Abstract
The begomoviruses are the largest and most economically important group of plant viruses transmitted exclusively by the whitefly Bemisia tabaci in a circulative, persistent manner. The circulation of the viruses within the insect vectors involves complex interactions between virus and vector components; however, the molecular mechanisms of these interactions remain largely unknown. Here we investigated the transcriptional response of the invasive B. tabaci Middle East-Asia Minor 1 species to Tomato yellow leaf curl China virus (TYLCCNV) using Illumina sequencing technology. Results showed that 1,606 genes involved in 157 biochemical pathways were differentially expressed in the viruliferous whiteflies. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that TYLCCNV can perturb the cell cycle and primary metabolism in the whitefly, which explains the negative effect of this virus on the longevity and fecundity of B. tabaci. Our data also demonstrated that TYLCCNV can activate whitefly immune responses, such as autophagy and antimicrobial peptide production, which might lead to a gradual decrease of viral particles within the body of the viruliferous whitefly. Furthermore, PCR results showed that TYLCCNV can invade the ovary and fat body tissues of the whitefly, and Lysotracker and Western blot analyses revealed that the invasion of TYLCCNV induced autophagy in both the ovary and fat body tissues. Surprisingly, TYLCCNV also suppressed the whitefly immune responses by downregulating the expression of genes involved in Toll-like signaling and mitogen-activated protein kinase (MAPK) pathways. Taken together, these results reveal the relationship of coevolved adaptations between begomoviruses and whiteflies and will provide a road map for future investigations into the complex interactions between plant viruses and their insect vectors.
Of the approximately 700 plant viruses that are officially recognized by the International Committee on Taxonomy of Viruses, more than 75% are transmitted by insect vectors (27). The interactions between plant viruses and insect vectors have received great attention worldwide because of their importance in both agriculture and scientific research (10, 24, 27, 31, 51). The most economically important insect vectors are restricted to insects of a few hemipteran families, such as aphids and whiteflies (27). The whitefly Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) is a cryptic species complex composed of at least 24 morphologically indistinguishable species (16, 17, 71, 78, 79, 84). In this species complex, the Middle East-Asia Minor 1 (herein called MEAM1) species, commonly referred to as the B “biotype,” has risen to international prominence since the 1980s due to its global invasion (15, 40, 54). B. tabaci causes excessive crop losses through direct feeding and transmission of plant viruses (6, 54). The most severe damage is its transmission of begomoviruses (family Geminiviridae, genus Begomovirus), the largest and most economically important group of plant viruses in the tropical and subtropical agroecosystems worldwide (20, 41, 48, 62, 76). B. tabaci can exclusively transmit 115 species of begomoviruses in a persistent-circulative mode (27), and epidemics of begomoviruses are usually associated with outbreaks of MEAM1 (10, 31, 76). In view of their importance, the begomoviruses and whiteflies have been used as an excellent model to study plant virus-insect vector interactions (10, 12, 31).
The relationships between begomoviruses and whiteflies are complex. For example, Tomato yellow leaf curl China virus (TYLCCNV) DNA can be transmitted horizontally and vertically by MEAM1, albeit with low frequency (77). This suggests that this virus has invaded the reproductive system of the viruliferous whiteflies. In addition, when the MEAM1 viruliferous whiteflies were allowed to feed on cotton, a non-host plant of the virus, the presence of TYLCCNV within the whitefly reduced the longevity and fecundity of the vector by 27% and 36%, and the proportion of adults with detectable TYLCCNV DNA decreased gradually but was still maintained at 50% after feeding on cotton for 960 h (31, 32). Collectively, these results indicate that TYLCCNV may have direct effects on whiteflies through inducing changes in the physiological activities and immune responses.
Begomoviruses are known to move through the insect vector, from the gut lumen into the hemolymph and finally into the salivary glands, from which these viruses are secreted back into the plant host during insect feeding (13, 27). During this process, a GroEL homolog protein produced by the B. tabaci endosymbiotic bacteria and the whitefly BtHSP16 protein have reportedly been involved in the viral transmission (49, 52). Moreover, both the virion stability and interactions with molecular components inside the salivary glands are necessary for begomoviral transmission (7). Thus, it is inevitable that during this long-standing virus-vector interaction, the whitefly has evolved strategies to protect itself against possible deleterious effects of the virus, and the begomovirus has also evolved to ensure both its survival and efficient transmission by the vector (13). However, up to now, the molecular mechanism underlying the complex interactions between begomovirus and whitefly are poorly understood and the global transcriptional response of the whitefly to begomoviruses remains unknown, either during circulative transmission or during long-term storage in the insect tissues (12, 24, 27). This largely limits our understanding of interactions between begomoviruses and whiteflies.
The recently available whitefly transcriptome sequences (80; unpublished data) in combination with the Illumina sequencing technology, which is far more precise and sensitive than other methods of measuring the levels of transcripts (43, 69, 74, 81), have provided us unprecedented opportunities to investigate the transcriptional response of B. tabaci to begomoviruses. Here we have shown that a number of genes involved in the cell cycle, primary metabolism, and immune response were regulated in the viruliferous whitefly. We also demonstrated for the first time, using Lysotracker staining and Western blotting, that TYLCCNV can induce autophagy in the whitefly ovary and fat body tissues. For persistence, begomoviruses can suppress whitefly immune responses. Overall, we present convincing evidence for the relationship of coevolved adaptations between begomoviruses and whiteflies. To our knowledge, this is the first report to study the direct effect of a plant virus on the global gene expression profile of its insect vector using a high-throughput sequencing method.
MATERIALS AND METHODS
Insect culture, virus, and plants.
Cotton (Gossypium hirsutum cv. Zhe-Mian 1793) is a non-host plant of TYLCCNV and was cultivated to the 7-8 true-leaf stage for experiments. A pair of virgin adults of B. tabaci MEAM1 (mtCO1 GenBank accession no. GQ332577) were released onto cotton plants to oviposit and develop for five generations. This culture of MEAM1 was maintained on cotton plants in climate chambers at 27 ± 1°C, a photoperiod of 14 h light/10 h darkness and 70% ± 10% relative humidity. The purity of the culture was monitored every 3 to 5 generations using the random amplified polymorphic DNA PCR technique combined with the sequencing of the mitochondrial cytochrome oxidase I gene, which has been used widely to differentiate B. tabaci genetic groups (16, 84).
Clones of TYLCCNV with its satellite DNA molecules, also known as DNAβ, were used as inocula (11). The Agrobacterium tumefaciens EHA105 culture was grown in yeast extract-peptone (YEP) medium containing kanamycin (50 mg/liter) and rifampin (50 mg/liter) at 28°C for 24 h. Then, the culture was centrifuged and resuspended in the mixture containing 10 mmol/liter MgCl2, 10 mmol/liter morpholineethanesulfonic acid (MES) and 200 mmol/liter acetosyringone, adjusted to an optical density at 600 nm of 0.8 to 1.0, and incubated at room temperature for 3 h.
To obtain virus-infected tobacco (Nicotiana tabacum cv. NC89), plants at the 3-4 true-leaf stage were inoculated with TYLCCNV and its DNAβ by agroinoculation as previously described (11). The uninfected tobacco was mock inoculated using the same Agrobacterium strain. TYLCCNV-infected and uninfected tobacco plants were cultivated to the 7-8 true-leaf stage when used in experiments. Virus infection of test plants was judged by the typical symptoms caused by the virus and further confirmed by PCR using the procedure described previously (57). All plants were grown in a greenhouse under natural lighting and controlled temperature.
Sample preparation and RNA isolation.
To prepare nonviruliferous and viruliferous whitefly samples, approximately 3,000 newly emerged adults of whitefly on cotton were released onto the leaves of two healthy tobacco plants in one cage and 3,000 whiteflies were transferred to two virus-infected tobacco plants in another cage. After 24 h of feeding, two cotton plants were placed near the tobacco plants in each of these two cages. The leaves of tobacco plants were gently turned upside-down, and thereby the majority of whiteflies flew onto the cotton plants within several minutes. After that, the tobacco plants and the remaining whiteflies on them were discarded immediately. The nonviruliferous and viruliferous whiteflies were allowed to feed on cotton plants in different cages for 120 h. Then, approximately 1,000 nonviruliferous and viruliferous female adults of whitefly were collected, respectively. All the experiments were conducted in climate chambers at 27 ± 1°C, a photoperiod of 14 h light/10 h darkness and 70% ± 10% relative humidity. PCR analyses showed that viral DNA was present in 100% of the whiteflies after a 24-h acquisition access period (AAP) on virus-infected tobacco plants and 80% of the whiteflies still carried viruses after 120 h of feeding on cotton (data not shown). Therefore, these two whitefly samples were referred to as the nonviruliferous and viruliferous whiteflies, respectively. Two biological replicates for both of the whitefly samples were conducted and processed independently. One replicate was used in the digital gene expression (DGE) library preparation and Illumina sequencing, and the other was used for quantitative PCR (qPCR) analysis. Total RNA was isolated using the SV total RNA isolation system (Promega) according to the manufacturer's protocol. RNA integrity was confirmed using the 2100 bioanalyzer (Agilent Technologies) with a minimum RNA integrated number value of 8.
Since previous studies have demonstrated that MEAM1 species could acquire virus after a 12-h period of feeding (32), to minimize the differential effects of healthy and infected tobacco plants on the gene expression profile of whiteflies, we only allowed the whiteflies to feed on tobacco plants for 24 h to acquire the viruses. After that, both the viruliferous and nonviruliferous whiteflies were allowed to fly freely onto cotton, a non-host plant of TYLCCNV, to feed for 120 h before RNA extraction. This 120-h period of feeding on cotton was set in order to diminish the differential effects of healthy and infected plants on whiteflies. The infected plants might differ from the healthy plants in nutrition, plant volatiles, and defense responses. Feeding for 120 h on cotton plants will significantly decrease the residual effects of tobacco volatiles and defense responses on the whitefly. Furthermore, the effect from the sap of infected plants will also be trivial after 5 days on cotton because the sap from infected tobacco will be used up within 2 days (this postulation is based on the well-known fact that all whiteflies will die within 2 days without a food supply). Moreover, after 5 days of feeding on cotton, 80% of the viruliferous whitefly adults still carried virions (32). Therefore, the regulation of viruliferous whitefly genes revealed here was likely to be caused mainly by TYLCCNV rather than the residual effect of healthy and infected tobacco plants. Another possible way of obtaining viruliferous whiteflies is to feed them with purified virions in artificial diets. However, artificial feeding with purified virions has several drawbacks: (i) it must put the whiteflies in an unnatural environment (feeding chamber), which would change whiteflies’ gene expression profile; (ii) the purified virions may contain proteins from the plant and chemicals from the reagents used to purify the virus; and (iii) the coat protein and the structure of the purified virion particles might be different from those of the viruses in the plants. A third possible way of obtaining viruliferous whiteflies is to inject virions into the whitefly body, but injections are likely to cause significant physical damages to the whitefly. Thus, neither of the last two methods is suitable for this study because they put the whiteflies in unnatural settings and would initiate a gene expression profile that is unrelated to vector-virus interactions.
DGE library preparation and sequencing.
Using the DGE method, which generates absolute rather than relative gene expression measurements and avoids many of the inherent limitations of microarray analysis, we analyzed the gene expression variations between the nonviruliferous and viruliferous whiteflies. First, mRNA was purified from 6 μg of the total RNA from each of the two samples with magnetic oligo(dT) beads. First- and second-strand cDNA was synthesized, and bead bound cDNA was subsequently digested with NlaIII, which recognizes the CATG sites. The cDNA fragments with 3′ ends were then purified with magnetic beads, and Illumina adapter 1 was added to their 5′ ends. The junction of Illumina adapter 1 and the CATG site is the recognition site of MmeI, which cuts 17 bp downstream of the CATG site, producing tags with adapter 1. After removing 3′ fragments with magnetic bead precipitation, Illumina adapter 2 was introduced at 3′ ends of tags, acquiring tags with different adapters at both ends to form a tag library. After 15 cycles of linear PCR amplification, 85 base strips were purified by 6% Tris-borate-EDTA (TBE)-PAGE gel electrophoresis. These strips were then digested, and the single-chain molecules were fixed on the Illumina chip for sequencing. Each molecule was grown into a single-molecule cluster sequencing template through in situ amplification. Then, four types of fluorescence-labeled nucleotides were added, and sequencing was performed using the method of sequencing by synthesis. Each tunnel generated millions of raw reads with sequencing lengths of 35 bp.
Tag annotation and data normalization for gene expression level.
Raw sequences were transformed into clean tags through removing adaptor sequence, low-quality sequences (tags with unknown sequences [“N”]), empty reads (sequence with only adaptor sequences but no tags), too-long or too-short tags, and tags with a copy number of 1 (probably a sequencing error). A preprocessed database of all possible CATG + 17-nucleotide tag sequences was created using our transcriptome reference database of the MEAM1 and Mediterranean species of whitefly (80; unpublished data). For annotation, all tags were mapped to the reference sequences and only allowed no more than 1 nucleotide mismatch. Clean tags mapped to reference sequences from multiple genes were filtered, and the remaining clean tags were designed as unambiguous tags. For gene expression analysis, the number of unambiguous clean tags for each gene was calculated and then normalized to the number of transcripts per million tags (TPM). The gene ontology (GO) classification system was used to determine the possible functions of all genes tagged.
Analysis of differential gene expression.
A rigorous algorithm was developed to identify differentially expressed genes (DEGs) between the nonviruliferous and viruliferous whiteflies, referring to the method described previously (2). False discovery rate (FDR) was used to determine the threshold of P value in multiple tests and analysis. We used an FDR of <0.001 and the absolute value of log2 ratio ≥1 as the threshold to judge the significance of the gene expression difference. The correlation between two libraries was statistically assessed by calculation of the Pearson correlation coefficient.
Pathway analysis.
Different genes usually cooperate with each other to exercise their biological functions. Pathway-based analysis helps in further understanding the biological functions of genes. All differentially expressed genes were mapped to terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and we looked for significantly enriched metabolic pathways or signal transduction pathways in DEGs using the same calculating formula as in GO analysis and the threshold of a Q value of ≤0.05.
qPCR analysis.
To confirm the results of the DGE analyses, the expression of 38 selected genes was measured using qPCR. cDNA was synthesized using the SYBR PrimeScript reverse transcription-PCR (RT-PCR) kit II (Takara). qPCRs were carried out on the ABI Prism 7500 fast real-time PCR system (Applied Biosystems) with SYBR green detection. Each gene was analyzed in triplicate, after which the average threshold cycle (CT) was calculated per sample. The relative expression levels were calculated using the 2−ΔΔCT method. As an endogenous control, the expression of β-actin was measured in parallel. In this study, both the Illumina sequencing and qPCR assay showed that the transcript of the β-actin gene is at the same level in both the viruliferous and nonviruliferous whiteflies (data not shown), indicating that the expression of the β-actin gene was unaffected by TYLCCNV.
Lysotracker assay.
The nonviruliferous and viruliferous female adults of whiteflies feeding on cotton plants for 120 h were collected as described above. Ovarioles of ovaries and fat bodies from the viruliferous and nonviruliferous whiteflies were dissected in PBS as described before (25, 33) and then incubated for 3 min in 100 μM LysoTracker Red DND-99 (Molecular Probes) and 2 μM Hoechst 33342 (Sigma) in PBS. Ovarioles and fat bodies were transferred to PBS on glass slides, covered, and immediately photographed on a confocal microscope (Leica). Three female adults were imaged for each of three independent experiments.
Western blotting.
When autophagy is induced, the conversion of protein Atg8-I to Atg8-II increases and the amount of Atg8-II correlates with the number of autophagosomes and can be monitored by immunoblotting (67). The nonviruliferous and viruliferous female adults of whiteflies were collected, respectively, from cotton plants at 0 h and 120 h as described above. Proteins in each sample were extracted, separated by 15% SDS-PAGE, and then blotted with Atg8 antibody. Antibodies were obtained from the following sources: anti-Atg8 from Abgent and anti-actin from Santa Cruz Biotechnology.
Detection of TYLCCNV DNA in organs and whole body of the viruliferous whitefly.
Viruliferous whiteflies were obtained through feeding whiteflies on virus-infected tobacco plants for 24 h. The viruliferous whiteflies were transferred to cotton plants as described. After 0 h and 120 h of feeding on cotton plants, female viruliferous whiteflies were collected. Ovaries and fat bodies from individual whiteflies were dissected in PBS, flushed four times with sterile double-distilled water, and collected according to the methods described before (23, 25, 33, 53). To prevent cross-contamination, we used new needles for each dissection. Nucleic acids from ovaries and fat bodies of 20 individual whiteflies were extracted as described previously (84). DNA was extracted from 20 whitefly individuals similarly. TYLCCNV DNA in these samples of individual whiteflies was detected using the primers and PCR procedures as described previously (57).
Microarray data accession number.
DGE library data sets obtained in this work are available at the NCBI Gene Expression Omnibus (GEO) under the accession number GSE24374.
RESULTS AND DISCUSSION
Basic quantitative parameters of DGE library sequencing.
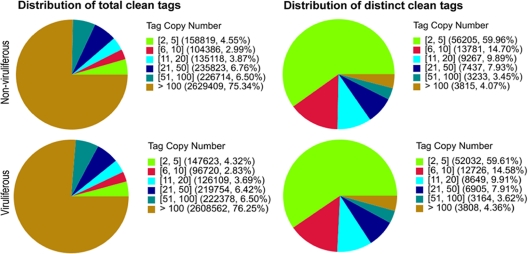
We sequenced two DGE libraries: the nonviruliferous and viruliferous whiteflies, which generated about 3.6 million raw tags for each sample (Table 1). After removal of the low-quality reads, the total number of tags per library ranged from 3.4 million to 3.5 million and the number of distinct tags ranged from 87,284 to 93,738. Since the copy number of a tag reflects the mRNA expression level, the distribution of tag expression was used to evaluate the normality of the DGE data (74). As shown in Fig. 1, the distribution of total tags and distinct tags over different tag abundance categories showed similar patterns for both DGE libraries, suggesting there was no bias in the construction of the libraries from the nonviruliferous and viruliferous whiteflies. However, under the distribution of total tags, high-expression tags with copy numbers larger than 100 were absolutely dominant whereas low-expression tags with copy numbers smaller than 5 occupied the majority of distinct tag distributions (Fig. 1), suggesting that while the majority of mRNA is expressed at low levels, a small proportion of mRNA is highly expressed.
TABLE 1.
Statistics of DGE sequencing
| Category | Parameter | Value for whitefly library |
|
|---|---|---|---|
| Nonviruliferous | Viruliferous | ||
| Raw tag | Total no. of tags | 3,665,554 | 3,581,661 |
| No. of distinct tags | 265,909 | 244,798 | |
| Clean tag | Total no. of tags | 3,490,269 | 3,421,146 |
| No. of distinct tags | 93,738 | 87,284 | |
| All tags mapping to gene | No. of distinct tags | 32,984 | 31,429 |
| Distinct tag % of clean tags | 35.19 | 36.01 | |
| All tag-mapped genes | No. of genes | 13,811 | 13,378 |
| % of ref. genesa | 39.56 | 38.32 | |
| Unambiguous tag- | No. of genes | 13,791 | 13,355 |
| mapped genes | % of ref. genes | 39.50 | 38.25 |
ref., reference.
FIG. 1.
Distribution of total tags and distinct tags over different tag abundance categories. Numbers in square brackets indicate the range of copy numbers for a specific category of tags. For example, “[2, 5]” means all the tags in this category have 2 to 5 copies. Numbers in parentheses show the total tag copy number for all the tags in that category.
Mapping tags to the reference transcriptome database.
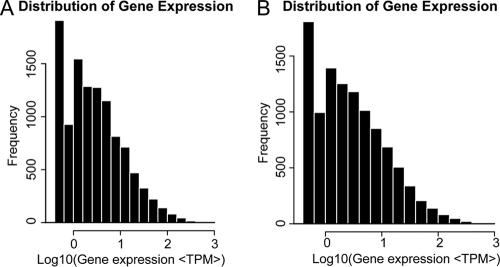
To reveal the molecular events behind DGE profiles, we mapped the tag sequences of these two DGE libraries to the transcriptome reference database of the MEAM1 and Mediterranean species of whitefly. This reference database contains 34,914 distinct sequences (80; unpublished data). Among the 87,284 to 93,738 distinct tags generated from the Illumina sequencing of the two libraries, 35% to 36% of distinct tags were mapped to a gene in the reference database and up to 39.50% (13,791) of the sequences in our transcriptome reference tag database could be unambiguously identified by unique tags (Table 1). To check whether the number of detected genes continued to increase as the total tag number increased, saturation analysis was performed. Results showed that when the total tag number reached 2 million or higher, the number of detected genes almost ceased to increase (data not shown). Tags mapped to a unique sequence are the most critical subset of the DGE libraries since the level of gene expression can be determined by calculating the number of unambiguous tags for each gene and then normalizing to the number of transcripts per million tags. Results show that mRNA transcribed from the major kinds of genes is represented in fewer than 10 copies and only a small proportion of genes are highly expressed (Fig. 2).
FIG. 2.
The level of gene expression. The level of gene expression was determined by calculating the number of unambiguous tags for each gene and then normalizing to the number of transcripts per million tags (TPM). (A) The level of gene expression of the nonviruliferous whiteflies. (B) The level of gene expression of the viruliferous whiteflies.
GO analysis.
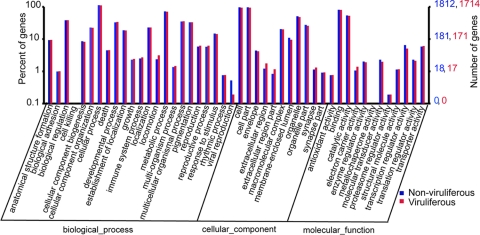
To investigate whether TYLCCNV affects the patterns of gene expression, we analyzed the number of expressed genes in a specific GO category for the nonviruliferous and viruliferous whiteflies. GO analysis showed that the distributions of gene functions for cDNA sequences from the nonviruliferous and viruliferous whiteflies were similar (Fig. 3). This result indicates that the number of genes expressed in each GO category was not significantly affected by TYLCCNV. For both libraries, the biological processes most represented are cellular process, whereas binding and catalytic activity are among the most represented molecular function categories. Interestingly, for the viruliferous whiteflies, locomotion and electron carrier activity were upregulated while viral reproduction and transcription regulator activity were significantly downregulated (Fig. 3; see Table S1 in the supplemental material).
FIG. 3.
Histogram presentations of GO classification of putative functions of genes from nonviruliferous and viruliferous whiteflies. The functions of genes identified cover three main categories: biological process, cellular component, and molecular function. The right y axis indicates the number of genes in a category. The left y axis indicates the percentage of a specific category of genes in that main category. GO analysis showed that the distributions of gene functions for the nonviruliferous and viruliferous whiteflies are similar.
Differentially expressed genes.
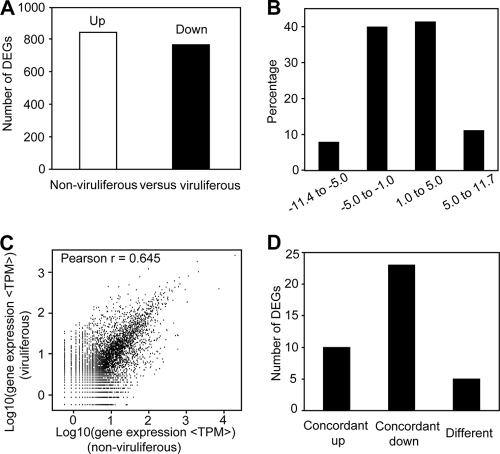
To identify genes showing significant changes in expression, we analyzed the differentially expressed tags between the nonviruliferous and viruliferous whiteflies. In total, 9,968 significantly changed tags were found between these two samples (see Table S2 in the supplemental material). They were mapped to 1,606 genes, with 840 genes upregulated and 766 genes downregulated in the viruliferous whiteflies (Fig. 4A; see also Table S3). The detected fold changes (log2 ratio) of gene expression ranged from −11.4 to 11.7, and more than 80% of the genes (1,303) were up- or downregulated between 1.0- and 5.0-fold (Fig. 4B). Correlation analysis of the two libraries reflects the extent to which the whitefly is affected by the TYLCCNV. The Pearson correlation coefficient of the two libraries was low (0.645), suggesting the large effect of virus on the gene expression profile of whitefly (Fig. 4C).
FIG. 4.
Analysis of differentially expressed genes between two libraries. (A) Summary of the numbers of differentially expressed genes in the Tomato yellow leaf curl China virus viruliferous whiteflies. “FDR < 0.001 and the absolute value of log2 ratio ≥ 1” were used as the threshold to judge the significance of gene expression difference. (B) Fold change distribution of differentially expressed genes. (C) Correlation analysis of two libraries. The Pearson correlation coefficient for two libraries is shown in the upper left corner of the plot. (D) Comparison of DGE data and qPCR results. “Concordant up” means that genes in the viruliferous whiteflies were upregulated for both DGE and qPCR analyses. “Concordant down” means that genes in the viruliferous whiteflies were downregulated for both DGE and qPCR analyses. “Different” means that, for DGE and qPCR analyses, the direction of change of gene expression in the viruliferous whiteflies was contrary.
To validate the DGE data, we compared the gene expression profiles of the nonviruliferous and viruliferous whiteflies using qPCR. Out of the 38 genes selected, 33 demonstrated a concordant direction of change for both DGE and qPCR (Fig. 4D). Moreover, the fold changes obtained by DGE were generally more extreme than those obtained by qPCR (see Tables S4 and S5 in the supplemental material). Similar results have been seen during the analysis of gene expression profile in mice using DGE and qPCR (74). The partial inconsistency may be caused by the lower sensitivity of qPCR than of DGE. When great similarity exists between two samples, the resulting small differences in gene expression are difficult to pick up with qPCR assays (74). Nevertheless, qPCR analysis confirmed the direction of change detected by DGE analysis, showing that DGE results are reliable (26, 74). Overall, our data demonstrate that the DGE technique can be used for gene expression analysis in an organism without genome information.
Perturbance of the cell cycle and primary metabolism in the whitefly by TYLCCNV.
Among the 1,606 differentially expressed genes, 298 genes were mapped to 157 pathways in the KEGG database. The pathway enrichment analysis showed that the proteasome pathway was significantly enriched in DEGs. For this pathway, 4 genes were upregulated and 7 genes downregulated (Table 2). Proteasomes can degrade protein substrates marked for degradation by the attachment of a ubiquitin moiety, forming the ubiquitin-proteasome pathway (3). Interestingly, we also found that 5 genes were upregulated and 7 genes downregulated in the ubiquitin-mediated proteolysis pathway (Table 2). The ubiquitin-proteasome pathway is involved in the regulation of the cell cycle, metabolic adaptation, and the immune response through degrading numerous short-lived proteins and regulatory proteins that control these cellular functions (3, 39, 50, 56, 58). Indeed, all of these pathways were significantly regulated in the viruliferous whitefly (see Table S6 in the supplemental material).
TABLE 2.
Expression profiles of genes involved in the ubiquitin-proteasome pathway in viruliferous whiteflies
| Category or gene ID | Homologous functiona | Species | Accession no. | FCb |
|---|---|---|---|---|
| Proteasome pathway | ||||
| BT_B_ZJU_Singletons96556 | Proteasome inhibitor | Nasonia vitripennis | XP_001602913.1 | 5.4 |
| BT_B_ZJU_Singletons100154 | Proteasome beta subunit | Tribolium castaneum | XP_968855.1 | 2.6 |
| BT_B_ZJU_Singletons98051 | Proteasome regulatory subunit 6B | Acyrthosiphon pisum | XP_001945593.1 | 2.5 |
| BT_B_ZJU_Singletons103983 | Proteasome regulatory complex component | Acyrthosiphon pisum | XP_001945112.1 | 3.7 |
| BT_Q_ZJU_Singletons8263 | D7-like protein | Perca flavescens | AF085252.1 | −1.8 |
| BT_B_ZJU_Singletons102473 | Proteasome alpha 4 subunit | Nasonia vitripennis | XP_001600177.1 | −2.6 |
| BT_B_ZJU_Singletons97160 | Proteasome beta 2 subunit-like protein | Nasonia vitripennis | XP_001600736.1 | −1.6 |
| BT_B_ZJU_Singletons96891 | Proteasome beta 5,8 subunit | Tribolium castaneum | XP_970194.1 | −1.3 |
| BT_B_ZJU_Singletons99343 | Proteasome activator pa28 beta subunit | Apis mellifera | XP_393883.2 | −2.0 |
| BT_B_ZJU_Singletons98630 | Proteasome regulatory subunit 6B | Pediculus humanus corporis | XP_002426965.1 | −8.8 |
| BT_B_ZJU_Singletons103025 | Proteasome non-ATPase regulatory subunit | Pediculus humanus corporis | XP_002432284.1 | −3.7 |
| Ubiquitin-mediated proteolysis pathway | ||||
| BT_Q_ZJU_Singletons27523 | Mdm2 | Acyrthosiphon pisum | XP_001948790.1 | 8.7 |
| BT_B_ZJU_Singletons100256 | Cytochrome B5 | Culicoides sonorensis | AAV84214.1 | 9.6 |
| BT_B_ZJU_Singletons26956 | Regulator of chromosome condensation | Pediculus humanus corporis | XP_002424836.1 | 1.2 |
| BT_B_ZJU_Singletons25427 | Ubiquitin-conjugating enzyme E2 | Drosophila grimshawi | XP_001990045.1 | 3.2 |
| BT_B_ZJU_Singletons98657 | Ubiquitin-conjugating enzyme E2 L3 | Pediculus humanus corporis | XP_002426196.1 | 1.6 |
| BT_B_ZJU_Singletons755 | Cdc20/fizzy1 | Tribolium castaneum | XP_970475.1 | −2.1 |
| BT_Q_ZJU_Singletons9609 | Cdc20/fizzy1 | Aedes aegypti | XM_001649172.1 | −8.8 |
| BT_B_ZJU_Singletons103092 | Regulator of chromosome condensation | Acyrthosiphon pisum | XM_001951632.1 | −2.9 |
| BT_B_ZJU_Singletons24867 | TRAF-type zinc finger containing 1 | Xenopus tropicalis | XP_001943669.1 | −3.4 |
| BT_B_ZJU_Singletons94263 | Anaphase-promoting complex subunit | Pediculus humanus corporis | XP_002431494.1 | −3.0 |
| BT_B_ZJU_Singletons25119 | Ubiquitin-activating enzyme 1 | Apis mellifera | XP_394434.2 | −2.6 |
| BT_B_ZJU_Singletons101831 | Ubiquitin conjugation factor E4 B | Pediculus humanus corporis | XP_002426709.1 | −3.3 |
The function of the homologous gene.
FC, fold change (log2 ratio) of gene expression.
For the cell cycle, most genes involved in the G1, S, and G2 phase were upregulated while most genes involved in the M phase were downregulated in the viruliferous whiteflies (Fig. 5A; see Table S6 in the supplemental material). We also noticed that one gene encoding cyclin B protein was downregulated. Cyclin B is a cell cycle regulatory protein, and its degradation by the ubiquitin-proteasome system is necessary for exit from telophase and entry into the interphase (G1, S, and G2 phases) of the next cell cycle (56). Thus, the regulation in the cell cycle interphase for the viruliferous whiteflies may be caused through degradation of the cyclin B protein by the ubiquitin-proteasome pathway. Perturbation of the cell cycle has been shown to be caused by many animal and plant viruses (1, 29, 36, 55). For example, Cabbage leaf curl virus, another member of the Begomovirus genus, can activate genes expressed during S and G2 phases and inhibit genes active in G1 and M phases of Arabidopsis (1). Beet severe curly top virus, in the same family, Geminiviridae, as TYLCCNV, can induce a RING finger ubiquitin E3 ligase, which is able to interact with cell cycle inhibitor ICK/KRP proteins and then regulate the plant cell cycle (36). The perturbation of the cell cycle caused by virus infection has been considered to be a unique strategy for the creation of a favorable intracellular environment to acquire maximized replication of viral DNA (29, 55). Therefore, cell cycle regulation by TYLCCNV might be required for its possible replication or circulation within the vector as well.
FIG. 5.
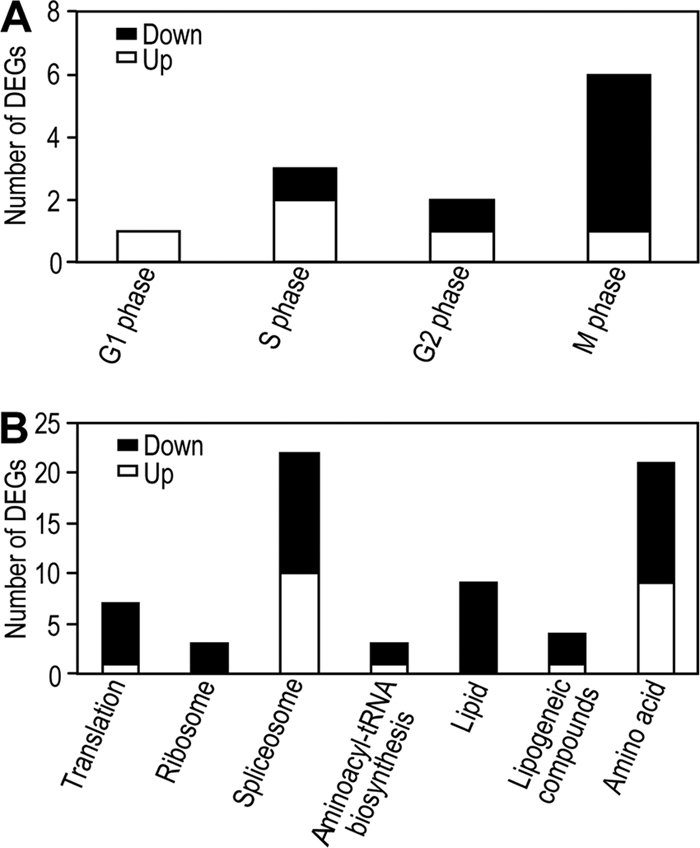
Regulated genes involved in the cell cycle and primary metabolism in the viruliferous whiteflies. (A) Regulated genes related to the cell cycle in viruliferous whiteflies. (B) Regulated genes involved in primary metabolism in viruliferous whiteflies.
For the primary metabolism, 22 genes were upregulated and 47 genes downregulated in the viruliferous whiteflies (Fig. 5B; see also Table S6 in the supplemental material). Interestingly, most genes involved in translation, ribosome, spliceosome, aminoacyl-tRNA biosynthesis, lipid metabolism, lipogeneic compound metabolism, and amino acid metabolism were downregulated in the viruliferous whiteflies, indicating that the protein synthesis and lipid and amino acid metabolism of the viruliferous whitefly were inhibited by TYLCCNV (Fig. 5B). Other virus infections also affect the protein production of insects. For example, polydnavirus infection inhibited translation of specific growth-associated host proteins, as well as synthesis of an insect plasma protein, arylphorin, and baculovirus infection caused the shutdown of protein synthesis in insect cells (18, 64, 65).
Taken together, the cell cycle and primary metabolism of whitefly can be perturbed by TYLCCNV, and the perturbation is probably caused by the invasion of insect tissues by virus. TYLCCNV can be transovarially transmitted by its vector MEAM1, demonstrating that it has invaded the reproductive system of the viruliferous whitefly (77). The perturbation of the cell cycle and primary metabolism in whitefly, combined with the invasion of insect tissues, might result in the reduced longevity and fecundity of viruliferous whiteflies on cotton plants (31). Our data also support the suggestion of other studies that Tomato yellow leaf curl virus (TYLCV) has several features of an insect pathogen (12, 14, 60). Besides TYLCCNV and TYLCV, a few other plant viruses have also been shown to exert cytological and metabolic changes in vector insects (42). The pathological effects upon their insect vectors of persistently transmitted plant viruses seem to be a common phenomenon, which probably induces an immune response in the vector insects. However, the role of the insect immune system in insect vector-plant virus interactions has received limited attention (24, 45).
Activation of the cellular and humoral immune responses of the whitefly by TYLCCNV.
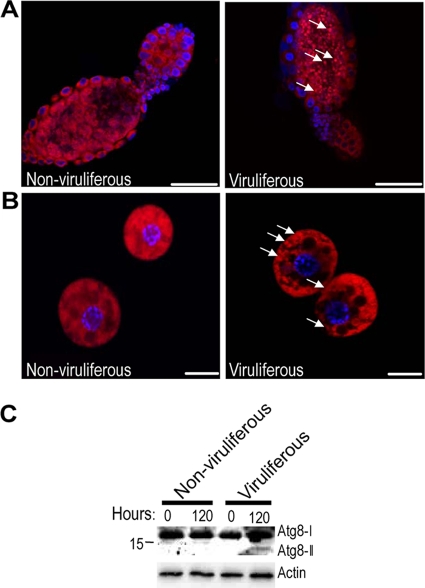
Cellular and humoral responses are the major effector response systems used by insects against microbial infection (8, 30, 44, 82). Interestingly, among the differentially expressed genes in the viruliferous whiteflies, many genes related to cellular and humoral immune response are upregulated, especially the genes in the autophagy-lysosome pathway (Table 3). Autophagy is an evolutionarily conserved intracellular process in which a bulk of the cytoplasm is enveloped inside a double-membraned vesicle and shuttled to the lysosome for degradation (35, 47). Autophagy can be utilized by the immune cells to combat viral infections (19, 35, 67, 68), and it has been shown to play a direct antiviral role against the mammalian viral pathogen Vesicular stomatitis virus in the fruit fly (67). For the viruliferous whiteflies, all genes involved in autophagy and most genes associated with lysosome function were significantly upregulated (Table 3; see Table S6 in the supplemental material), strongly suggesting that this pathway is activated in the viruliferous whitefly. To further demonstrate that TYLCCNV induces autophagy in vivo, we monitored autophagy in the viruliferous whitefly with Lysotracker staining and Western blotting. These two methods also have been used in monitoring autophagy in fruit flies (28, 61, 67).
TABLE 3.
Upregulation of genes involved in cellular and humoral immune responses in viruliferous whiteflies
| Category or gene IDa | Homologous functionb | Species | Accession no. | FCc |
|---|---|---|---|---|
| Autophagy | ||||
| BT_Q_ZJU_Singletons163362 | Atg3 | Tribolium castaneum | XP_967286.2 | 4.2 |
| BT_Q_ZJU_Singletons11737 | Atg9 | Apis mellifera | XP_395581.3 | 9.0 |
| BT_B_ZJU_Singletons96409 | Atg12 | Bombyx mori | NM_001142491.1 | 3.8 |
| Lysosome | ||||
| BT_B_ZJU_Singletons103389 | AP-1 | Pediculus humanus corporis | XP_002423855.1 | 3.8 |
| BT_B_ZJU_Singletons26635 | Cathepsin B | Acyrthosiphon pisum | NP_001119613.2 | 1.8 |
| BT_Q_ZJU_Singletons14336 | Iduronate 2-sulfatase | Tribolium castaneum | XP_967324.1 | 10.0 |
| BT_B_ZJU_Singletons16195 | Vacuolar ATP synthase subunit S1 | Pediculus humanus corporis | XM_002429902.1 | 9.3 |
| BT_B_ZJU_Singletons99868 | Protein tyrosine phosphtase | Tribolium castaneum | XP_968930.1 | 1.3 |
| BT_B_ZJU_Singletons102872 | Saposin | Tribolium castaneum | XP_966852.1 | −1.4 |
| BT_B_ZJU_Singletons102637 | Cathepsin D | Armigeres subalbatus | EU205672.1 | −2.6 |
| BT_B_ZJU_Singletons23076 | Phosphatidylcholine acyltransferase | Tribolium castaneum | XP_966553.1 | −2.5 |
| Complement and coagulation | ||||
| BT_B_ZJU_Singletons104106 | Papilin | Apis mellifera | XP_396472.3 | 3.7 |
| Antimicrobial peptide | ||||
| BT_B_ZJU_Singletons20384 | Antimicrobial knottin protein Btk-3 | Bemisia tabaci | ABC40571.1 | 3.7 |
| Melanization | ||||
| BT_B_ZJU_Singletons23021 | Dopa decarboxylase | Acyrthosiphon pisum | XP_001950143.1 | 9.3 |
Cellular response includes autophagy and lysosomes. Humoral response includes antimicrobial peptides, melanization, complement, and coagulation.
The function of the homologous gene.
FC, fold change (log2 ratio) of gene expression.
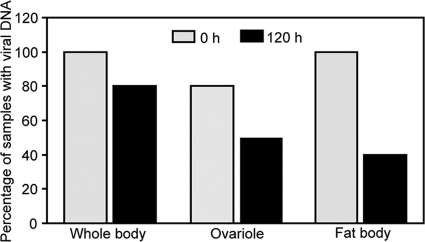
In the nonviruliferous whiteflies, Lysotracker staining is weak and diffuse. In contrast, both the ovariole and fat body from the viruliferous whiteflies display intense, punctate lysotracker staining (Fig. 6A and B). These results indicate that TYLCCNV can induce autophagy in the ovary and fat body tissues of the whitefly. Atg8-II is an autophagosome-specific protein marker upon autophagy induction. We monitored the production of Atg-II for the viruliferous whiteflies at different time points. For both the nonviruliferous and viruliferous whiteflies freshly transferred onto cotton plants (0 h), no Atg8-II protein was detected; however, the Atg8-II protein was induced in the viruliferous whiteflies after 120 h (Fig. 6C), indicating that autophagy can occur only after TYLCCNV has reached inside the whiteflies for some time. To investigate whether TYLCCNV invades the ovary and fat body tissues of the whitefly, we examined the presence of viral DNA in these tissues by PCR. As shown in Fig. 7, TYLCCNV DNA can be detected in ovaries from 80% of adult whiteflies and fat bodies from 100% of adult whiteflies after a 24-h AAP on virus-infected tobacco plants. Interestingly, after 120 h of feeding on cotton, ovaries from only 50% of adult whiteflies and fat bodies from 40% of adult whiteflies carried viral DNA (Fig. 7). These results mean that TYLCCNV can invade the ovary and fat body tissues of whitefly and induce autophagy, which probably leads to the gradual decrease of the virions.
FIG. 6.
Detection of autophagy in adult whiteflies. Ovariole of ovary (A) and fat body (B) tissues stained with the lysosome-specific fluorescent dye Lysotracker Red (red) and the nuclear dye Hoechst 33342 (blue). Compared with the tissues from the nonviruliferous whitefly (left), there is increased punctate Lysotracker staining (white arrows) in the viruliferous whiteflies (right). Scale bar in panels A and B, 50 μm and 10 μm, respectively. Similar findings were observed in three independent experiments. (C) Western blotting of the Atg8 protein. Arabic numbers above each lane indicate the time points (in hours) after the nonviruliferous and viruliferous whiteflies were transferred to healthy cotton plants. A molecular size markers (in kilodaltons) is shown on the left of the panel. Atg8-I (∼16 kDa) is observed in all whiteflies, and Atg8-II (∼14 kDa) is induced only in the viruliferous whiteflies feeding on cotton plants for 120 h.
FIG. 7.
Percentages of viruliferous whiteflies (obtained by feeding for 24 h on infected tobacco plants) with detectable TYLCCNV DNA in the whole body, ovaries, and fat bodies after 0 h and 120 h of feeding on healthy cotton plants.
Antimicrobial peptide production is the major humoral reaction of insects. The gene encoding a putative antimicrobial knottin protein, Btk-3, was found to be upregulated in the viruliferous whitefly (Table 3). The Btk-1 and Btk-3 proteins were reported to be more abundant in cDNA libraries from the Tomato mottle virus and Tomato yellow leaf curl virus viruliferous MEAM1 whiteflies through comparison of the expressed sequence tag (EST) sequences (38, 63). Therefore, the observed activation of genes encoding the Btk-3 protein in our study is consistent with previous reports on the interactions between different begomoviruses and whiteflies using other technologies. This agreement seems to demonstrate the reliability of results obtained using the Illumina sequencing method.
Apart from the upregulation of most genes involved in autophagy and antimicrobial peptide production, the genes involved in the humoral response, such as complement-, coagulation-, and melanization-related genes, were also upregulated (Table 3). This suggests that TYLCCNV can activate both the humoral and cellular immune responses of whiteflies, which may result in the degradation of virions and explains the decreased proportion of viruliferous whiteflies on healthy cotton plants (Fig. 7). Thus, the activation of an immune response is probably the evolved strategy of the whitefly to protect itself from the deleterious effect of begomovirus. Nevertheless, the decreased proportion of viruliferous whiteflies may also be caused by secretion of the virions into the phloem of plants during the feeding of viruliferous whiteflies on cotton plants. Both of these factors could contribute to the decreased proportion of viruliferous whiteflies.
Downregulation of genes involved in signal transduction of the immune response and apoptosis.
Viruses can also evade or antagonize the host immune response through a complex combination of such processes as signaling interference, effector modulation, and continual viral genetic variation, which supports persistent infection and spread of the virus (4, 21, 22, 59). Interestingly, in the viruliferous whitefly, a number of genes involved in the Toll-like signaling, mitogen-activated protein kinase (MAPK), RIG-I-like receptor, Notch, and transforming growth factor beta (TGF-β) pathways were downregulated, suggesting that the TYLCCNV might interfere with the signaling transduction of the whitefly. The Toll pathway plays important roles in the antiviral responses of insects (83, 85). In thrips, the Toll pathway can be activated by Tomato spotted wilt virus infection (45). Surprisingly, in viruliferous whiteflies, genes involved in the Toll-like signaling pathway were downregulated, indicating that begomovirus may suppress this pathway (Table 4). Several studies have implicated the involvement of MAPKs in medfly and mosquito innate immune responses (37, 72), and the activation of MAPK pathways is required for efficient infection by baculoviruses (34). Here we found that most genes involved in the MAPK pathway were downregulated (Table 4).
TABLE 4.
Downregulation of genes involved in signal transduction and apoptosis in viruliferous whiteflies
| Category or gene ID | Homologous functiona | Species | Accession no. | FCb |
|---|---|---|---|---|
| Toll-like signaling pathway | ||||
| BT_B_ZJU_Singletons96892 | Transmembrane trafficking protein | Tribolium castaneum | XP_971914.1 | −9.5 |
| BT_B_ZJU_Singletons24867 | TRAF-type zinc finger containing 1 | Xenopus tropicalis | XP_001943669.1 | −3.4 |
| BT_B_ZJU_Singletons101328 | Leucine-rich transmembrane protein | Pediculus humanus corporis | XP_002422869.1 | −1.3 |
| MAPK pathway | ||||
| BT_B_ZJU_Singletons103594 | MAPKK4 | Nasonia vitripennis | XP_001603456.1 | 1.9 |
| BT_B_ZJU_Singletons101082 | MAPKK1-interacting protein 1 | Apis mellifera | XP_001121654.1 | 2.4 |
| BT_B_ZJU_Singletons102809 | Calcium-binding protein p22 | Maconellicoccus hirsutus | ABM55600.1 | −9.5 |
| BT_B_ZJU_Singletons99736 | MAPK activating protein | Apis mellifera | XP_624118.1 | −4.4 |
| BT_B_ZJU_Singletons98676 | MAP-binding protein-interacting protein | Nasonia vitripennis | XP_001605487.1 | −2.4 |
| RIG-I-like receptor pathway | ||||
| BT_B_ZJU_Singletons103928 | DEAD box ATP-dependent RNA helicase | Acyrthosiphon pisum | XP_001951890.1 | −1.7 |
| Notch pathway | ||||
| BT_B_ZJU_Singletons26945 | Deltex | Pediculus humanus corporis | XM_002426000.1 | −2.4 |
| TGF-beta pathway | ||||
| BT_B_ZJU_Singletons101016 | DNA-binding protein inhibitor | Ixodes scapularis | XM_002414872.1 | −2.9 |
| Apoptosis | ||||
| BT_Q_ZJU_Singletons13955 | Apoptosis inhibitor | Pediculus humanus corporis | XP_002423394.1 | 8.9 |
| BT_B_ZJU_Singletons11470 | Programmed cell death protein 11 | Acyrthosiphon pisum | XP_001951020.1 | 3.1 |
| BT_Q_ZJU_Singletons27465 | Cyclin B | Nasonia vitripennis | NP_001154932.1 | −3.3 |
| BT_B_ZJU_Singletons103804 | Cyclin K | Pediculus humanus corporis | XP_002428532.1 | −3.0 |
| BT_B_ZJU_Singletons102809 | Calcium-binding protein p22 | Maconellicoccus hirsutus | ABM55600.1 | −9.5 |
| BT_B_ZJU_Singletons97698 | Programmed cell death protein 2 | Nasonia vitripennis | XP_001603635.1 | −2.5 |
| BT_B_ZJU_Singletons100224 | Proliferating cell nuclear antigen | Fenneropenaeus chinensis | ABM66815.1 | −3.1 |
The function of the homologous gene.
FC, fold change (log2 ratio) of gene expression.
Apoptosis is another powerful effector response to virus infection and can reduce viral replication, infectivity, and spread within the insect host (9, 73). However, to date apoptosis is largely unexplored in insects (9). For the viruliferous whiteflies, the 5 of 7 genes involved in the apoptosis pathway were downregulated (Table 4). Among the two upregulated genes, one has the function of putative apoptosis inhibitor. Therefore, the begomovirus seems to inhibit the apoptosis pathway of whitefly to ensure its replication and spread.
The suppression of the host immune system is common in the interaction and coevolution between endoparasites and their insect hosts, and this suppression of insect immunity is mediated by polydnaviruses symbiotically associated with parasitoid wasps (66, 70, 75). However, plant virus immune evasion in invertebrate hosts is not well studied (24). To our knowledge, this is the first report that plant virus can repress the immune response of a vector insect. This suppression may favor plant virus persistence in the vector as the result of long-term coadaption and coevolution of plant viruses and vector insects.
Because of the high expense of Illumina sequencing, only one biological replicate at one time point was examined. To confirm the Illumina sequencing results, we did qPCR analysis for 38 specific genes from a second biological replicate. This does provide some confirmatory information for the reproducibility of this study. Hopefully, as the cost of Illumina sequencing continues to drop, future experiments can investigate multiple biological replicates. The selection of a time point for sample collection depends on a trade-off between reducing the secondary effects from infected plants and having a high percentage of viruliferous whiteflies. A longer period of feeding (e.g., 10 days) on cotton plants will decrease the percentage of viruliferous whiteflies to 40% (32). However, a shorter feeding time on cotton plants (e.g., 2 days) might not be enough to eliminate the residual effects of infected plants on whiteflies. A similar method has been used to detect the gene expression of viruliferous whiteflies regulated by TYLCV (14). In that study, whiteflies that had access to infected tomato plants for 48 h were subsequently reared on eggplants. Five days later, whiteflies were collected for RNA extraction and cloning of differentially expressed genes using a PCR differential display technique (14). The 120 h of feeding on cotton plant may reduce the chance of finding genes responsive to the virus at an early time point, but this time frame would allow detection of insect responses when the virus has invaded various tissues and triggered sustained reactions in the whitefly. Indeed, autophagy in ovary and fat body tissues could not be detected in the viruliferous whiteflies just after a 24-h AAP; however, it was induced in the viruliferous whiteflies after 120 h of feeding on cotton (Fig. 6).
Recently, the transcriptional response of aphid intestine to luteoviruses has been reported. In this study, the viruliferous aphids were obtained through feeding on the virus-infected pea plants, and a comparative transcriptomic analysis was conducted using a cDNA microarray. A total of 128 genes were significantly regulated (5). Compared to our study, the relatively low number of regulated genes obtained in that study was caused by many factors. First, their array represents only about 20% of genes present in the pea aphid genome. Second, the luteovirus used has no negative effects on the longevity and fecundity of the aphid. Third, this study investigated the transcriptional response of the intestine, not the whole body of the aphid, to luteoviruses; therefore, it cannot detect differentially expressed genes in other tissues. Furthermore, we examined the transcriptional response of the whole body of whiteflies using Illumina sequencing technology, which is more sensitive than microarray snalysis. Nonetheless, both studies have provided a great deal of discovery data for the response of insect vectors to circulatively transmitted plant viruses that can now be used to better target specific genes and pathways for functional studies.
In summary, we present, for the first time, an analysis of the global transcriptional response of whiteflies to begomoviruses using high-throughput sequencing. Our data show that TYLCCNV can perturb the cell cycle and primary metabolism of whiteflies. This virus can activate the humoral and cellular immune responses of vector insects, especially the autophagy pathway. It can also suppress the immune response of the whitefly by interfering with the Toll-like and MAPK signaling pathways for its persistence. Overall, our results reveal the relationship of coevolved adaptations between begomoviruses and whiteflies. This study will provide a road map for future investigations of these fascinating interactions between plant viruses and vector insects and may offer hints for the development of novel antiviral strategies in the future.
Supplementary Material
Acknowledgments
We thank Yun-Lin Su, Xiao-Li Bing, Dan-Mei Yao, and Jin-Hua Liu for assistance in collecting the whitefly samples and Xue-Ping Zhou, Institute of Biotechnology, Zhejiang University, for providing clones of TYLCCNV inocula.
This work was supported by the National Natural Science Foundation of China (30730061 and 31021003), the National Basic Research Program of China (2009CB119203), and the earmarked fund for the Modern Agro-industry Technology Research System in China.
Footnotes
Published ahead of print on 26 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ascencio-Ibañez, J. T., et al. 2008. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting the pathogen response and cell cycle controls during geminivirus infection. Plant Physiol. 148:436-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audic, S., and J. M. Claverie. 1997. The significance of digital gene expression profiles. Genome Res. 7:986-995. [DOI] [PubMed] [Google Scholar]
- 3.Bochtler, M., L. Ditzel, M. Groll, C. Hartmann, and R. Huber. 1999. The proteasome. Annu. Rev. Biophys. Biomol. Struct. 28:295-317. [DOI] [PubMed] [Google Scholar]
- 4.Bowie, A. G., and L. Unterholzner. 2008. Viral evasion and subversion of pattern-recognition receptor signaling. Nature 8:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brault, V., et al. 2010. Transcriptomic analysis of intestinal genes following acquisition of pea enation mosaic virus by the pea aphid Acyrthosiphon pisum. J. Gen. Virol. 91:802-808. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. K., D. R. Frohlich, and R. C. Rosell. 1995. The sweetpotato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annu. Rev. Entomol. 40:511-534. [Google Scholar]
- 7.Caciagli, P., et al. 2009. Virion stability is important for the circulative transmission of Tomato yellow leaf curl sardinia virus by Bemisia tabaci, but virion access to salivary glands does not guarantee transmissibility. J. Virol. 83:5784-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christophides, G. K., et al. 2002. Immunity-related genes and gene families in Anopheles Gambiae. Science 298:159-165. [DOI] [PubMed] [Google Scholar]
- 9.Clem, R. J. 2005. The role of apoptosis in defense against baculovirus infection in insects. Curr. Top. Microbiol. Immunol. 289:113-130. [DOI] [PubMed] [Google Scholar]
- 10.Colvin, J., et al. 2006. Host-plant viral infection effects on arthropod-vector population growth, development and behaviour: management and epidemiological implications. Adv. Virus Res. 67:419-452. [DOI] [PubMed] [Google Scholar]
- 11.Cui, X. F., X. R. Tao, Y. Xie, C. M. Fauquet, and X. P. Zhou. 2004. A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78:13966-13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czosnek, H. 2007. Tomato yellow leaf curl virus disease, management, molecular biology, breeding for resistance. Springer, Dordrecht, Netherlands.
- 13.Czosnek, H., M. Ghanim, and M. Ghanim. 2002. The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci—insights from studies with tomato yellow leaf curl virus. Ann. Appl. Biol. 140:215-231. [Google Scholar]
- 14.Czosnek, H., et al. 2001. Whiteflies: vectors and victims (?) of geminiviruses. Adv. Virus Res. 57:291-322. [DOI] [PubMed] [Google Scholar]
- 15.Dalton, R. 2006. The Christmas invasion. Nature 443:898-900. [DOI] [PubMed] [Google Scholar]
- 16.De Barro, P. J., S. S. Liu, L. M. Boykin, and A. B. Dinsdale. 2011. Bemisia tabaci: a statement of species status. Annu. Rev. Entomol. 56:1-19. [DOI] [PubMed] [Google Scholar]
- 17.Dinsdale, A., L. Cook, C. Riginos, Y. M. Buckley, and P. J. De Barro. 2010. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 133:196-208. [Google Scholar]
- 18.Du, X., and S. M. Thiem. 1997. Responses of insect cells to baculovirus infection: protein synthesis shutdown and apoptosis. J. Virol. 71:7866-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espert, L., P. Codogno, and M. Biard-Piechaczyk. 2007. Involvement of autophagy in viral infections: antiviral function and subversion by viruses. J. Mol. Med. 85:811-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fauquet, C. M., and J. Stanley. 2005. Revising the way we conceive and name viruses below the species level: a review of geminivirus taxonomy calls for new standardized isolate descriptors. Arch. Virol. 150:2151-2179. [DOI] [PubMed] [Google Scholar]
- 21.Finlay, B. B., and G. McFadden. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767-782. [DOI] [PubMed] [Google Scholar]
- 22.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 23.Ghanim, M., S. Morin, and H. Czosnek. 2001. Rate of Tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 91:188-196. [DOI] [PubMed] [Google Scholar]
- 24.Gray, S., and F. E. Gildow. 2003. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41:539-566. [DOI] [PubMed] [Google Scholar]
- 25.Guo, J. Y., G. Y. Ye, S. Z. Dong, and S. S. Liu. 2010. An invasive whitefly feeding on a virus-infected plant increased its egg production and realized fecundity. PLoS One 5:e11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegedus, Z., A. Zakrzewska, W. C. Ágoston, et al. 2009. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol. Immunol. 46:2918-2930. [DOI] [PubMed] [Google Scholar]
- 27.Hogenhout, S. A., El-Desouky Ammar, A. E. Whitfield, and M. G. Redinbaugh. 2008. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46:327-359. [DOI] [PubMed] [Google Scholar]
- 28.Hou, Y. C., S. Chittaranjan, S. G. Barbosa, K. McCall, and S. M. Gorski. 2008. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182:1127-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda, M., and M. Kobayashi. 1999. Cell-cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology 258:176-188. [DOI] [PubMed] [Google Scholar]
- 30.Jiravanichpaisal, P., B. L. Lee, and K. Soderhall. 2006. Cell-mediated immunity in arthropods: hematopoiesis, coagulation, melanization and opsonization. Immunobiology 211:213-236. [DOI] [PubMed] [Google Scholar]
- 31.Jiu, M., et al. 2007. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS One 2:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiu, M., X. P. Zhou, and S. S. Liu. 2006. Acquisition and transmission of two begomoviruses by the B and a non-B biotype of Bemisia tabaci from Zhejiang, China. J. Phytopathol. 154:587-591. [Google Scholar]
- 33.Juhasz, G., and T. P. Neufeld. 2008. Experimental control and characterization of autophagy in Drosophila, p. 125-133. In V. Deretic (ed.), Methods in molecular biology, vol. 445: Autophagosome and phagosome. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 34.Katsuma, S., K. Mita, and T. Shimada. 2007. ERK- and JNK-dependent signaling pathways contribute to Bombyx mori nucleopolyhedrovirus infection. J. Virol. 81:13700-13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudchodkar, S. B., and B. Levine. 2009. Viruses and autophagy. Rev. Med. Virol. 19:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai, J. B., et al. 2009. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 57:905-917. [DOI] [PubMed] [Google Scholar]
- 37.Lamprou, I., et al. 2007. Distinct signalling pathways promote phagocytosis of bacteria, latex beads and lipopolysaccharide in medfly haemocytes. Immunology 121:314-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leshkowitz, D., et al. 2006. Whitefly (Bemisia tabaci) genome project: analysis of sequenced clones from egg, instar, and adult (viruliferous and non-viruliferous) cDNA libraries. BMC Genomics 7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, C. Y. 2004. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 22:81-127. [DOI] [PubMed] [Google Scholar]
- 40.Liu, S. S., et al. 2007. Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318:1769-1772. [DOI] [PubMed] [Google Scholar]
- 41.Mansoor, S., R. W. Briddon, Y. Zafar, and J. Stanley. 2003. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8:128-134. [DOI] [PubMed] [Google Scholar]
- 42.Maramorosch, K., and D. D. Jensen. 1963. Harmful and beneficial effects of plant viruses in insects. Annu. Rev. Microbiol. 17:495-530. [DOI] [PubMed] [Google Scholar]
- 43.Marioni, J. C., C. E. Mason, S. M. Mane, M. Stephens, and Y. Gilad. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18:1509-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marmaras, V. J., and M. Lampropoulou. 2009. Regulators and signalling in insect haemocyte immunity. Cell Signal. 21:186-195. [DOI] [PubMed] [Google Scholar]
- 45.Medeiros, R. B., R. D. O. Resende, and A. C. de Avila. 2004. The plant virus Tomato spotted wilt Tospovirus activates the immune system of its main insect vector, Frankliniella occidentalis. J. Virol. 78:4976-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reference deleted.
- 47.Mizushima, N., B. Levine, A. M. Cuervo, and D. J. Klionsky. 2008. Autophagy fights disease through cellular self-digestion. Nature 451:1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffat, A. S. 1999. Geminiviruses emerge as serious crop threat. Science 286:1835. [Google Scholar]
- 49.Morin, S., M. Ghanim, I. Sobol, and H. Czosnek. 2000. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast two-hybrid system. Virology 276:404-416. [DOI] [PubMed] [Google Scholar]
- 50.Mykles, D. L. 1999. Structure and functions of arthropod proteasomes. Mol. Biol. Rep. 26:103-111. [DOI] [PubMed] [Google Scholar]
- 51.Ng, J. C. K., and B. W. Falk. 2006. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44:183-212. [DOI] [PubMed] [Google Scholar]
- 52.Ohnesorge, S., and E. R. Bejarano. 2009. Begomovirus coat protein interacts with a small heatshock protein of its transmission vector (Bemisia tabaci). Insect Mol. Biol. 18:693-703. [DOI] [PubMed] [Google Scholar]
- 53.Ohnishi, J., T. Kitamura, F. Terami, and K. Honda. 2009. A selective barrier in the midgut epithelial cell membrane of the nonvector whitefly Trialeurodes vaporariorum to Tomato yellow leaf curl virus uptake. J. Gen. Plant Pathol. 75:131-139. [Google Scholar]
- 54.Oliveira, M. R. V., T. J. Henneberry, and P. Anderson. 2001. History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot. 20:709-723. [Google Scholar]
- 55.Op De Beeck, A., and P. Caillet-Fauquet. 1997. Viruses and the cell cycle. Prog. Cell Cycle Res. 3:1-19. [DOI] [PubMed] [Google Scholar]
- 56.Pagano, M. 1997. Cell cycle regulation by the ubiquitin pathway. FASEB J. 11:1067-1075. [DOI] [PubMed] [Google Scholar]
- 57.Qian, Y. J., and X. P. Zhou. 2005. Pathogenicity and stability of a truncated DNAβ associated with Tomato yellow leaf curl China virus. Virus Res. 109:159-163. [DOI] [PubMed] [Google Scholar]
- 58.Reinstein, E. 2004. Immunologic aspects of protein degradation by the ubiquitin-proteasome system. Isr. Med. Assoc. J. 6:420-424. [PubMed] [Google Scholar]
- 59.Rowe, M., et al. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U. S. A. 104:3366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubinstein, G., and H. Czosnek. 1997. Long-term association of Tomato yellow leaf curl virus with its whitey vector Bemisia tabaci: effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 78:2683-2689. [DOI] [PubMed] [Google Scholar]
- 61.Scott, R. C., G. Juhász, and T. P. Neufeld. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seal, S. E., F. vandenBosch, and M. J. Jeger. 2006. Factors influencing Begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 25:23-46. [Google Scholar]
- 63.Shatters, R. G., Jr., et al. 2008. A knottin-like putative antimicrobial gene family in the whitefly Bemisia tabaci biotype B: cloning and transcript regulation. Fourth International Bemisia Workshop International Whitefly Genomics Workshop. J. Insect Sci. 8:53. [Google Scholar]
- 64.Shelby, K. S., and B. A. Webb. 1994. Polydnavirus infection inhibits synthesis of an insect plasma protein, Arylphorin. J. Gen. Virol. 75:2285-2292. [DOI] [PubMed] [Google Scholar]
- 65.Shelby, K. S., and B. A. Webb. 1997. Polydnavirus infection inhibits translation of specific growth-associated host proteins. Insect Biochem. Mol. Biol. 27:263-270. [DOI] [PubMed] [Google Scholar]
- 66.Shelby, K. S., and R. A. Webb. 1999. Polydnavirus-mediated suppression of insect immunity. J. Insect Physiol. 45:507-514. [DOI] [PubMed] [Google Scholar]
- 67.Shelly, S., N. Lukinova, S. Bambina, A. Berman, and S. Cherry. 2009. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shoji-Kawata, S., and B. Levine. 2009. Autophagy, antiviral immunity, and viral countermeasures. Biochim. Biophys. Acta 1793:1478-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simon, S. A., et al. 2009. Short-read sequencing technologies for transcriptional analyses. Annu. Rev. Plant Biol. 60:305-333. [DOI] [PubMed] [Google Scholar]
- 70.Summers, M. D., and S. D. Dib-Hajj. 1995. Polydnavirus-facilitated endoparasite protection against host immune defenses. Proc. Natl. Acad. Sci. U. S. A. 92:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun, D. B., J. Xu, J. B. Luan, and S. S. Liu. 1 November 2010. Reproductive incompatibility between the B and Q biotypes of the whitefly Bemisia tabaci: genetic and behavioural evidence. Bull. Entomol. Res. 101. doi: 10.1017/S0007485310000416. [DOI] [PubMed]
- 72.Surachetpong, W., N. Singh, K. W. Cheung, and S. Luckhart. 2009. MAPK ERK signaling regulates the TGF-b1-dependent mosquito response to Plasmodium falciparum. PLoS Pathog. 5:e1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terenius, O. 2008. Hemolin-A lepidopteran anti-viral defense factor? Dev. Comp. Immunol. 32:311-316. [DOI] [PubMed] [Google Scholar]
- 74.'t Hoen, P. A. C., et al. 2008. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 36:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thoetkiattikul, H., M. H. Beck, and M. R. Strand. 2005. Inhibitor κB-like proteins from a polydnavirus inhibit NF-κB activation and suppress the insect immune response. Proc. Natl. Acad. Sci. U. S. A. 102:11426-11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Varma, A., and V. G. Malathi. 2003. Emerging geminivirus problems: a serious threat to crop production. Ann. Appl. Biol. 142:145-164. [Google Scholar]
- 77.Wang, J., et al. 2010. The low frequency of horizontal and vertical transmission of two begomoviruses through whiteflies exhibits little relevance to the vector infectivity. Ann. Appl. Biol. 157:125-133. [Google Scholar]
- 78.Wang, P., D. B. Sun, B. L. Qiu, and S. S. Liu. 2011. The presence of six cryptic species of the whitefly Bemisia tabaci complex in China as revealed by crossing experiments. Insect Sci. 18:66-77. [Google Scholar]
- 79.Wang, P., Y. M. Ruan, and S. S. Liu. 2010. Crossing experiments and behavioral observations reveal reproductive incompatibility among three putative species of the whitefly Bemisia tabaci. Insect Sci. 17:508-516. [Google Scholar]
- 80.Wang, X. W., et al. 2010. De novo characterization and comparison of the whitefly transcriptomes reveals genes associated with development and insecticide resistance. BMC Genomics 11:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang, Z., M. Gerstein, and M. Snyder. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10:57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waterhouse, R. M., et al. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316:1738-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xi, Z., J. L. Ramirez, and G. Dmopoulos. 2008. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog. 4:e1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu, J., P. J. De Barro, and S. S. Liu. 2010. Reproductive incompatibility among genetic groups of Bemisia tabaci supports the proposition that the whitefly is a cryptic species complex. Bull. Entomol. Res. 100:359-366. [DOI] [PubMed] [Google Scholar]
- 85.Zambon, R. A., M. Nandakumar, V. N. Vakharia, and L. P. Wu. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 102:7257-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.