Abstract
HIV-1-specific T lymphocyte responses in individuals exposed to HIV-1 but who remain persistently seronegative (HESNs) have been reported in some but not all previous studies. This study was designed to resolve unequivocally the question of whether HESNs make HIV-1-specific T cell responses. We performed a blind investigation to measure HIV-1-specific T cell responses in both HIV-1-serodiscordant couples and HIV-1-unexposed seronegative controls (HUSNs). We found low-frequency HIV-1-specific T cells in both HESNs and HUSNs but show that the response rates were higher over time in the former (P = 0.01). Furthermore, the magnitudes of the HIV-1-specific T cell responses were significantly higher among responding HESNs than among HUSNs over time (P = 0.002). In both groups, responses were mediated by CD4 T cells. The responses were mapped to single peptides, which often corresponded to epitopes restricted by multiple HLA-DR types that have previously been detected in HIV-1-infected patients. HIV-1-specific T cell responses in HUSNs and some HESNs likely represent cross-reactivity to self or foreign non-HIV-1 antigens. The significantly greater T cell responses in HESNs, including in two who were homozygous for CCR5Δ32, demonstrates that HIV-1-specific T cell responses can be induced or augmented by exposure to HIV-1 without infection.
The development of a vaccine against HIV-1 remains one of the most pressing scientific challenges of the modern age. An improved understanding of the interaction between the virus and the human host is paramount to the development of an efficacious vaccine. The repeated demonstration that there are people who remain resistant to HIV-1 infection, HIV-1-exposed seronegative individuals (HESNs), might give clues as to whether immune-mediated protection is possible. In nearly all other viral infections, naturally occurring examples of immunity have provided the rationale for vaccine design.
Previous characterizations of HESNs have focused on several different types of cohorts. These include commercial sex workers living within regions of HIV-1 endemicity (2, 26, 39, 40, 63, 64, 72), HIV-1-serodiscordant couples practicing unprotected sex (7, 31, 41, 58, 71, 74), intravenous drug users (50, 67, 76, 77), men who have sex with men who engage in unsafe sex practices (13, 23, 31, 34, 42), hemophiliacs exposed to contaminated blood products (4, 24, 75), and newborn HIV-negative babies whose mothers are HIV positive (65). From these studies has come one clear genetic protective factor, the Δ32 deletion in the CCR5 gene, which, in homozygotes, results in nonexpression of this HIV-1 coreceptor (18, 46). Several of these studies have suggested that HESNs, independently of their CCR5 genotype, make T cell responses to HIV-1, but other results have been negative (36, 62). Furthermore, responses in HESNs have often been of low magnitude and transient in nature (23, 31, 36, 39, 70), and in earlier studies, the assays used were not as well standardized as now. Thus, the significance of previous findings has been questioned, but the importance of the question remains paramount to vaccine design.
A central issue in all HESN studies is the definition and recruitment of participants. Infection with HIV-1 from a single exposure is a rare event (27), and many individuals with a reported HESN phenotype may represent chance and not have any immunological or genetic resistance factors. The ability of a study to detect factors associated with resistance to HIV-1 infection therefore hinges on a well-defined cohort of HESN participants. To maximize the proportion of true HESNs recruited into the Center for HIV-AIDS Vaccine Immunology 002 (CHAVI 002) study reported herein, the enrollment criteria for HESNs included unprotected sex with an HIV-1-positive partner on at least 25 occasions in the previous year.
Controls are also of great importance in these studies, and an absence of sufficient numbers of appropriately recruited HIV-1-unexposed seronegative individuals (HUSNs) in some studies has contributed to skepticism over the existence and significance of HESNs and the immune parameters associated with them. The recruitment of HUSNs is again a complex issue, with assumptions, such as lab workers being HUSNs, sometimes being misplaced. In this study, HUSNs were couples who reported monogamous behavior over the past year and were documented as negative for both HIV-1 and other sexually transmitted infections.
In order to address these issues and discern whether there is an increased rate of HIV-1-specific T cell responses in HESNs, we set up a cohort of serodiscordant couples (HESNs with HIV-1-seropositive partners) and HUSN couples at St. Mary's Hospital, London, United Kingdom, whose identities were concealed at the laboratory level. Exposure to HIV-1 and infection risk were quantified through detailed sexual behavior questionnaires (SBQs); each participant was asked to respond to the questions independently, and responses from couples were analyzed for concordance (A. J. Ritchie et al., unpublished data). A cultured gamma interferon (IFN-γ) enzyme-linked immunospot (ELISpot) assay, which has >20-fold-increased sensitivity relative to that of the conventional ex vivo ELISpot assay (10, 32), was used to ensure that low-frequency HIV-1-specific T cells were detected. T cell responses were analyzed at 4 different time points, 3 months apart, to determine whether responses were sustained or were transient. Finally, having shown that nearly all responses were mediated by CD4 T cells, a subset of responses from both the HESN and HUSN donors were mapped to individual peptide epitopes.
MATERIALS AND METHODS
HPTN 052 cohort.
Peripheral blood mononuclear cells (PBMCs) from 16 Malawian HIV-1-serodiscordant couples who were enrolled under the HIV Prevention Trials Network 052 (HPTN 052) V3.0 study protocol (21) were analyzed. Couples reported that the first occasion of high-risk sex occurred at least 3 months prior to being screening. At enrollment, HIV-1-infected participants were antiretroviral therapy (ART) naive, with CD4 counts ranging from 350 to 550 cells/mm3. HESN partners were required to have returned negative HIV-1 serology tests up to 14 days prior to enrollment. Serodiscordant couples reported at least 3 vaginal or anal sex acts in the 3 months prior to enrollment.
CHAVI 002, St. Mary's cohort.
Twenty-four serodiscordant and 14 HIV-1-negative-concordant couples were enrolled at St. Mary's Hospital, London, United Kingdom. Serodiscordant couples were eligible for enrollment if they reported at least 25 unprotected anal, vaginal, and/or oral sex acts in the preceding 12 months, with the first sex act occurring at least 10 months prior to the screening. Documented evidence that the HIV-infected partner was seropositive throughout this period was also required. For practical reasons, ART was not an exclusion criterion (Ritchie et al., unpublished data). At enrollment, 7/24 HIV-1-infected participants were not on ART and had plasma viral loads (pVLs) ranging from 1,656 to 34,0581 copies/ml. Of those on ART at enrollment, 15 had pVLs below the limit of detection (50 copies/ml) and the remaining 2 had pVLs of 56 and 170 copies/ml. Sex outside the primary relationship was regularly reported by both the HIV-1-infected participants and their HESN partners (50 to 78% of individuals in each group at each visit).
Enrollment of HUSN couples required negative HIV-1 serology and no sexually transmitted infections (STIs) at screening, with both partners self-reporting that they had been sexually monogamous for at least 12 months (Ritchie et al., unpublished data). T cell data from one HUSN was excluded from analysis due to the self-reporting of one occasion of sex outside the relationship occurring between screening and enrollment.
The study protocol asked participants to make 4 visits at 3-month intervals, at which times blood was collected. Some participants were lost to follow-up; 12/14 HUSN and 17/24 serodiscordant couples completed all 4 visits. At all visits attended, participants answered a detailed SBQ relating to the previous 3 months. The study protocol was approved by local and NIH AIDS ethics committees.
Study design.
The CHAVI 002 study was powered to detect differences in the overall ELISpot response rates between HESNs and HUSNs after each time point. Power calculations and statistical design considered (i) HESN and HUSN response rate assumptions and (ii) the number of participants in each group that could reasonably be recruited. With 20 participants per group, there was a 90% power of detecting a difference in ex vivo IFN-γ ELISpot results if response rates in HUSNs and HESNs were 5% and 24%, respectively. Previous studies with low-risk, seronegative subjects had suggested a cultured ELISpot response rate of 25% in HUSNs (32). To accommodate this higher HUSN response rate, cumulative repeat response rates (i.e., a positive T cell response at 2 or more visits) in cultured assays were compared. This approach provided a 90% power of detecting a difference in the cumulative repeat response rates if the response rate at each visit was 25% in HUSNs and 54% in HESNs. A Pocock stopping boundary was used to account for the multiple comparisons due to repeated testing of data at each time point to determine study futility. Analysis of data supported continued follow-up of study participants.
CCR5Δ32 gene expression.
The presence of the CCR5Δ32 mutation was detected using a PCR-based method as described previously (81). Homo- and heterozygosity was found in 2 and 6 of 24 HESNs and in 1 and 4 of 27 HUSNs, respectively. These differences are not significant, but CCR5Δ32 homozygosity most likely explains why these 2 HESNs remained uninfected (18, 46).
Highly sensitive HIV-1 RNA detection assays.
Validated transcription-mediated amplification (TMA) assays (Aptima; Gen-Probe, San Diego, CA) were used to detect HIV-1 RNA in the plasma of all CHAVI 002 subjects. One-half-milliliter volumes of plasma were tested per assay run, giving a sensitivity of 5 copies of RNA/ml (50% limit of detection) or 13 copies of RNA/ml (95% limit of detection) (35). The use of TMA in quadruplicate assays improves the limit of detection to fewer than 3.5 HIV RNA copies/ml (35). In this study, each plasma sample was tested a median of 6 times. A TMA signal-to-cutoff (S/Co) ratio of <1 is considered negative, while an S/Co ratio of ≥1 is considered positive.
Peptides.
Peptides were synthesized by Sigma, St. Louis, MO, and/or the Medical Research Council Human Immunology Unit, WIMM, Oxford, United Kingdom. For the HPTN 052 study, a pool of consensus, clade C 18-mer peptides overlapping by 10 nucleotides, spanning GAG and NEF (2 μg/ml), and a pool of 204 known HIV-1 cytotoxic T lymphocyte (CTL) epitopes (44) (0.7 μg/ml) were used. For the CHAVI 002 study, 18-mer peptides overlapping by 10 nucleotides and representing the consensus clade B proteome were pooled into proteins (ENV, GAG, POL, NEF, and a single pool including all accessory [ACC] proteins, VIF, TAT, VPR, VPU, and reverse transcriptase [RT]) and tested in quadruplicate in ex vivo and cultured assays (1 to 2 μg/ml) (44). The NEF and ACC peptide pools were combined into a single pool for cultured assays. The same group of 204 CTL epitopes (0.7 μg/ml) was also used in T cell assays but divided into two pools, CTLA and CTLB, for cultured assays. Positive controls included a pool of influenza virus, Epstein-Barr virus (EBV), and cytomegalovirus (CMV) (FEC) peptides (1.5 μg/ml), consisting of known CTL epitopes from those viruses (17), phytohemagglutinin (PHA; 10 μg/ml; Sigma), and/or staphylococcal enterotoxin B (SEB; 2 μg/ml; Sigma). All peptide pools and negative controls (6 replicates for ex vivo and quadruplicate experiments in the cultured ELISpot assay) contained ≤0.45% dimethyl sulfoxide (DMSO).
For all ex vivo, cultured, and mapping ELISpots, peptide plates were preprepared in round-bottom 96-well plates (Corning Inc., Corning, NY). Peptide pools plus positive and negative controls were prepared at 2× the final concentration in R10 (10% fetal bovine serum [FBS; Gemini Bio-Products, West Sacramento, CA], 86% RPMI 1640 [Sigma], 2 mM l-glutamine [Sigma], 1× penicillin-streptomycin [Sigma], 10 mM HEPES [Sigma], 1 mM sodium pyruvate [Sigma]) or RAB-10 (10% human AB serum [Sigma], 86% RPMI 1640, 2 mM l-glutamine, 1× penicillin-streptomycin, 10 mM HEPES, 1 mM sodium pyruvate) as required. Peptide plates were sealed with microtiter plate sealers (Diversified Biotech, Dedham, MA) and stored at −80°C for up to 2 years.
Generation of STCLs.
PBMCs (2 × 106 to 4 × 106) were cultured at 37°C and 5% CO2 for 10 days in RAB-10, peptide pools, and recombinant interleukin 7 (r-IL-7; 25 ng/ml; R & D Systems, Minneapolis, MN). Cells were supplemented with 1,800 U/ml r-IL-2 (Novartis, Camberley, United Kingdom) on days 3 and 7 and additional RAB-10 on day 7. For cultured ELISpots, cells were collected on day 10. Excess cells were either cryopreserved or given additional IL-2 and RAB-10 and incubated for a further 2 to 3 days and then used for peptide mapping and intracellular cytokine stain (ICS) assays. Prior to use in assays, all short-term cell lines (STCLs) were washed four times and rested at 37°C and 5% CO2 for 29 to 34 h in fresh RAB-10. STCLs were used in cultured ELISpot, mapping ELISpot, and ICS experiments.
ELISpots.
MSIPS4510 polyvinylidene difluoride (PVDF) membrane plates (Millipore, Bedford, MA) were coated with mouse anti-human IFN-γ 1-D1K antibody (Mabtech, Nacka Strand, Sweden) and blocked with R10. Cells in R10 or RAB-10 were added and allowed to rest for 30 to 60 min at 37°C and 5% CO2, and then peptide and controls were added. Plates were incubated for 18 to 20 h at 37°C and 5% CO2, cells were discarded, and plates were washed six times. One hundred microliters per well of 1-μg/ml biotinylated mouse anti-human IFN-γ 7-B6-1 (Mabtech) in PBS (Sigma) containing 0.5% bovine serum albumin (BSA; Sigma) was added, and the plates were incubated at 4°C for 18 to 72 h. Plates were washed six times, and 100 μl/well of peroxidase avidin-biotin complex (Vector Laboratories, Burlingame, CA) was added for 1 h at room temperature. Next, plates were washed six times and developed with 100 μl/well of a 3-amino-9-ethylcarbazole (AEC) substrate solution for 5 min and then washed liberally with water. Plates were read using a calibrated AID machine and AID EliSpot 3.1.1 HR software (Autoimmun Diagnostika, Strasburg, Germany).
ELISpot positivity criteria.
A positive T cell response in both ex vivo and cultured ELISpots for the CHAVI 002 study was defined by the distribution-free resampling (DFR) (2×) method described by Moodie et al. (56), with the addition of filters for positive and negative controls. Peptide pools with adjusted one-sided P values of ≤0.05 were declared positive. The quantitative responses were then vetted for biological significance such that, in addition to a significant P value, mean background-subtracted responses for ex vivo ELISpots of >6 spot-forming units (SFU)/200,000 PBMCs and for cultured assays of >12 SFU/40,000 cells were required.
An additional criterion was applied to cultured ELISpots. In these assays, STCLs were tested using the main pool against which they were generated and three subpools, each containing one-third of the peptides from the main pool. A response against the main pool was considered positive only if there was also a response in one or more of the subpools. Subpool positivity was defined as for ELISpots, with a mean background-subtracted response of 10 SFU/40,000 cells being required. Magnitude was defined as the sum of the magnitudes of the positive subpools for an individual, calculated at each time point.
Statistical analysis of cultured ELISpot data.
Cumulative repeat probabilities were estimated by nonparametric maximum-likelihood methods (37) to account for the missing data. Generalized estimating equations (GEE) (45) for binary data, with an assumption of working independence, were used to model the overall positive/negative responses over time and to test for differences in these between HESNs and HUSNs. A similar GEE model was also used to examine the effect of gender on HUSN responses over time. Lastly, a GEE model assuming a Gaussian response and working independence was used to model the log10-transformed total magnitude over time as a function of time and group and as an indicator of a positive response and the interaction of a positive response with the group (with group comparisons allowed for positive responders only).
ICS and flow cytometry.
Cells (0.5 × 106 to 1 × 106) were incubated for 1 h at 37°C and 5% CO2 with no antigen, peptide pools, individual peptides, or SEB. Golgi Plug (1 μl/ml; BD Biosciences, Franklin Lakes, NJ) was added, and cells were incubated for a further 5 h. Cells were washed and resuspended in 1 ml of 0.5% BSA-PBS. One microliter per tube of LIVE/DEAD fixable, near-infrared (IR), dead cell stain (Molecular Probes, Eugene, OR) was added, and cells were incubated for 30 min at 4°C, washed twice, and resuspended in 100 μl of 0.1% BSA-PBS. Preoptimized volumes of the following mouse anti-human antibodies were then added to each tube: CD3 Pacific Blue, CD4 phycoerythrin (PE), CD14 PE-Cy7, CD19 PE-Cy7, CD56 PE-Cy7 (all from BD Biosciences), and CD8 PE-Texas Red (ECD) (Beckman Coulter, High Wycombe, United Kingdom). Cells were incubated for 15 min at room temperature, washed, fixed, and permeabilized (Fix & Perm fixation medium; Caltag, Little Balmer, United Kingdom), and a preoptimized volume of mouse anti-human IFN-γ fluorescein isothiocyanate (FITC) antibody (BD Biosciences) was added. Cells were incubated for 15 min at room temperature, washed, and resuspended in 100 μl of Cytofix fixation buffer (BD Biosciences). Samples were run on a CyAn analyzer (Beckman Coulter), and data were analyzed using FlowJo version 8.8.6 (Tree Star, Ashland, OR). The following gating strategy was used: pulse width/single events/lymphoblasts/alive CD14−, CD19−, and CD56−/CD3+ cells. CD4+ and CD8+ cells were then plotted against IFN-γ. A positive T cell response required the acquisition of ≥50,000 CD3+ lymphocytes, and the background-subtracted frequency and number of IFN-γ-producing T cells in antigen-stimulated cells had to be at least twice that of nonstimulated cells and ≥30, respectively.
RESULTS
Conventional IFN-γ ELISpot assays failed to detect T cell responses in HESN and HUSN donors.
Standardized ex vivo IFN-γ ELISpot assays were performed on PBMCs obtained at visit 1 from members of the CHAVI 002 cohort. Multiple HIV-1-specific T cell responses were detected in all 20 HIV-1-seropositive patients tested, while no HIV-1-specific T cell responses were detected in the 16 HESNs or 21 HUSNs tested (data not shown). These results appeared consistent with the more negative previous reports (36, 62).
Preliminary study of HIV-1-specific T cell responses detected in HESNs using the cultured IFN-γ ELISpot assay.
We have previously demonstrated that the sensitivity of detection of IFN-γ-secreting T cells as measured by the ELISpot assay can be increased greater than 20-fold following specific expansion of T cells over a 10-day in vitro culture (10, 32). In a preliminary study, we used this cultured ELISpot assay to measure HIV-1-specific T cell responses at two time points in Malawian HIV-1-serodiscordant couples recruited in the HPTN 052 study. T cell responses were measured using overlapping peptides representing the clade C GAG and NEF consensus amino acid sequence and also a pool of common optimal HIV-1 peptide epitopes presented by HLA class I to CD8 T cells. Two previously published approaches, the “empirical” (32) and DFR(2×) (56) methods, were applied to define positivity (Table 1). Both methods found that the majority of HIV-1-seropositive partners had T cell responses to both HIV-1 CTL epitopes and GAG and NEF peptides (Table 1). In the HESN partners, positive HIV-1-specific T cell responses of less than 1,000 spot-forming units (SFU)/106 cells (data not shown) were detected by both methods in approximately 25% of HESN partners at both visits (Table 1). Although no appropriate HUSN controls were available, this small pilot confirmed that low-frequency HIV-1-specific T cell responses could be detected in seronegative individuals. These data and data from published studies (32) informed the inclusion of cultured ELISpot assays and guided the power calculations for the main CHAVI 002 study. The DFR(2×) statistical method with additional laboratory-defined criteria (see Materials and Methods) to increase stringency was chosen to define positivity in cultured ELISpot assays.
TABLE 1.
Summary of HIV-1-specific T cell responses detected in HPTN 052 serodiscordant couples
| Group | Short-term cell linea | Visit | No. positive by DFR(2×) assay/totalb | No. positive by empirical assay/totalc |
|---|---|---|---|---|
| HIV-positive individuals | GAG/NEF | 1 | 9/11 | 9/11 |
| 7 | 6/12 | 8/12 | ||
| HESN partners | 1 | 3/12 | 2/12 | |
| 7 | 4/12 | 3/12 | ||
| HIV-positive individuals | HIV-1 CTL | 1 | 10/10 | 10/10 |
| 7 | 8/9 | 7/9 | ||
| HESN partners | 1 | 2/12 | 1/12 | |
| 7 | 3/12 | 3/12 |
PBMCs were stimulated with either a pool of previously defined HIV-1 CTL epitopes or a clade C GAG/NEF peptide pool and cultured for 10 days, and responses were measured by an IFN-γ ELISpot assay. Negative and antigen-positive wells were tested in quadruplicate.
The DFR(2×) method is described in reference 56 and in Materials and Methods. In addition to having a significant P value, the mean background-subtracted response must exceed 250 SFU/106 for positivity.
The empirical method defines positivity as a value 4 times the mean value for negative wells, with mean background-subtracted responses of antigen-stimulated wells being ≥300 SFU/106 cells and the mean of negative wells being ≤250 SFU/106 (32).
HESNs maintain higher rates of HIV-1-specific T cell responses than HUSNs across multiple visits.
Across the 4 visits in this study, anti-HIV-1 T cell responses were detected in the HESNs and, more surprisingly, in the HUSNs by cultured ELISpot assay. Responses were found in 53% to 61% of HESNs and occurred more frequently than in HUSNs, of whom 26% to 44% responded (Table 2).
TABLE 2.
HIV-1-specific T cell responses measured in HESNs and HUSNsa
| Group | No. with positive T cell response/total (%) at visitb: |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| HESNs | 13/24 (54) | 10/19 (53) | 11/18 (61) | 10/18 (56) |
| HUSNs | 9/27 (33) | 7/23 (30) | 10/23 (44) | 6/23 (26) |
Using a generalized estimating equation (GEE) model for these data, a significant difference was observed (Wald P = 0.01) where the odds of an HIV-1-specific T cell response in an average HESN was 2.5 times higher than in an average HUSN.
Data are presented as the fractions (and percentages) of participants producing a positive HIV-1-specific T cell response in a cultured ELISpot assay at the indicated visits. Positivity was determined by the DFR(2×) method described in reference 56, which was applied to both the antigen pool and one or more subpools (see Materials and Methods).
A longitudinal analysis was performed to test for group differences over time. A significant difference was observed (Wald P = 0.01), where the odds of an HIV-1-specific T cell response in an average HESN was 2.5 times higher than in an average HUSN (Table 2). There was no significant interaction between group and time (Wald P = 0.71), nor was there a significant effect of time on T cell responses in the HESN group (Wald P = 0.78), indicating that the significant difference in T cell response rates observed between HUSNs and HESNs was constant over time and that HESN responses were well maintained over time. The members of the HESN group were all male, whereas 9/28 HUSNs were female, prompting an additional analysis to examine the potential confounding factor of gender. Overall, the cultured ELISpot response rate over time was significantly higher in female HUSNs than in male HUSNs (Wald P = 0.001), indicating that the significantly higher response rate observed in the HESN group did not result from gender differences.
Individual HESNs maintained HIV-1-specific T cell responses across multiple visits more often than HUSNs. Although this study was not powered to statistically analyze these differences, this trend was observed in the maintenance of T cell responses against any HIV-1 peptide pool (Fig. 1) as well as in the maintenance of T cell responses against the same HIV-1 peptide subpool (see Table S1 in the supplemental material).
FIG. 1.
HESNs better maintain HIV-1 T cell responses than HUSNs. A participant was considered to have an HIV-1-specific T cell response at a visit if a positive ELISpot response to one or more HIV-1 proteins was observed. Data pertaining to the maintenance of responses against a single HIV-1 protein are contained in Table S1 in the supplemental material. Data represent the percentages of HUSNs (left panel) and HESNs (right panel) having detectable T cell responses against any HIV-1 protein during 0 to 4 of the 4 study visits.
HESNs have a higher magnitude of HIV-1-specific T cell responses than HUSNs.
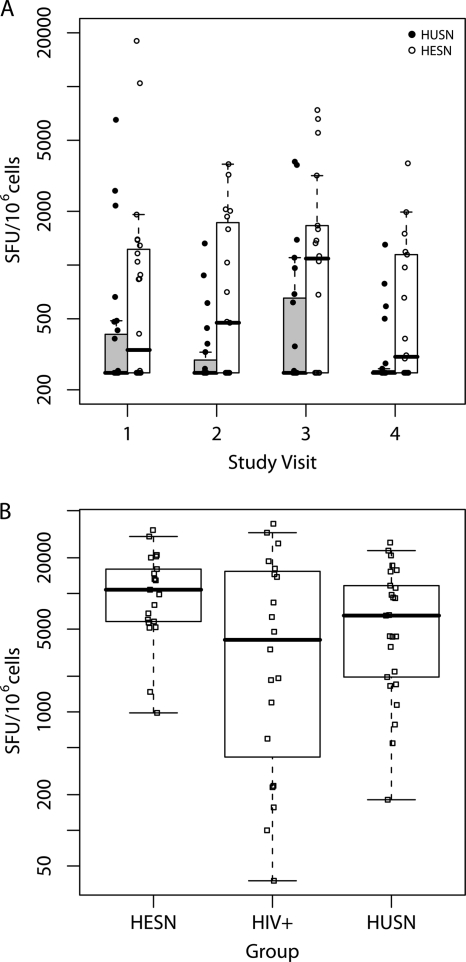
There was also a significant difference between HESN and HUSN groups in the total magnitudes of HIV-1-specific T cell responses among positive responders over time (Wald P = 0.002) (Fig. 2 A). We considered the possibility that the difference in magnitude of HIV-1-specific T cell responses in HESN and HUSN groups reflected a general difference in the abilities of T cells to expand in culture. Therefore, T cells were also expanded and tested against a pool of influenza virus, EBV, and CMV (FEC) peptides (17). Comparisons of the responses of both HESN and HUSN participants at visit 1 revealed no significant difference in the magnitudes of FEC peptide-specific T cell responses when they were measured following 10 days of culture (Wilcoxon rank sum test P = 0.08) (Fig. 2B).
FIG. 2.
HESNs have higher-magnitude HIV-1 T cell responses than HUSNs. PBMCs were cultured against 6 pools of HIV-1 and 1 pool of FEC (control) peptides for 10 days and responses quantified using the cultured IFN-γ ELISpot assay. (A) All HESNs and HUSNs with responses to HIV-1 peptides by the DFR(2×) method described in the text. Generalized estimating equations (GEE) were used to model the log10-transformed total magnitudes over time of HIV-1 responders in the HESN and HUSN groups, revealing a higher magnitude of responses in the HESN group across time (Wald P = 0.002). (B) All visit 1 responses to the FEC peptide pool were quantified and found to be not significantly different across the 3 study groups (Kruskal-Wallis P = 0.12) or between the HESN and HUSN groups (Wilcoxon P = 0.08). Box plots are superimposed on the raw data for each individual, with a midline and box used to represent the median ± the 25th and 75th percentiles, respectively; whiskers extend to the extreme data points that are ≤1.5 times the interquartile range.
Responses in HESNs homozygous for CCR5Δ32.
We considered whether seronegative participants with high cultured ELISpot responses might have high precursor frequencies of cells, sufficient to be detected ex vivo. Although all visit 1 samples in the uninfected groups were negative, ex vivo ELISpot analysis of visits 2 to 4 was performed on those participants who produced a cultured response of >2,000 SFU/106 cells: 9 from among the HESNs and 1 from among the HUSNs. One HESN (S048) had a detectable ex vivo response against NEF at the second time point, with a magnitude of 83 SFU/106 cells. This participant maintained T cell responses against the NEF pool in cultured ELISpots across all 4 visits at >1,000 SFU/106 cells (see Table S1 in the supplemental material), reported persistent unprotected sex with his HIV-1-positive partner, and is homozygous for CCR5Δ32, rendering his leukocytes highly resistant to primary HIV-1 infection (18, 46). A second HESN was homozygous for CCR5Δ32 (S050); he did not have measurable ex vivo T cell responses but maintained HIV-1-specific T cell responses in cultured assays across multiple visits (see Table S1 in the supplemental material).
Further characterization and specificity of the T cell responses in HESNs and HUSNs.
Where PBMCs were available, HESN and HUSN participants who produced an HIV-1-specific T cell response by cultured ELISpot assay were also examined by flow cytometry for peptide-stimulated intracellular IFN-γ production and CD3, CD4, or CD8 surface marker staining. Over 90% of HIV-1-specific T cell responses that were positive in the cultured ELISpot assay were also positive in this assay. IFN-γ-producing T cells that responded to the peptide pools were nearly all CD4+ (see Table S2 in the supplemental material). One HESN, the CCR5Δ32-homozygous participant S048, produced a CD8-restricted T cell response that was mapped to a peptide (see Table S2 in the supplemental material). Two other CD8-restricted responses were detected in a single HUSN, S079 (see Table S2 in the supplemental material).
Where either PBMCs or STCLs were available for the responding seronegative participants, the T cell responses were mapped at the peptide level using the cultured IFN-γ ELISpot assay. In the 25 participants tested, T cell responses were targeted to 24 different HIV-1 peptides, 19 of which matched epitopes and peptides previously described for HIV-1-seropositive patients and several other experimental settings (Table 3). Fourteen of these epitopes have been described as binding to multiple HLA-DR types (Table 3). Identification of the epitope specificities of these T cell responses to epitopes previously described for HIV-seropositive patients strongly validates the basic finding here that HESNs and HUSNs do make bona fide T cell responses to HIV-1 peptides.
TABLE 3.
Epitope details of mapped peptides in HESNs and HUSNs
| No. of participants responding | Group | Patient identifier(s) | ICS | Peptidea | Information (reference[s])b |
|---|---|---|---|---|---|
| 1 | HESN | S074 | CD4 | HPVHAGPIA | GAG aa 216-224 |
| EWDRV---------PGQMRE | HLA-DR promiscuous (9) | ||||
| 1 | HESN | S074 | CD4 | TSTLQEQIGW | GAG aa 240-249 |
| PRGSDIAGT----------M | HLA-DR promiscuous (9) | ||||
| 3 | HESN | S069 | CD4 | LGLNKIVRMYSPTSILDI | GAG aa 268-285 |
| S050 | I------------------R | 8/24 SPs responded (1) | |||
| S063 | I---------- | HLA-DR promiscuous (38) | |||
| WII--------------- | HLA-DR promiscuous (38); 9/36 SPs responded (38) | ||||
| 1 | HUSN | S079 | CD8 | AEAMSQVTNSATIMMQRG | GAG aa 364-381; responding HUSN is HLA-A2 |
| --------------- | Induces responses in HLA-A2 SPs (22) | ||||
| --------- | Induces responses in HLA-A2 SPs (79) | ||||
| 1 | HESN | S069 | CD4 | KELYPLASLRSLFGNDPS | GAG aa 481-498 |
| ------------------SQ | 3/36 SPs responded (38) | ||||
| D-------------- | HLA-DR promiscuous (25); 8/32 SPs responded (25) | ||||
| 1 | HESN | S019 | CD4 | IPHPAGLKKKKSVTVLDV | POL aa 249-266 |
| WEVQLG--------- | 1/3 mouse strains responded (19) | ||||
| 1 | HUSN | S042 | CD4 | IKKEKVYLAWVPAHKGIG | POL aa 681-698 |
| --------------- | HLA-DR promiscuous (80); 6/22 SPs responded (80) | ||||
| 1 | HUSN | S042 | CD4 | PVVAKEIVASCDKCQLKG | POL aa 745-762 |
| 1 | HESN | S057 | CD4 | NSDIKVVPRRKAKIIRDY | POL aa 969-986 |
| VIQD-------------- | 4/36 SPs responded (38) | ||||
| 1 | HUSN | S079 | CD4 | PRISSEVHIPLGDARLVI | VIF aa 49-66 |
| CD8 | |||||
| 1 | HUSN | S084 | CD4 | VSPRCEYQAGHNKVGSLQ | VIF aa 129-146 |
| 1 | HUSN | S017 | CD4 | AGHNKVGSLQYLALAALI | VIF aa 137-154 |
| -------V--VAPKK | HLA-DR promiscuous (25); 7/32 SPs responded (25) | ||||
| 1 | HESN | S042 | NDc | LQYLALAALITPKKIKPP | VIF aa 145-162; responding HESN is HLA-A2 |
| S------V---VA--- | HLA-DR promiscuous (25); 7/32 SPs responded (25) | ||||
| --------- | Strong HLA-A2 binder (16) | ||||
| 1 | HUSN | S017 | CD4 | HFPRIWLHGLGQHIYETY | VPR aa 33-50 |
| R-------------- | HLA-DR promiscuous (12) | ||||
| 1 | HESN | S052 | CD4 | QQLLFIHFRIGCQHSRIG | VPR aa 64-81 |
| ------------R----- | HLA-DR promiscuous (25); 9/32 SPs responded (25) | ||||
| -------R-----VT | HLA-DR promiscuous (12) | ||||
| 1 | HESN | S028 | CD4 | SFNCGGEFFYCNTTQLFN | ENV aa 375-392 |
| -------S-----S | 4/15 SPs responded (30) | ||||
| 1 | HUSN | S004 | CD4 | KYKVVKIEPLGVAPTKAK | ENV aa 485-502 |
| SELYL------------- | HLA-DR promiscuous (25); 7/32 SPs responded (25) | ||||
| ELY----------------- | 3/36 SPs responded (30) | ||||
| 1 | HESN | S028 | CD4 | MEWEREIDNYTSLIYTLI | ENV 629-646 |
| 1 | HUSN | S032 | CD4 | LELDKWASLWNWFDITNW | ENV 661-678 |
| -------N----LWY | 48/99 SPs responded (78) | ||||
| 1 | HUSN | S059 | CD4 | DRDRSGRLVDGFLALIWD | ENV 741-758 |
| 1 | HUSN | S032 | CD4 | NLLQYWSQELKNSAVSLL | ENV 798-815 |
| --------------- | Murine MHC class promiscuous (33) | ||||
| RIVELLGRRGWEALKYWW---------------- | Murine MHC class promiscuous (8); 17/29 SPs responded (8) | ||||
| 2 | HESN | S027 | CD4 | IEVVQRACRAILHIPRRI | ENV 830-847 |
| S073 | DRV-----G-Y---R | Murine MHC class promiscuous (33); 6/11 responded (13), 6/10 responded (14) SPs; 4/5 HESNs responded (13) | |||
| 1 | HESN | S048 | CD4 | QVPLRPMTYKGALDLSHF | NEF 73-90; responding HESN is HLA-A3 |
| CD8 | VGFPVRP----------A-V--- | 5/13 SPs responded (20) | |||
| VGFPVTP----------A-V-----LKEKGGL | Predicted as HLA-DR promiscuous (59); 5/10 vaccinees responded (29) | ||||
| ----------A-V----- | HLA-DR promiscuous (66); CTL responses in multiple ethnicities (28) | ||||
| ---------- | Induces responses in HLA-A3 seropositive individuals (3, 82) | ||||
| 1 | HESN | S048 | CD4 | EREVLVWKFDSRLAFHHM | NEF 177-194 |
| CD8 | P-K--------------- | HLA-DR promiscuous (38); 13/36 SPs responded (38) | |||
| -----------ARELH | HLA-DR promiscuous (38); 9/36 SPs responded (38) | ||||
| P-K--------------- | CTL responses found in multiple ethnicities (28) |
The top row of each group shows the HIV-1 peptide that induced the T cell response in one or more subjects. Below are aligned HIV-1 sequences that either have been experimentally defined or are predicted to be HIV-1 epitopes.
The top row of each group denotes the HXB2 position of the mapped peptide and, where appropriate, the participant's HLA class I genotype. The lower row(s) provides details of the corresponding sequence in the adjacent peptide column. SPs, HIV-1-seropositve patients; aa, amino acids; MHC, major histocompatibility complex.
ND, assay not done.
Failure to detect HIV in HUSNs and HESNs by the TMA assay.
Finally, we retested HUSN and HESN samples for low-level HIV-1 infection. Over the course of the study, all HUSNs remained seronegative by standard HIV-1 clinical tests (Ritchie et al., unpublished data); however one HESN seroconverted at visit 2 and from then on was excluded from analysis. The highly sensitive transcription-mediated amplification (TMA) assays run on samples from the HIV-1-seropositive participants were positive in 210 of 351 assays. All HIV-1-positive participants with a detectable pVL of over 50 copies/ml were positive in all replicate assays runs, while those on ART with a pVL of <50 copies/ml returned varied TMA results. One of 557 TMA assays run on HUSN participants was positive, compared with 3 of 359 assays run on HESN participants (P = 0.31, Fisher's two-sided exact test). Given that the positive results for HESNs were rare and not greater than for HUSNs, we concluded that these results were likely to be false positives and that HIV-1 was not reliably detected in HESNs.
DISCUSSION
The primary goal of this study was to resolve the uncertainty about whether HESNs make T cell responses to HIV-1. We compared the frequencies of HIV-1-specific T cell responses over time between HESN and HUSN groups in a powered, blind study with independent data analysis. In these donors, no T cell responses were detectable without culture, except, notably, in an HESN participant who was homozygous for CCR5Δ32 and therefore genetically resistant to HIV-1 infection (18, 46). Using the more sensitive cultured ELISpot assay, T cell responses were found in both HESNs and HUSNs. Significant differences were found in the frequencies of T cell responses over time, with HESNs more likely to have positive T cell responses than HUSNs. HESNs more often maintained HIV-1-specific T cell responses across visits than HUSNs. Also, among positive responders, T cell responses were of significantly higher magnitude in HESNs than in HUSNs. Given that the cultured ELISpot assay expands antigen-specific cells before quantifying them, these differences between the groups could represent increased precursor frequencies in the HESNs and/or an increased proliferative capacity of the HIV-1-specific T cells in the HESNs. Previous studies have revealed differences in the activation (5, 11, 43) and memory (48, 69) phenotypes of T cells in HESNs from those of unexposed controls, while increased proliferation of cells in response to HIV-1 peptides has been reported on several occasions (6, 31).
The relatively high frequency of HIV-1-specific T cell responses detected in the HUSN group of the CHAVI 002 study is surprising. They clearly represent true antigen-induced responses, despite these individuals having no identifiable exposure to HIV-1. These were CD4 T cell responses which typically mapped to a single HIV-1 peptide. Detection of HIV-1-specific T cell responses in unexposed individuals is consistent with previous work by our group (32) and others (36). These responses are very unlikely to reflect poor recruitment of HUSNs given the thorough SBQ and persistent HIV-seronegative status of both HUSN partners. One question that this finding raises is whether these T cell responses were primed in vitro. However, the assay conditions used for the cultured ELISpot assay were very unlikely to be adequate to do this. Specifically, the assays were of short duration, 12 days, with no antigen costimulation, no exogenous cytokines apart from IL-2, no mitogenic stimulation, and no enrichment of dendritic cells, all of which are necessary for in vitro primary responses (49, 57, 68). The HIV-1-specific T cell responses in HUSNs instead are likely to reflect low-frequency antigen-experienced T cells previously induced by exposure to environmental antigens, where the T cell receptor cross-reacts with HLA-bound HIV-1 peptides. Mason (51) estimated that TCRs must be inherently cross-reactive, capable of reacting with 104 to 107 different peptide antigens, to accommodate all possible epitope peptides with a finite number of T cell receptors, so such cross-reactions should be expected. Moreover, cross-reactivity does not require linear sequence similarity between the two antigens; similarities in solvent-accessible side chains when bound to the presenting HLA molecule are sufficient (15, 47, 52). The existence of an almost unlimited range of environmental antigens makes identification of the source of cross-reactivity difficult.
The HESN and HUSN HIV-1-specific responses were mapped to peptides that often contained T cell epitopes previously reported in various studies, including those of natural HIV-1 infection (Table 3). This striking concordance of epitope specificity with findings for HIV-1-infected patients in particular strongly reinforces the validity of the T cell responses observed in this study. Sixty-one percent of the CD4-restricted epitopes mapped in this study have been previously described as “promiscuous”—that is, able to complex with multiple HLA class II molecules. It is not surprising that there should be a bias toward such promiscuous epitopes because they will be presented by many of the HLA class II types in the donor population. The presence of HIV-1-specific immune responses in uninfected individuals may have a significant impact on the specificity of the CD4 T cell response following seroconversion, resulting in the immunodominance of those responses. Such “prepriming” might ensure better T and B cell responses when HIV-1 infection occurs; CD4 T cells are central to adaptive immunity, particularly the generation of high-affinity antibodies and the maintenance of CD8 T cell memory (73). However, the possible beneficial effects could be counterbalanced by harmful effects; HIV-1-specific CD4 T cells, when activated by the incoming infection, could be primary targets for the virus. In a follow-up study, we are currently assessing the impact of preexisting HIV-1-specific T cell response in vaccine trial participants on HIV-1 seroconversion and T lymphocyte immunodominance following seroconversion.
Although exposure to HIV-1 in the HESN partners is well quantified in this cohort, it is relatively low compared to the exposure that may be expected in some other HESN groups, such as commercial sex workers in areas with high rates of infection who report multiple daily exposures (64). The high levels of antiretroviral drug use in the infected partners in this cohort and their associated decreases in viral load further limit the potential exposure of these HESNs (Ritchie et al., unpublished data). However, over the course of the study, more than half of HESNs reported additional sex partners outside the primary relationship, but this additional risk was unquantifiable because of the unknown HIV-1 serostatus of extraneous partners. Additionally, oral sex with the study partner was not included in risk estimates, and sex acts using condoms were assumed to carry no risk. Therefore, the calculated HIV-1 risk indices (Ritchie et al., unpublished data) reflect minimal estimates of HIV-1 infection risk for the HESN participants in this cohort. It is well recognized that HIV-1 infection is a low-probability event for any single sex act (27) and that the actual exposure risk is much higher than the infection risk. The HESNs in this study are likely to have been exposed to virus-infected cells from the partner, to free antigen, and to defective as well as infectious virus. We did not find any correlation between the calculated risk index (Ritchie et al., unpublished data) and the presence of a cultured ELISpot T cell response (data not shown).
It is not unexpected that noninfectious exposures are more likely to induce a CD4+ than a CD8+ response, as found in this study. The strong favoring of CD4+ T cell responses in HESNs could reflect that low antigen thresholds are required for the activation of CD4 T cells (61) or a greater likelihood that precursor CD4 T cells detect promiscuously binding CD4 epitopes. For humans, this view is supported by vaccine studies using attenuated viral vectors such as the MVA and NYVAC strains, which preferentially stimulate vaccine-specific CD4+ T cells, in contrast to the more immunogenic adenovirus vectors that strongly induce CD8 T cells (32). In this light, our results are not inconsistent with earlier studies of commercial sex workers (2, 39, 40, 63, 64) or of individuals exposed to unusually high levels of the virus, in which studies the detection of HIV-1-specific CD8 T cells was reported for a higher proportion of HESNs than in the present study (55, 60).
While our data clearly demonstrate that HIV-1-specific T cell responses really do exist in many HESNs, as well as HUSNs, the immunological significance of these responses remains uncertain. The enhanced responses in HESNs compared to those in HUSNs must be a marker of exposure but may play no role in maintaining the HESN phenotype. This is exemplified by two HESNs who had strong HIV-1 T cell responses detected at 3 or 4 visits and had a CCR5Δ32-homozygous genotype. These are clear examples of individuals who are highly resistant to HIV-1 infection through a non-T cell-related mechanism, having HIV-1-specific T cell responses maintained across multiple visits due to exposure. This finding does not exclude the possibility that HIV-1-specific T cells may offer some protection in other HESNs. However, the low-frequency T cell responses (undetectable in ex vivo assays) reported in this study seem unlikely to offer protective immunity given that T cell responses were detected at higher frequencies in ex vivo assays in the nonprotective STEP vaccine trial (53). While HIV-1-specific CD4 T cell responses which afforded modest protection from infection were detected in uninfected vaccinees in the RV144 vaccine trial, T cell responses were again detectable at a higher frequency in ex vivo ELISpot assays (54).
The detection of other immune responses, including mucosal IgA (34), in other HESN studies shows that immunological markers of exposure are not limited to T cell responses. It is possible therefore that a combination of immune responses and/or genetic factors may decrease the likelihood of infection, and this needs exploration. However, such combinations may vary greatly between individuals, making it difficult to discern which are protective. What should now be feasible is to retrospectively measure these preinfection immune responses in stored PBMC samples from vaccine trial recipients who did and did not become infected to determine whether they influence positively or negatively the acquisition of infection. In future vaccine trials, the possibility that these prevaccination immune responses, which are present in quite high numbers of people, could influence the outcome needs to be considered.
In conclusion, the current study shows unequivocally that low-level, ongoing exposure to HIV-1 induces HIV-1-specific T cell responses that are significantly more frequent and of a higher magnitude in HESNs than in unexposed controls. These T cell responses, more often detected in HESNs than in HUSNs across multiple visits, were made primarily by CD4+ cells and mapped to single peptides, often including epitopes that are commonly immunogenic after HIV-1 infection. The data may be consistent with most of the previous studies of immune responses in HESNs in whom relatively low exposures, such as in our study groups, fail to give positive responses in a standard ELISpot assay but can be revealed by amplification of T cell memory. It is quite possible that very high levels of exposure in sex workers, for example, also prime or boost CD8 T cell responses.
Supplementary Material
Acknowledgments
This work was supported by the Center for HIV-AIDS Vaccine Immunology (CHAVI) (NIAID grant A1067854).
We thank the SCHARP database management team, the CHAVI management and repository teams, and Family Health International for invaluable support. We also thank Stuart Shapiro for useful discussions, Bette Korber for assistance in peptide design, and the following individuals for technical assistance: Victoria Whale, Arjun Krishnakumar, Rachel Tanner, Robert King, and Lucy Li.
Footnotes
Published ahead of print on 26 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adams, S. L., R. A. Biti, and G. J. Stewart. 1997. T-cell response to HIV in natural infection: optimized culture conditions for detecting responses to gag peptides. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:257-263. [DOI] [PubMed] [Google Scholar]
- 2.Alimonti, J. B., et al. 2006. Characterization of CD8 T-cell responses in HIV-1-exposed seronegative commercial sex workers from Nairobi, Kenya. Immunol. Cell Biol. 84:482-485. [DOI] [PubMed] [Google Scholar]
- 3.Appay, V., et al. 2002. Persistent HIV-1-specific cellular responses despite prolonged therapeutic viral suppression. AIDS 16:161-170. [DOI] [PubMed] [Google Scholar]
- 4.Barretina, J., et al. 2000. Evaluation of the putative role of C-C chemokines as protective factors of HIV-1 infection in seronegative hemophiliacs exposed to contaminated hemoderivatives. Transfusion 40:461-467. [DOI] [PubMed] [Google Scholar]
- 5.Begaud, E., et al. 2006. Reduced CD4 T cell activation and in vitro susceptibility to HIV-1 infection in exposed uninfected Central Africans. Retrovirology 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beretta, A., et al. 1996. HIV-1-specific immunity in persistently seronegative individuals at high risk for HIV infection. Immunol. Lett. 51:39-43. [DOI] [PubMed] [Google Scholar]
- 7.Bernard, N. F., C. M. Yannakis, J. S. Lee, and C. M. Tsoukas. 1999. Human immunodeficiency virus (HIV)-specific cytotoxic T lymphocyte activity in HIV-exposed seronegative persons. J. Infect. Dis. 179:538-547. [DOI] [PubMed] [Google Scholar]
- 8.Berzofsky, J. A., et al. 1991. Construction of peptides encompassing multideterminant clusters of human immunodeficiency virus envelope to induce in vitro T cell responses in mice and humans of multiple MHC types. J. Clin. Invest. 88:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boritz, E., E. L. Rapaport, T. B. Campbell, J. R. Koeppe, and C. C. Wilson. 2007. CD4+ T cell targeting of human immunodeficiency virus type 1 (HIV-1) peptide sequences present in vivo during chronic, progressive HIV-1 disease. Virology 361:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campion, S., M. S. Cohen, A. J. McMichael, S. Galvin, and N. Goonetilleke. 2010. Improved detection of latent Mtb infection in HIV-1 seropositive individuals using cultured cellular assays. Eur. J. Immunol. doi: 10.1002/eji.201040296. [DOI] [PMC free article] [PubMed]
- 11.Card, C. M., et al. 2009. Decreased immune activation in resistance to HIV-1 infection is associated with an elevated frequency of CD4(+)CD25 (+)FOXP3(+) regulatory T cells. J. Infect. Dis. 199:1318-1322. [DOI] [PubMed] [Google Scholar]
- 12.Castelli, F. A., et al. 2008. Immunoprevalence of the CD4+ T-cell response to HIV Tat and Vpr proteins is provided by clustered and disperse epitopes, respectively. Eur. J. Immunol. 38:2821-2831. [DOI] [PubMed] [Google Scholar]
- 13.Clerici, M., et al. 1992. Cell-mediated immune response to human immunodeficiency virus (HIV) type 1 in seronegative homosexual men with recent sexual exposure to HIV-1. J. Infect. Dis. 165:1012-1019. [DOI] [PubMed] [Google Scholar]
- 14.Clerici, M., et al. 1989. Interleukin-2 production used to detect antigenic peptide recognition by T-helper lymphocytes from asymptomatic HIV-seropositive individuals. Nature 339:383-385. [DOI] [PubMed] [Google Scholar]
- 15.Colf, L. A., et al. 2007. How a single T cell receptor recognizes both self and foreign MHC. Cell 129:135-146. [DOI] [PubMed] [Google Scholar]
- 16.Corbet, S., et al. 2003. Optimization and immune recognition of multiple novel conserved HLA-A2, human immunodeficiency virus type 1-specific CTL epitopes. J. Gen. Virol. 84:2409-2421. [DOI] [PubMed] [Google Scholar]
- 17.Currier, J. R., et al. 2002. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods 260:157-172. [DOI] [PubMed] [Google Scholar]
- 18.Dean, M., et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 19.De Groot, A. S., et al. 1991. Human immunodeficiency virus reverse transcriptase T helper epitopes identified in mice and humans: correlation with a cytotoxic T cell epitope. J. Infect. Dis. 164:1058-1065. [DOI] [PubMed] [Google Scholar]
- 20.De Groot, A. S., et al. 2005. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine 23:2136-2148. [DOI] [PubMed] [Google Scholar]
- 21.Division of AIDS, National Institute of Allergy and Infectious Diseases. 20 November 2006, posting date. HPTN 052. A randomized trial to evaluate the effectiveness of antiretroviral therapy plus HIV primary care versus HIV primary care alone to prevent the sexual transmission of HIV-1 in serodiscordant couples. HIV Prevention Trials Network. http://www.hptn.org/research_studies/hptn052.asp.
- 22.Dyer, W. B., et al. 2008. Mechanisms of HIV non-progression; robust and sustained CD4+ T-cell proliferative responses to p24 antigen correlate with control of viraemia and lack of disease progression after long-term transfusion-acquired HIV-1 infection. Retrovirology 5:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erickson, A. L., et al. 2008. Potentially exposed but uninfected individuals produce cytotoxic and polyfunctional human immunodeficiency virus type 1-specific CD8(+) T-cell responses which can be defined to the epitope level. Clin. Vaccine Immunol. 15:1745-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabio, G., et al. 1990. Susceptibility to HIV infection and AIDS in Italian haemophiliacs is HLA associated. Br. J. Haematol. 75:531-536. [DOI] [PubMed] [Google Scholar]
- 25.Fonseca, S. G., et al. 2006. Identification of novel consensus CD4 T-cell epitopes from clade B HIV-1 whole genome that are frequently recognized by HIV-1 infected patients. AIDS 20:2263-2273. [DOI] [PubMed] [Google Scholar]
- 26.Fowke, K. R., et al. 1996. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 348:1347-1351. [DOI] [PubMed] [Google Scholar]
- 27.Fox, J., and S. Fidler. 2010. Sexual transmission of HIV-1. Antiviral Res. 85:276-285. [DOI] [PubMed] [Google Scholar]
- 28.Frahm, N., et al. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gahery-Segard, H., et al. 2000. Multiepitopic B- and T-cell responses induced in humans by a human immunodeficiency virus type 1 lipopeptide vaccine. J. Virol. 74:1694-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geretti, A. M., C. A. Van Baalen, J. C. Borleffs, C. A. Van Els, and A. D. Osterhaus. 1994. Kinetics and specificities of the T helper-cell response to gp120 in the asymptomatic stage of HIV-1 infection. Scand. J. Immunol. 39:355-362. [DOI] [PubMed] [Google Scholar]
- 31.Goh, W. C., et al. 1999. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J. Infect. Dis. 179:548-557. [DOI] [PubMed] [Google Scholar]
- 32.Goonetilleke, N., et al. 2006. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J. Virol. 80:4717-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hale, P. M., et al. 1989. T cell multideterminant regions in the human immunodeficiency virus envelope: toward overcoming the problem of major histocompatibility complex restriction. Int. Immunol. 1:409-415. [DOI] [PubMed] [Google Scholar]
- 34.Hasselrot, K., et al. 2009. Oral HIV-exposure elicits mucosal HIV-neutralizing antibodies in uninfected men who have sex with men. AIDS 23:329-333. [DOI] [PubMed] [Google Scholar]
- 35.Hatano, H., et al. 2009. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J. Virol. 83:329-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hladik, F., et al. 2003. Most highly exposed seronegative men lack HIV-1-specific, IFN-gamma-secreting T cells. J. Immunol. 171:2671-2683. [DOI] [PubMed] [Google Scholar]
- 37.Hudgens, M. G. 2003. Estimating cumulative probabilities from incomplete longitudinal binary responses with application to HIV vaccine trials. Stat. Med. 22:463-479. [DOI] [PubMed] [Google Scholar]
- 38.Kaufmann, D. E., et al. 2004. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J. Virol. 78:4463-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaul, R., et al. 2001. CD8(+) lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J. Clin. Invest. 107:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaul, R., et al. 2000. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J. Immunol. 164:1602-1611. [DOI] [PubMed] [Google Scholar]
- 41.Kebba, A., et al. 2004. HIV type 1 antigen-responsive CD4+ T-lymphocytes in exposed yet HIV type 1 seronegative Ugandans. AIDS Res. Hum. Retroviruses 20:67-75. [DOI] [PubMed] [Google Scholar]
- 42.Koning, F. A., et al. 2004. Correlates of resistance to HIV-1 infection in homosexual men with high-risk sexual behaviour. AIDS 18:1117-1126. [DOI] [PubMed] [Google Scholar]
- 43.Koning, F. A., et al. 2005. Low-level CD4+ T cell activation is associated with low susceptibility to HIV-1 infection. J. Immunol. 175:6117-6122. [DOI] [PubMed] [Google Scholar]
- 44.Leitner, T., et al. 2005. HIV sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 45.Liang, K.-Y., and S. L. Zeger. 1986. Longitudinal data analysis using generalized linear models. Biometrika 73:13-22. [Google Scholar]
- 46.Liu, R., et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 47.Lo, W. L., et al. 2009. An endogenous peptide positively selects and augments the activation and survival of peripheral CD4+ T cells. Nat. Immunol. 10:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo Caputo, S., et al. 2003. Mucosal and systemic HIV-1-specific immunity in HIV-1-exposed but uninfected heterosexual men. AIDS 17:531-539. [DOI] [PubMed] [Google Scholar]
- 49.Lubong Sabado, R., et al. 2009. In vitro priming recapitulates in vivo HIV-1 specific T cell responses, revealing rapid loss of virus reactive CD4 T cells in acute HIV-1 infection. PLoS One 4:e4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makedonas, G., et al. 2002. HIV-specific CD8 T-cell activity in uninfected injection drug users is associated with maintenance of seronegativity. AIDS 16:1595-1602. [DOI] [PubMed] [Google Scholar]
- 51.Mason, D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19:395-404. [DOI] [PubMed] [Google Scholar]
- 52.Mason, R. D., M. I. Bowmer, C. M. Howley, and M. D. Grant. 2005. Cross-reactive cytotoxic T lymphocytes against human immunodeficiency virus type 1 protease and gamma interferon-inducible protein 30. J. Virol. 79:5529-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McElrath, M. J., et al. 2008. HIV-1 vaccine-induced immunity in the test-of-concept step study: a case-cohort analysis. Lancet 372:1894-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michael, N. 2010. Correlates of immunity: RV144—lessons learned. Presented at the AIDS Vaccine Conference, Atlanta, GA. http://www.hivvaccineenterprise.org/conference_archive/2010/pdf-presentations/Friday/Plenary-03/MichaelN.pdf.
- 55.Missale, G., et al. 2004. Parenteral exposure to high HIV viremia leads to virus-specific T cell priming without evidence of infection. Eur. J. Immunol. 34:3208-3215. [DOI] [PubMed] [Google Scholar]
- 56.Moodie, Z., et al. 2010. Response definition criteria for ELISPOT assays revisited. Cancer Immunol. Immunother. 59:1489-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moser, J. M., et al. 2010. Optimization of a dendritic cell-based assay for the in vitro priming of naive human CD4+ T cells. J. Immunol. Methods 353:8-19. [DOI] [PubMed] [Google Scholar]
- 58.Nicastri, E., et al. 1999. Reduction of IFN-gamma and IL-2 production by peripheral lymphocytes of HIV-exposed seronegative subjects. AIDS 13:1333-1336. [DOI] [PubMed] [Google Scholar]
- 59.Pancre, V., et al. 2007. Presence of HIV-1 Nef specific CD4 T cell response is associated with non-progression in HIV-1 infection. Vaccine 25:5927-5937. [DOI] [PubMed] [Google Scholar]
- 60.Perez, C. L., K. Hasselrot, G. Bratt, K. Broliden, and A. C. Karlsson. 2010. Induction of systemic HIV-1-specific cellular immune responses by oral exposure in the uninfected partner of discordant couples. AIDS 24:969-974. [DOI] [PubMed] [Google Scholar]
- 61.Pettersson, F. E., and K. O. Gronvik. 2003. Long-term CD4+ and CD8+ memory T cells developed in severe combined immunodeficiency mice during homoeostasis exhibit differences in sensitivity to antigen. Scand. J. Immunol. 57:311-318. [DOI] [PubMed] [Google Scholar]
- 62.Rosenberg, E. S., et al. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. [DOI] [PubMed] [Google Scholar]
- 63.Rowland-Jones, S., et al. 1995. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 64.Rowland-Jones, S. L., et al. 1998. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Invest. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rowland-Jones, S. L., et al. 1993. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet 341:860-861. [DOI] [PubMed] [Google Scholar]
- 66.Rychert, J., S. Saindon, S. Placek, D. Daskalakis, and E. Rosenberg. 2007. Sequence variation occurs in CD4 epitopes during early HIV infection. J. Acquir. Immune Defic. Syndr. 46:261-267. [DOI] [PubMed] [Google Scholar]
- 67.Saez-Cirion, A., et al. 2006. Persistent resistance to HIV-1 infection in CD4 T cells from exposed uninfected Vietnamese individuals is mediated by entry and post-entry blocks. Retrovirology 3:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sallusto, F., and A. Lanzavecchia. 2009. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur. J. Immunol. 39:2076-2082. [DOI] [PubMed] [Google Scholar]
- 69.Schenal, M., et al. 2005. Distinct patterns of HIV-specific memory T lymphocytes in HIV-exposed uninfected individuals and in HIV-infected patients. AIDS 19:653-661. [DOI] [PubMed] [Google Scholar]
- 70.Schmechel, S. C., et al. 2001. Immune defence against HIV-1 infection in HIV-1-exposed seronegative persons. Immunol. Lett. 79:21-27. [DOI] [PubMed] [Google Scholar]
- 71.Shacklett, B. L., et al. 2002. Dendritic cell amplification of HIV type 1-specific CD8+ T cell responses in exposed, seronegative heterosexual women. AIDS Res. Hum. Retroviruses 18:805-815. [DOI] [PubMed] [Google Scholar]
- 72.Simonsen, J. N., et al. 1990. HIV infection among lower socioeconomic strata prostitutes in Nairobi. AIDS 4:139-144. [DOI] [PubMed] [Google Scholar]
- 73.Stockinger, B., C. Bourgeois, and G. Kassiotis. 2006. CD4+ memory T cells: functional differentiation and homeostasis. Immunol. Rev. 211:39-48. [DOI] [PubMed] [Google Scholar]
- 74.Suy, A., et al. 2007. Immunological profile of heterosexual highly HIV-exposed uninfected individuals: predominant role of CD4 and CD8 T-cell activation. J. Infect. Dis. 196:1191-1201. [DOI] [PubMed] [Google Scholar]
- 75.Tenenbaum, S. A., et al. 2005. Evidence of HIV exposure and transient seroreactivity in archived HIV-negative severe hemophiliac sera. Virol. J. 2:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomescu, C., et al. 2010. Increased plasmacytoid dendritic cell maturation and natural killer cell activation in HIV-1 exposed, uninfected intra-venous drug users. AIDS 24:2151-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Truong, L. X., et al. 2003. CD4 cell and CD8 cell-mediated resistance to HIV-1 infection in exposed uninfected intravascular drug users in Vietnam. AIDS 17:1425-1434. [DOI] [PubMed] [Google Scholar]
- 78.Wahren, B., et al. 1989. HIV-1 peptides induce a proliferative response in lymphocytes from infected persons. J. Acquir. Immune Defic. Syndr. 2:448-456. [PubMed] [Google Scholar]
- 79.Wang, S., et al. 2007. Identification of HLA-A11-restricted HIV-1-specific cytotoxic T-lymphocyte epitopes in China. Curr. HIV Res. 5:119-128. [DOI] [PubMed] [Google Scholar]
- 80.Wilson, C. C., et al. 2001. Identification and antigenicity of broadly cross-reactive and conserved human immunodeficiency virus type 1-derived helper T-lymphocyte epitopes. J. Virol. 75:4195-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, L., et al. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu, X. G., et al. 2002. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J. Virol. 76:8690-8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.