FIG. 7.
Proposed model of the events during viral transcription, replication, and packaging, which may be impacted by the mutants described herein. These may require contact between Z and viral RNA, viral NP, and viral L or coordination of all these interactions.
On the other hand, the K68A mutation showed much stronger activity in downregulating viral RNA synthesis and did not produce infectious VLPs. Notably, it has been shown that K68 in LASV Z participates in the formation of a positively charged groove that is distinct from the region recently identified as an eIF4E binding site (37). Our results have provided for the first time experimental evidence that this groove plays a critical role in the control of viral RNA synthesis. This groove may mediate Z's interaction with the RNP during transcription and replication or with a cellular factor required for these processes. Further experiments are required to dissect and functionally characterize these interactions.
Our findings strongly support the idea that residues R16, P21, D22, G27, P28, K68, P72, and T73 in LASV Z play critical roles in the formation of infectious viral particles by modulating viral RNA synthesis directed by the viral RNP rather than by acting at late stages of viral egress. These residues might mediate the interaction with the L polymerase and modulate its activity. Alternatively, these residues may be involved in the packaging of RNP into viral particles through direct interaction with either viral NP or viral RNA; however, interactions with NP or RNA need not be mutually exclusive (Fig. 7). Direct binding of Z to RNA has not been experimentally documented, but the matrix proteins of several negative-strand RNA viruses, including influenza virus and human respiratory syncytial virus, have been shown to interact directly with either RNA or NP (24, 29). Further research is warranted to fully elucidate the specific interactions and functional consequences mediated by these Z residues.
Acknowledgments
We thank members of the Buchmeier laboratory for helpful discussions and critical reading of the manuscript.
This work was supported by NIH grant AI065359 to M. J. Buchmeier and NIH/NIAID grant RO1 AI047140 to J. C. de la Torre.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1.Aguilar, P. V., et al. 2009. Reemergence of Bolivian hemorrhagic fever, 2007-2008. Emerg. Infect. Dis. 15:1526-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amman, B. R., et al. 2007. Pet rodents and fatal lymphocytic choriomeningitis in transplant patients. Emerg. Infect. Dis. 13:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonthius, D. J., B. Nichols, H. Harb, J. Mahoney, and B. Karacay. 2007. Lymphocytic choriomeningitis virus infection of the developing brain: critical role of host age. Ann. Neurol. 62:356-374. [DOI] [PubMed] [Google Scholar]
- 4.Bonthius, D. J., et al. 2007. Congenital lymphocytic choriomeningitis virus infection: spectrum of disease. Ann. Neurol. 62:347-355. [DOI] [PubMed] [Google Scholar]
- 5.Borio, L., et al. 2002. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287:2391-2405. [DOI] [PubMed] [Google Scholar]
- 6.Briese, T., et al. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 5:e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchmeier, M. J., J.-C. de la Torre, and C. Peters. 2007. Arenaviridae: the viruses and their replication, p. 1792-1827. In D. M. Knipe et al. (ed.), Fields virology, fifth ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 8.Buchmeier, M. J., H. A. Lewicki, O. Tomori, and K. M. Johnson. 1980. Monoclonal antibodies to lymphocytic choriomeningitis virus react with pathogenic arenaviruses. Nature 288:486-487. [DOI] [PubMed] [Google Scholar]
- 9.Bui, H. H., et al. 2007. Protein sequence database for pathogenic arenaviruses. Immunome Res. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capul, A. A., et al. 2007. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J. Virol. 81:9451-9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casabona, J. C., J. M. Levingston Macleod, M. E. Loureiro, G. A. Gomez, and N. Lopez. 2009. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J. Virol. 83:7029-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornu, T. I., and J. C. de la Torre. 2002. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J. Virol. 76:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichler, R., et al. 2004. Characterization of the Lassa virus matrix protein Z: electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP). Virus Res. 100:249-255. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, S. A., et al. 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 354:2235-2249. [DOI] [PubMed] [Google Scholar]
- 16.Groseth, A., S. Wolff, T. Strecker, T. Hoenen, and S. Becker. 2010. Efficient budding of the Tacaribe virus matrix protein Z requires the nucleoprotein. J. Virol. 84:3603-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacamo, R., N. Lopez, M. Wilda, and M. T. Franze-Fernandez. 2003. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J. Virol. 77:10383-10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitching, A., et al. 2009. A fatal case of Lassa fever in London, January 2009. Euro Surveill. 14(6):pii=19117. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19117. [PubMed] [Google Scholar]
- 19.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, K. J., M. Perez, D. D. Pinschewer, and J. C. de la Torre. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J. Virol. 76:6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenard, J. 1996. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology 216:289-298. [DOI] [PubMed] [Google Scholar]
- 22.Neuman, B. W., et al. 2005. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 79:3822-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 24.Noton, S. L., et al. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 88:2280-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U. S. A. 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez, M., and J. C. de la Torre. 2003. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 77:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez, M., D. L. Greenwald, and J. C. de la Torre. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78:11443-11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radoshitzky, S. R., et al. 2010. Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J. Virol. 84:10569-10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez, L., I. Cuesta, A. Asenjo, and N. Villanueva. 2004. Human respiratory syncytial virus matrix protein is an RNA-binding protein: binding properties, location and identity of the RNA contact residues. J. Gen. Virol. 85:709-719. [DOI] [PubMed] [Google Scholar]
- 30.Safronetz, D., et al. 2010. Detection of Lassa virus, Mali. Emerg. Infect. Dis. 16:1123-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shtanko, O., et al. 2010. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein-induced virus-like particles. J. Virol. 84:5415-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strecker, T., et al. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J. Virol. 77:10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strecker, T., et al. 2006. The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol. J. 3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Traub, E. 1936. The epidemiology of lymphocytic choriomeningitis in white mice. J. Exp. Med. 64:183-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urata, S., T. Noda, Y. Kawaoka, H. Yokosawa, and J. Yasuda. 2006. Cellular factors required for Lassa virus budding. J. Virol. 80:4191-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volpon, L., M. J. Osborne, and K. L. Borden. 2008. NMR assignment of the arenaviral protein Z from Lassa fever virus. Biomol. NMR Assign. 2:81-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volpon, L., M. J. Osborne, A. A. Capul, J. C. de la Torre, and K. L. Borden. 2010. Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc. Natl. Acad. Sci. U. S. A. 107:5441-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilda, M., N. Lopez, J. C. Casabona, and M. T. Franze-Fernandez. 2008. Mapping of the Tacaribe arenavirus Z-protein binding sites on the L protein identified both amino acids within the putative polymerase domain and a region at the N terminus of L that are critically involved in binding. J. Virol. 82:11454-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]