Abstract
Different human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV) vaccine vectors expressing the same viral antigens can elicit disparate T-cell responses. Within this spectrum, replicating variable vaccines, like SIVmac239Δnef, appear to generate particularly efficacious CD8+ T-cell responses. Here, we sequenced T-cell receptor β-chain (TRB) gene rearrangements from immunodominant Mamu-A*01-restricted Tat28-35SL8-specific CD8+ T-cell populations together with the corresponding viral epitope in four rhesus macaques during acute SIVmac239Δnef infection. Ultradeep pyrosequencing showed that viral variants arose with identical kinetics in SIVmac239Δnef and pathogenic SIVmac239 infection. Furthermore, distinct Tat28-35SL8-specific T-cell receptor (TCR) repertoires were elicited by SIVmac239Δnef compared to those observed following a DNA/Ad5 prime-boost regimen, likely reflecting differences in antigen sequence stability.
Vaccination of nonhuman primates with a live-attenuated strain of simian immunodeficiency virus (SIV), such as SIVmac239Δnef, consistently protects against pathogenic SIV challenge (1, 4, 11). Furthermore, evidence from depletion experiments indicates that this protection is mediated, at least in part, by CD8+ T cells (9, 15, 18). However, the mechanisms that underlie protective CD8+ T-cell immunity in this model remain unclear.
In rhesus macaques, immunodominant SIV-specific CD8+ T-cell responses that are restricted by the same major histocompatibility complex (MHC) class I molecule are composed of T-cell receptor (TCR) repertoires with various levels of complexity based on the viral antigen being presented. CD8+ T-cell clonotypes specific for the Mamu-A*01-restricted Gag181-189CM9 epitope, which is biologically constrained and therefore unable to mutate freely, expand to form diverse TCR repertoires (13, 14). In contrast, CD8+ T-cell clonotypes specific for the Mamu-A*01-restricted Tat28-35SL8/TL8 epitope converge on a prevalent TCRβ motif and consequently exhibit limited diversity. This restricted antigen recognition profile, combined with a tolerance for mutation in this region of the virus, facilitates immune escape by mutation at potential TCR contact residues (14). Understanding how different vaccines affect TCR repertoire formation, therefore, has potential implications for the design of a prophylactic human immunodeficiency virus (HIV) vaccine. In particular, vaccines that evolve within hosts, such as SIVmac239Δnef, may elicit different TCR repertoires than traditional DNA prime-virus boost regimens.
The advent of ultradeep pyrosequencing has provided unparalleled precision in the characterization of viral quasispecies in HIV-infected humans and SIV-infected nonhuman primates (2, 3, 8, 10, 12, 19). Thus, for the first time, the effects of viral variation on the cognate CD8+ T-cell repertoire can be assessed quantitatively. Here, we examined the kinetics of Tat28-35SL8 mutation and conducted a comprehensive assessment of the contemporaneous Tat28-35SL8-specific CD8+ T-cell repertoires in SIVmac239Δnef-vaccinated rhesus macaques.
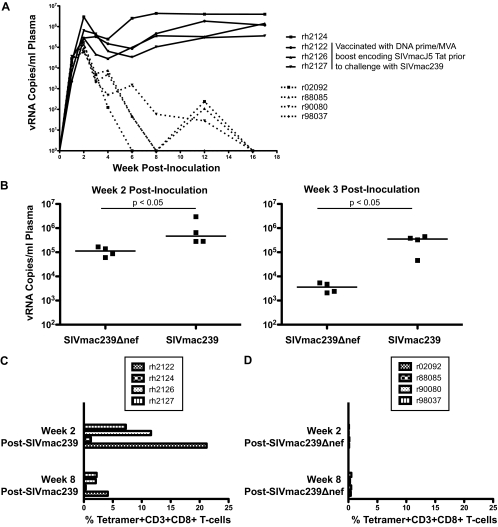
Four Mamu-A*01+ SIVmac239Δnef-vaccinated rhesus macaques were selected for this study; details of their vaccination were published previously (15). Initially, we compared viral replication levels and magnitudes of Tat28-35SL8-specific CD8+ T-cell responses between this group and a group of four Mamu-A*01+ SIVmac239-infected rhesus macaques (Fig. 1; see also Table 1 at https://xnight.primate.wisc.edu:8443/labkey/project/WNPRC/WNPRC_Laboratories/oconnor/public/publications/begin.view?) (15, 21). SIVmac239Δnef-vaccinated macaques exhibited lower peak and postpeak viremias, and these viral load differences were significant at both week 2 and week 3 postvaccination (week 2 medians, 1.1 × 105 versus 4.7 × 105 viral RNA [vRNA] copies/ml; week 3 medians, 3.6 × 103 versus 3.5 × 105 vRNA copies/ml [P = 0.0143 at both time points; one-tailed Mann-Whitney U test]) (Fig. 1A and B). Additionally, all four animals controlled viral replication to undetectable levels within the first 18 weeks of infection. Tat28-35SL8 responses also differed between groups, with the SIVmac239-infected macaques exhibiting much larger responses than the SIVmac239Δnef-vaccinated macaques (Fig. 1C and D). Three of the SIVmac239-infected macaques (rh2122, rh2126, and rh2127) were vaccinated with a DNA prime-recombinant-modified-vaccinia-virus-Ankara-boost regimen prior to intrarectal SIVmac239 infection, thereby explaining the enhanced magnitude of their week 2 postinfection Tat28-35SL8 responses.
FIG. 1.
(A) Viral loads (numbers of viral RNA copies/ml plasma) for each macaque following infection with SIVmac239 (unbroken lines) or vaccination with SIVmac239Δnef (broken lines). These viral loads have been published previously (15, 21). (B) Comparison of plasma viral loads between SIVmac239Δnef-vaccinated and SIVmac239-infected rhesus macaques at weeks 2 and 3 postinoculation. Significance was assessed using a one-tailed Mann-Whitney test (P = 0.0143 at both time points). (C) Percentages of tetramer-binding Tat28-35SL8-specific cells within the CD3+ CD8+ T-cell population for four rhesus macaques at weeks 2 and 8 after SIVmac239 infection. These results have been published previously (21). (D) Percentages of tetramer-binding Tat28-35SL8-specific cells within the CD3+ CD8+ T-cell population for four rhesus macaques at weeks 2 and 8 after SIVmac239Δnef vaccination.
Viral variation driven by CD8+ T cells in the context of a live-attenuated SIV vaccine has not been reported previously. Our group has used ultradeep pyrosequencing to show rapid and widespread viral variation within the Tat28-35SL8 epitope by week 3 after SIVmac239 infection, with some macaques showing low frequency mutations as early as week 2 postinfection (2). Therefore, we performed ultradeep pyrosequencing of the Tat28-35SL8 epitope at weeks 2 and 3 after SIVmac239Δnef vaccination in the four selected Mamu-A*01+ rhesus macaques by use of previously published methods (2).
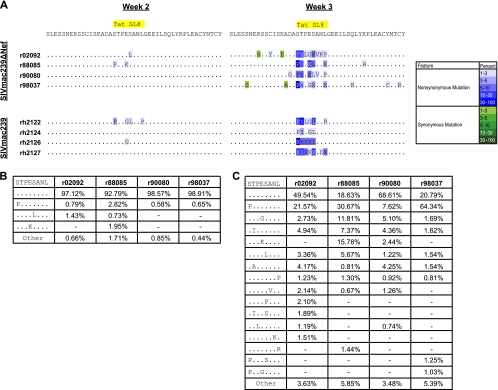
Ultradeep pyrosequencing resulted in an average of 2,940 sequence reads per sample, with a range of 2,700 to 3,784 sequence reads. We compared SIVmac239Δnef Tat28-35SL8 variation with our previously published sequence data from the four SIVmac239-infected rhesus macaques (2). Rapid viral variation was observed within the Tat28-35SL8 epitope in SIVmac239Δnef-vaccinated Mamu-A*01+ macaques, despite their lower acute-phase viral loads and smaller Tat28-35SL8-specific CD8+ T-cell responses relative to those of macaques infected with SIVmac239 (Fig. 1 and 2; see also raw data at https://xnight.primate.wisc.edu:8443/labkey/project/WNPRC/WNPRC_Laboratories/oconnor/public/publications/begin.view?) (Sequence Read Archive accession no. SRA028442.1). The pattern of viral variation within the Tat28-35SL8 epitope was similar to our previous findings in SIVmac239-infected macaques (2). Additionally, we observed a similar level of interindividual variability in terms of the positions and quantities of specific mutations (Fig. 2A and C). The overall percentage of viral variants circulating within each macaque differed at week 3 postvaccination. In r90080, only 31.4% of the peripheral viral population showed variation within Tat28-35SL8. In contrast, r88085 and r98037 had higher proportions of viral variants (81.4% and 79.2%, respectively). Taken together, these data demonstrate that Tat28-35SL8 differs with similar kinetics and levels of breadth in SIVmac239Δnef-vaccinated and SIVmac239-infected rhesus macaques.
FIG. 2.
Ultradeep pyrosequencing analysis of the Tat28-35SL8 region from four SIVmac239Δnef-vaccinated Mamu-A*01+ rhesus macaques. (A) Consensus sequence from each sample. The ultradeep pyrosequencing consensus contains any mutation present in 1% or more of the total reads. Changes are indicated using the single-letter amino acid code for substitutions where only 1 residue was found at that position with a frequency of >1%. All other changes are indicated with the letter X. Nonsynonymous and synonymous mutations are colored according to prevalence, indicated by the color key on the right. Data from SIVmac239-infected rhesus macaques are shown for comparison and were published previously (2). (B) Percentage of each variant observed within Tat28-35SL8 at week 2. Only variants present at frequencies of 1% or greater in at least one macaque are shown. Variants present at less than 1% were summed, and their combined frequency is listed as “Other.” Dashes indicate that a variant was not present in that sample. (C) Percentage of each variant observed within Tat28-35SL8 at week 3. The details are the same as those for panel B.
It is possible that SIVmac239Δnef epitope variation affects the protective efficacy of this vaccine, given that coevolution of the host immune system with the diversifying virus may influence the TCR repertoire within virus-specific T-cell responses. In addition, a diversifying live-attenuated virus may elicit immune responses that differ from those generated by nonmosaic, static immunogens, such as SIV genes packaged into adenovirus type 5 (Ad5) viral vectors. To investigate these possibilities, we undertook a comprehensive analysis of all expressed TRB gene rearrangements within Tat28-35SL8-specific CD8+ T-cell populations by use of an unbiased template-switch anchored reverse transcription-PCR (RT-PCR) as described previously (6, 14).
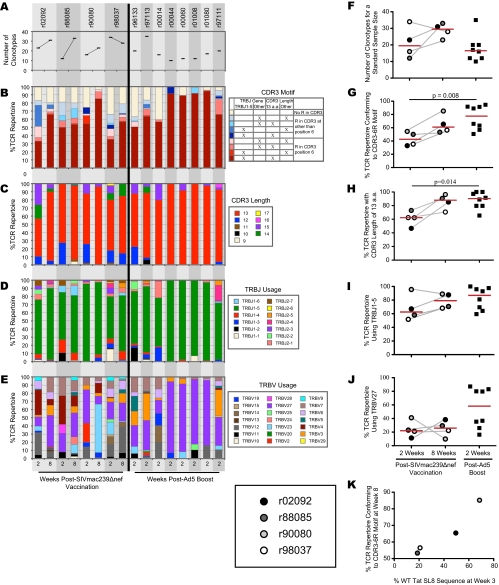
CD8+ T cells specific for Tat28-35SL8 were identified directly ex vivo using fluorochrome-conjugated SL8/Mamu-A*01 tetrameric complexes and sorted at >98% purity by flow cytometry (14). At week 2 postvaccination, before substantial variation occurred within the Tat28-35SL8 epitope, common trends were observed within the four Tat28-35SL8-specific TCRβ repertoires (Fig. 3 and 4 A to E). Although these early SIVmac239Δnef Tat28-35SL8-specific CD8+ T-cell populations featured diverse TRBV gene usage (Fig. 4E) and were polyclonal, with a median of 21 (range, 12 to 36) unique TCRβ clonotypes per repertoire, many clonotypes conformed to a common CDR3β motif that comprised a central arginine residue at position 6, the usage of the TRBJ1-5 gene, and an overall length of 13 amino acids (CDR3-6R). Additionally, previously described “public” clonotypes were sequenced from each animal (Fig. 3). These findings are in line with previous reports and provide further evidence that the characteristics of the mobilized antigen-specific T-cell repertoire are determined by the nature of the antigen rather than the context of presentation (13, 14, 16).
FIG. 3.
CDR3β amino acid sequences, TRBV and TRBJ gene usage, and relative frequencies of Tat28-35SL8-specific CD8+ T-cell clonotypes for all four macaques at weeks 2 and 8 after SIVmac239Δnef vaccination. Clonotypes shared between time points are color coded in the frequency column, and public clonotypes are color coded in the CDR3 sequence column. Public clonotypes were identified as TCRβ amino acid sequences observed in more than one macaque with reference to an extensive database including data from previous studies (13, 14). Sequences were aligned against the rhesus macaque TRB genes (7), for which international Immunogenetics (IMGT) information system nomenclature is used. The asterisk indicates potential allelic variations within the TRBV13-1 (G at nucleotide position 7 from the 3′ gene end) and TRBJ1-6 (T at nucleotide position 20 from the 5′ gene end) gene-encoded portions of the CDR3 protein.
FIG. 4.
(A) Numbers of unique Tat28-35SL8-specific CD8+ T-cell clonotypes in macaques following vaccination with SIVmac239Δnef or DNA prime-Ad5 boost with Tat-encoding vectors, estimated for a standard sample size of 70 TCRβ sequences across all samples (20). (B to E) Frequencies of Tat28-35SL8-specific CD8+ T-cell clonotypes in macaques following vaccination with SIVmac239Δnef or DNA prime-Ad5 boost with Tat-encoding vectors that feature particular CDR3 amino acid (a.a.) motif characteristics (B), CDR3 lengths (C), TRBJ gene usage (D), and TRBV gene usage (E). (F to J) Comparisons of TCR repertoire parameters between macaques at week 2 after SIVmac239Δnef vaccination, week 8 after SIVmac239Δnef vaccination, and week 2 after Ad5 boost; number of unique Tat28-35SL8-specific CD8+ T-cell clonotypes (F), CDR3-6R motif frequency (G), frequency of CDR3s with lengths of 13 amino acids (H), frequency of TRBJ1-5 gene usage (I), and frequency of TRBV27 gene usage (J). (K) Relationship between the frequency of Tat28-35SL8-specific CD8+ T-cell clonotypes with the CDR3-6R motif at week 8 after SIVmac239Δnef vaccination and the frequency of wild-type (WT) Tat28-35SL8 sequences observed at week 3 after SIVmac239Δnef vaccination.
Next, we determined the changes that occurred in the Tat28-35SL8-specific CD8+ T-cell repertoires following the emergence of substantial variation within the Tat28-35SL8 epitope (Fig. 3). Overall, TRBV gene usage remained diverse at 8 weeks after SIVmac239Δnef vaccination (Fig. 4E). However, marked differences in TRBV gene usage over time were apparent. For example, TRBV23-1 was not used at week 2 but comprised 59.2% of sequences in r90080 at week 8. The polyclonality of these responses was greater at week 8 postvaccination than at week 2, with a median of 31 unique TCRβ clonotypes (range, 24 to 33 clonotypes) per repertoire. Despite this diversification, the CDR3-6R motif remained prevalent and public clonotypes were observed at week 8 in all four Tat28-35SL8-specific CD8+ T-cell repertoires (Fig. 3 and 4B). These data suggest that the CDR3β core and TRBJ1-5 play determinative roles in Tat28-35SL8 antigen recognition, but structural data are required to elucidate more fully the nature of the TCR/SL8/Mamu-A*01 interaction.
It is a widely held belief that vaccine-elicited CD8+ T-cell responses should cross-recognize epitope variants to exert optimal efficacy against variable viruses. Such cross-reactivity could be generated either by incorporating multiple different clonotypes or by preferential recruitment of specific clonotypes with highly degenerate antigen recognition profiles (5, 13). To assess whether TCR repertoires elicited by SIVmac239Δnef vaccination differed from those elicited by a prime-boost vaccination, we compared the values for different TCR repertoire parameters between our cohort and a previously studied cohort of rhesus macaques that received a DNA prime-Ad5 boost vaccine containing SIVmac239 Tat (22). For each macaque, we measured TCR diversity, motif frequency, CDR3 length, and TRBJ and TRBV gene usage (Fig. 4A to E); these features were compared between the two cohorts (Fig. 4F to J). Estimated for a standard sample size of 70 TCRβ sequences, the number of unique clonotypes from each macaque at week 2 after SIVmac239Δnef vaccination was similar to the number of unique clonotypes sequenced at week 2 after Ad5 boost (Fig. 4F). However, there were significant differences between the two cohorts in the features of the TCR repertoires, with the CDR3-6R motif and an overall CDR3 length of 13 amino acids being significantly more prevalent in the Ad5-boosted cohort than in the SIVmac239Δnef-vaccinated cohort at week 2 postvaccination (P = 0.008 and P = 0.014, respectively; Mann-Whitney U test) (Fig. 4G and H). Tat28-35SL8-specific TCRs often use the TRBV27 gene; indeed, TRBV27 gene expression was observed in all four SIVmac239Δnef-vaccinated rhesus macaques at both week 2 and week 8 postvaccination. Compared to that in the SIVmac239Δnef-vaccinated cohort, TRBV27 gene expression in the Ad5-boosted cohort had increased complexity, with half of the macaques exhibiting similar frequencies and the other half exhibiting much higher frequencies (Fig. 4J). Interestingly, the four macaques in the Ad5-boosted cohort with the highest frequencies of the CDR3-6R motif also had the highest frequencies of TRBV27 gene usage; indeed, a median of 81.6% (range, 78.4 to 85.7%) of all clonotypes in these macaques expressed the TRBV27 gene and contained the CDR3-6R motif. Structural data will be required to elucidate the underlying basis of this associative bias.
The kinetics of viral escape within Tat28-35SL8 could affect the emergence of novel CD8+ T-cell clonotypes in SIVmac239Δnef-vaccinated rhesus macaques. To assess this possibility, we compared the frequency of wild-type Tat28-35SL8 sequences at week 3 after SIVmac239Δnef vaccination with the prevalence of the CDR3-6R motif in the TCR repertoires sequenced from each macaque at week 8 after SIVmac239Δnef vaccination (Fig. 4K). Intriguingly, we observed an association between the proportion of the TCR repertoire featuring CDR3-6R clonotypes sequenced at week 8 postvaccination and the proportion of wild-type Tat28-35SL8 reads observed at week 3 postvaccination within each macaque. Importantly, the differences in CDR3-6R frequencies in the week 8 postvaccination TCR repertoires between the macaques (Fig. 4G) could not be accounted for by viral sequence differences at week 2 postvaccination (Fig. 2B). This result suggests that continued presence of wild-type Tat28-35SL8 antigen drives a more focused CDR3-6R motif-bearing CD8+ T-cell response. Such a process would be in keeping with previous reports and, by extension, could help to explain Tat28-35SL8-specific repertoire diversity patterns in the presence of a complex mixture of epitope variants, each of which carries the potential to productively engage distinct TCR structures and expand the corresponding cognate clonotypes (13, 14). However, other factors clearly affect TCR usage in response to the Tat28-35SL8 epitope, and further investigation is required to confirm this tentative conclusion given the limited number of macaques and time points in this study.
Many HIV vaccines are under investigation, and comprehensive analyses of the CD8+ T-cell responses that they elicit will guide future studies. Recently, the idea of vaccinating macaques with mosaic vaccines containing multiple HIV variants has been posited and tested, with encouraging results (17). SIVmac239Δnef exhibits viral variation in rhesus macaques and is replication competent, lending an advantage over mosaic prime-boost vaccines. Ultradeep pyrosequencing gives a nuanced view of such viral variation in live-attenuated SIV-vaccinated primates, and for the first time, quantitative comparisons of viral variation with other measurements of adaptive immunity are possible. Here, we show that the Tat28-35SL8-specific TCRβ repertoire changes over the course of acute SIVmac239Δnef vaccination, trending toward greater diversity than for macaques vaccinated with a DNA prime-Ad5 boost regimen. Ancillary studies are warranted to determine the extent to which these findings can be generalized. However, the current study provides both a testable hypothesis, according to which epitope sequence complexity and cognate repertoire diversity are intimately linked, and the tools with which to unravel this interrelationship. Additional studies comparing prevaccine boost, postvaccine boost, and postinfection TCR repertoires may further elucidate the complex relationship of clonotype selection and viral variation.
Constituent TCRs determine the specificity and cross-reactivity profile of a CD8+ T-cell population, and it is becoming abundantly clear that not all TCRs are created equal. Consequently, a detailed understanding of HIV/SIV coevolution with emerging T-cell responses at the clonotypic level may provide key information for the design of an effective prophylactic vaccine.
Acknowledgments
We thank David Watkins for invaluable advice and for the animal samples utilized in the study.
This publication was made possible by grant number P51 RR000167 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), to the Wisconsin National Primate Research Center, University of Wisconsin—Madison; the research was conducted at a facility constructed with support from the Research Facilities Improvement Program, grant numbers RR15459-01 and RR020141-01. Additional funding was provided by grant numbers 1R01AI084787, R01 AI077376, and 144PRJ23CG from the NIH. D.A.P. is a Medical Research Council (United Kingdom) Senior Clinical Fellow; V.V. is an Australian Research Council Future Fellow.
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or the NIH.
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1.Almond, N., et al. 1995. Protection by attenuated simian immunodeficiency virus in macaques against challenge with virus-infected cells. Lancet 345:1342-1344. [DOI] [PubMed] [Google Scholar]
- 2.Bimber, B. N., et al. 2009. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J. Virol. 83:8247-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman, F. D., et al. 2008. Massively parallel pyrosequencing in HIV research. AIDS 22:1411-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 5.Davenport, M. P., D. A. Price, and A. J. McMichael. 2007. The T cell repertoire in infection and vaccination: implications for control of persistent viruses. Curr. Opin. Immunol. 19:294-300. [DOI] [PubMed] [Google Scholar]
- 6.Douek, D. C., et al. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168:3099-3104. [DOI] [PubMed] [Google Scholar]
- 7.Greenaway, H. Y., et al. 2009. Extraction and characterization of the rhesus macaque T-cell receptor beta-chain genes. Immunol. Cell Biol. 87:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le, T., et al. 2009. Low-abundance HIV drug-resistant viral variants in treatment-experienced persons correlate with historical antiretroviral use. PLoS One 4:e6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzner, K. J., et al. 2000. Effects of in vivo CD8(+) T cell depletion on virus replication in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Exp. Med. 191:1921-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsuya, Y., et al. 2008. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J. Virol. 82:10747-10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norley, S., B. Beer, D. Binninger-Schinzel, C. Cosma, and R. Kurth. 1996. Protection from pathogenic SIVmac challenge following short-term infection with a nef-deficient attenuated virus. Virology 219:195-205. [DOI] [PubMed] [Google Scholar]
- 12.Poon, A. F., et al. 2010. Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol. Biol. Evol. 27:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price, D. A., et al. 2009. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J. Exp. Med. 206:923-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price, D. A., et al. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 21:793-803. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds, M. R., et al. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 205:2537-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudd, B. D., et al. 2010. Diversity of the CD8+ T cell repertoire elicited against an immunodominant epitope does not depend on the context of infection. J. Immunol. 184:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santra, S., et al. 2010. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat. Med. 16:324-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz, J. E., et al. 2005. Effect of CD8+ lymphocyte depletion on virus containment after simian immunodeficiency virus SIVmac251 challenge of live attenuated SIVmac239delta3-vaccinated rhesus macaques. J. Virol. 79:8131-8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varghese, V., et al. 2009. Minority variants associated with transmitted and acquired HIV-1 nonnucleoside reverse transcriptase inhibitor resistance: implications for the use of second-generation nonnucleoside reverse transcriptase inhibitors. J. Acquir. Immune Defic. Syndr. 52:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venturi, V., K. Kedzierska, S. J. Turner, P. C. Doherty, and M. P. Davenport. 2007. Methods for comparing the diversity of samples of the T cell receptor repertoire. J. Immunol. Methods 321:182-195. [DOI] [PubMed] [Google Scholar]
- 21.Vogel, T. U., et al. 2003. Multispecific vaccine-induced mucosal cytotoxic T lymphocytes reduce acute-phase viral replication but fail in long-term control of simian immunodeficiency virus SIVmac239. J. Virol. 77:13348-13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson, N. A., et al. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]