Abstract
Novel Epstein-Barr Virus (EBV) strains with deletion of either EBER1 or EBER2 and corresponding revertant viruses were constructed and used to infect B lymphocytes to make lymphoblastoid cell lines (LCLs). The LCLs were used in microarray expression profiling to identify genes whose expression correlates with the presence of EBER1 or EBER2. Functions of regulated genes identified in the microarray analysis include membrane signaling, regulation of apoptosis, and the interferon/antiviral response. Although most emphasis has previously been given to EBER1 because it is more abundant than EBER2, the differences in cell gene expression were greater with EBER2 deletion. In this system, deletion of EBER1 or EBER2 had little effect on the EBV transformation frequency of primary B cells or the growth of the resulting LCLs. Using the recombinant viruses and novel EBER expression vectors, the nuclear redistribution of rpL22 protein by EBER1 in 293 cells was confirmed, but in LCLs almost all of the cells had a predominantly cytoplasmic expression of this ribosomal protein, which was not detectably changed by EBER1. The changes in LCL gene expression identified here will provide a basis for identifying the mechanisms of action of EBER RNAs.
Epstein-Barr virus (EBV) is a ubiquitous gammaherpesvirus that persistently infects more than 90% of the world's population and contributes to the development of several lymphoid and epithelial malignancies, including Burkitt's lymphoma (BL), Hodgkin's lymphoma (HL), nasal T/NK lymphomas, nasopharyngeal carcinoma (NPC), and some gastric carcinoma cases (19).
The most abundant viral transcripts in latent EBV infection are functional RNAs, which may allow the virus to influence the infected cell while avoiding immune surveillance. EBV uses several forms of small noncoding RNAs (43), expressing small nucleolar RNAs (15), numerous miRNAs (3, 33, 34), and two noncoding EBV-encoded RNAs, EBER1 and EBER2 (27). EBERs are nontranslated, nonpolyadenylated, RNA polymerase III-transcribed RNAs of 167 and 172 nucleotides (27, 37). EBERs are abundantly expressed during persistent infection, including EBV-associated cancers, but their expression is reduced in the virus lytic cycle, implying an important role of EBERs during EBV latency (27, 39). EBER1 and EBER2 form similar stable stem-loops by intramolecular base pairing, which enables interaction with several cellular proteins (12).
EBER1 interacts strongly with ribosomal protein L22 (rpL22), a constituent of the large ribosomal subunit (47). Up to three L22 proteins can be bound by one EBER1 molecule simultaneously on binding sites in stem-loops 1, 3, and 4 (7, 9, 11, 13, 46). L22 can become delocalized from nucleoli and cytoplasm into the nucleoplasm in the presence of EBV (45) or EBER1 (13). Absence of L22 from nucleoli might result in a depletion of the protein from ribosomes, as shown in EBV-infected Akata cells (13). In contrast, L22 has previously been found to be present in ribosomes of EBV-positive Raji cells (45). The physiological function of L22 is unclear since it is not essential for survival or overall development of L22 knockout mice or for protein synthesis in vitro (1, 23). Interestingly, L22 deletion in mice results in a p53-dependent apoptosis of αβ-T lymphocyte precursors selectively, while γδ-T cell precursors are spared (1). Interaction of L22 with human telomerase RNA (24) and histone H1 (31) might indicate other possible roles of EBER1-L22 interaction. L22 also binds to herpes simplex virus 1-infected cell proteins ICP4 and ICP22 (25, 26), equine herpesvirus 1 immediate-early gene protein IE (20), and the 3′X-untranslated region of hepatitis C virus (49).
EBERs also interact with retinoic acid-inducible gene I (RIG-I) protein (40), which detects viral double-stranded RNA and activates NF-κB and interferon regulatory factor 3 to initiate the production of type I interferon and inflammatory cytokines. RIG-I-mediated signaling enhances the expression of interleukin-10 (IL-10) and has been reported to thus promote growth in BL cells in response to EBER presence (21, 41). In addition, EBERs were found to trigger the induction of autocrine growth factor insulin-like growth factor IGF-1 in nasopharyngeal carcinoma and gastric carcinoma-derived epithelial cells (16, 17), in addition to IL-9 in T cells (53).
Interferon-inducible serine/threonine kinase PKR can also be bound by EBERs to modify its phosphorylation and activation in vitro, potentially preventing alpha interferon (IFN-α)-induced apoptosis and inhibition of protein translation (4, 28, 29). However, EBERs are mainly localized in the nucleus (11, 14), making an interference with the cytoplasmic function of PKR improbable in vivo. Consistent with this, EBERs were found to have no effect on the phosphorylation status of PKR or its direct substrate, eukaryotic initiation factor 2α, in EBV-infected BL cells, but were nevertheless required for protection from IFN-induced apoptosis (38).
EBER1 has been reported to be secreted from EBV-infected cells in complex with La (11, 27), a protein generally involved in biogenesis and maturation of polymerase III transcripts, and was shown to consequently be able to induce an IFN response by stimulating Toll-like receptor 3 signaling in adjacent cells (18). EBER2 was not found to be secreted in that study.
EBER2 has been reported to be required for efficient transformation of B lymphocytes and maximum growth potential of lymphoblastoid cell lines (LCLs), enhancing IL-6 expression to assist LCL growth (50, 51). However, other studies have found EBERs not to be essential for primary infection, viral replication, or B-cell immortalization (44). EBERs have been linked to the establishment of malignant phenotypes and tumor formation in immunodeficient SCID mice (22, 42, 52) and to resistance to apoptotic inducers and upregulation of BCL-2 oncoprotein, which can protect BL-derived Akata cells from c-Myc-induced apoptosis (22, 38). Transgenic mice expressing EBERs in the lymphoid compartment were also found to develop lymphoid hyperplasia and, in some cases, lymphoma (36).
Although there has been considerable progress, the exact roles of EBER1 or EBER2 during EBV infection and their individual mechanisms of action are still not clear. In the long term, a better understanding of EBER function might lead to novel therapeutic approaches for the treatment of EBV-associated cancers based on the presence of EBERs in the tumor cells. With this in mind, we have therefore focused on the consequences of EBER expression that are intrinsic to EBV-infected cells. In the present study we have created novel deletion mutants of EBER1 or EBER2 in the B95-8 EBV BAC. Deletion of EBER1 or EBER2 did not affect transformation efficiency or growth rate of LCLs in this system, but we confirmed the relocation of L22 protein in 293 cells using the EBV mutants and revertants. Microarray analysis identified host cell genes whose expression in LCLs correlates with EBER1 or EBER2 expression. Although most attention thus far has been given to EBER1 (because it is more abundant), the alterations in LCL gene expression caused by the absence of EBER2 were clearer than those associated with the absence of EBER1.
MATERIALS AND METHODS
Construction and characterization of EBER BAC mutants.
The EBER genes were deleted individually from the B95-8 EBV BAC (5) by RecA-mediated homologous recombination. Targeting constructs were cloned between the BamHI and SalI sites of pKov-Kan-ΔCm (p4415.3, kanamycin resistant, sucrose sensitive) as described previously (48). In each case, EBER sequence was replaced with an XbaI site for monitoring the deletion. The EBER1 deletion replaced EBV bases 6599 to 6798 inclusive with TCTAGA; the targeting plasmid contained EBV bases 6066 to 7322 as a flanking sequence (BamHI was added at the 6066 end). The EBER2 deletion replaces bases 6950 to 7130 inclusive with TCTAGA; this targeting plasmid contained EBV bases 6669 to 7413 as a flanking sequence (BamHI was added at the 6669 end). For reversion to parental sequence, the EBER1 deletion was reverted with a targeting plasmid containing wild-type (wt) EBV bases 6066 to 7322; for the EBER2 deletion, a targeting plasmid containing wt EBV bases 6669 to 7413 was used for the revertant, again with cloned BamHI to SalI in p4415.3.
B95-8 EBV BAC (p2089, chloramphenicol resistant) in Escherichia coli strain DH10B was kindly provided by W. Hammerschmidt (5). For the production of EBER deletion mutants, targeting plasmids were transformed into HB9 bacteria with pDF2.5-tet (expressing RecA, tetracycline resistant, ts replication origin, only grows at 30°C). Transformed cells were selected on chloramphenicol (Cm), kanamycin (Kan), and tetracycline (Tet) at 30°C overnight, and small pools of colonies were then cured of RecA by growth on Cm and Kan at 42°C overnight. Correct cointegrants were identified by PCR and restriction digestion analysis on pulsed-field gels. Competent bacteria from these characterized cointegrants were then resolved by again transforming pDF2.5, selecting on Cm and Tet at 30°C overnight and curing small pools of colonies of p4415.3 vector sequence by liquid growth in sucrose, Cm, and Tet at 30°C, followed by plating on sucrose and Cm and growth at 42°C overnight to remove the RecA plasmid. Correctly resolved deletion mutants (which were now sensitive to Kan on replica plating) were identified by PCR and restriction digestion analysis on pulsed-field gels. Reversion of the mutant bacterial artificial chromosome (BAC) to wt sequence was accomplished by a similar procedure, starting with competent E. coli containing the deletion mutant and using the indicated reversion targeting plasmid.
Restriction digests of BAC DNA were analyzed by pulsed-field gel electrophoresis using a Bio-Rad CHEF DR II apparatus on 1% agarose gels in 0.5× Tris-borate-EDTA at 14°C. The electrophoresis was for 14 h at 6 V/cm, with a start switch of 1 and an end switch of 10.
Production of 293 cells and LCLs containing BAC EBV with EBER deletion.
The BAC vector contains a hygromycin resistance gene and green fluorescent protein (GFP). BAC DNA was purified on Qiagen columns and transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen). Hygromycin-resistant colonies were isolated, and cell lines were grown out. 293 cells were grown in Dulbecco modified Eagle medium with 10% fetal calf serum (FCS) and 50 U of penicillin-streptomycin/ml, and 293 EBV-BAC cells were maintained with the addition of 0.1 μg of hygromycin B/μl. The BAC DNA in these lines was recovered into E. coli and analyzed by restriction enzyme digestion and PCR to ensure that there were no detectable rearrangements or deletions apart from the desired EBER mutations. The lines were also tested for proper EBER expression and the ability to produce infectious EBV. EBV from the 293 cell lines that passed all of these quality controls was then used for infection of human B lymphocytes purified from peripheral blood to test the transformation efficiency and produce LCLs. LCLs were grown in RPMI supplemented with 10% FCS and penicillin-streptomycin. Infectious virus was produced by transfecting 293 cell lines containing EBV-BACs with BZLF1 and BALF4 expression plasmids (6, 30). Supernatant was harvested, filtered through a 0.45-μm-pore-size membrane, and infectivity was assessed by counting GFP-positive Raji cells (green Raji units [GRU]). The resulting LCLs used for subsequent microarray analysis were again checked for correct EBER expression and normal levels of EBNA proteins and LMP1.
Northern blotting for EBERs.
Total cell RNA was treated with glyoxal and electrophoresed on agarose gels in 10 mM sodium phosphate (pH 7.0) prior to blotting on to Biodyne A nylon membrane (Pall) and probing with 32P-labeled PCR products corresponding to EBER1 or EBER2 RNA sequence made with the primers P0364 and P0365 or the primers P4112 and P4114, respectively (10). The probes were labeled with a Megaprime DNA labeling system (GE Healthcare).
B-cell transformation and growth assays.
Primary B cells were isolated from buffy coats by negative selection with the RosetteSep system (Stem Cell Technologies). A total of 105 B cells per well were infected with 2-fold serial dilutions of recombinant EBVs, starting with 4,000 GRU/ml. Cells were maintained in RPMI supplemented with 20% FCS, with half of the medium being exchanged every 3 days. To exclude any possible contribution of spontaneous transformation when growing out LCLs from low-input amounts of EBV, 0.15 μg of hygromycin B/μl was added temporarily after 3 weeks for approximately 2 weeks to ensure that only BAC EBV LCLs would survive. No hygromycin was added while cells were under experimental analysis or during the growth transformation assays.
For the growth transformation assay, 1.5 × 105 primary B cells per well were plated in 96-well plates and then infected with serial 5-fold dilutions of each EBV type. Half of the culture medium was replaced with fresh medium every 2 to 3 days, and the number of wells with proliferating B cells was counted 4 weeks postinfection. The titers for transformation (50% transforming doses [TD50]) were calculated by the Reed-Muench method (35). For the LCL growth assay, 2-fold serially diluted LCLs were plated in 96-well plates starting with 1,000 cells/well. Half of the culture medium was replaced with fresh medium every 3 days. The number of wells with proliferating B cells was counted 4 weeks after the start of the assay. In addition, 5 × 104 LCLs were cultured in 1 ml of medium for 10 days to examine their growth. The number of viable cells was counted every 2 days with trypan blue.
RNA extraction and reverse transcription-PCR (RT-PCR).
Total cell RNA was extracted with TRIzol (Invitrogen). Cytoplasmic RNA was extracted with the RNeasy kit and treated on the column with RNase-free DNase (Qiagen). cDNA was prepared with a Protoscript first-strand cDNA synthesis kit (New England Biolabs) using oligo(dT). To detect the expression of genes regulated with EBER expression, a GoTaq PCR system (Promega) was used with the primers listed in Fig. S1 in the supplemental material.
Microarray analysis.
For the main experiment (the 2010 experiment) presented here, microarray analysis was performed with Agilent G4112F ID 014850 whole human genome oligonucleotide microarrays. Four to six samples for each of the EBV wt, mutant, or revertants were used for the analysis. Synthesis of Cy3-labeled cRNA was performed with a Quick Amp labeling kit (one color, catalog no. 5190-0442; Agilent Technologies) according to the manufacturer's recommendations. cRNA fragmentation, hybridization, and washing steps were also carried out as recommended by the manufacturer's one-color microarray-based gene expression analysis protocol (v5.7; Agilent Technologies). Slides were scanned on an Agilent MicroArray Scanner G2565CA at two different photomultiplier tube (PMT) settings (100 and 5%) to increase the dynamic range of the measurements (extended-dynamic-range mode). The data extraction was performed in accordance with Agilent's feature extraction software v9.5.3.1 by using the recommended default extraction protocol file (GE1-v5_95_Feb07.xml).
Data were processed using Agilent's feature extraction software v9.5.3.1. The data were normalized using an inter-array approach (global linear scaling to one arbitrary reference array, according to the 75th percentiles of intensity distribution). In addition, a lower-intensity threshold of 47 was used as a surrogate value for all measurements that fell below the intensity threshold.
For the 2008 experiment shown in the present study, the same microarrays were used in a dual-color mode with a B95-8 LCL as the reference RNA. Synthesis of Cy3- or Cy5-labeled cRNA was performed with a QuickAmp labeling kit (two color, catalog no. 5190-0444; Agilent Technologies). Three independent samples were used for the EBER2 mutant or parental BAC LCL; in this case, tonsil B cells were used to make the LCLs. Subsequent steps were carried out according to the manufacturer's two-color microarray-based gene expression analysis protocol (v5.7; Agilent Technologies). The data extraction was as described above using the extraction protocol file GE2-v5_95_Feb07.xml.
Microarray data analysis was performed with Genomics Suite software (Partek) and Genedata Expressionist software (Genedata AG). The principal component analysis shown in Fig. 3B was performed with Partek software using the covariance method. Differences in gene expression levels according to EBER expression were defined with analysis of variance (ANOVA) using both Partek and Genedata software. Differentially regulated genes were identified by comparing deletion mutant samples with the wt and revertant samples, requiring a fold change of at least 2, with a P value of <0.001. A false discovery rate of 1% was applied to the P values for the comparisons, which resulted in the lists of EBER1 and EBER2 regulated genes in the LCLs shown in Fig. S2 and S3 in the supplemental material.
Western blotting.
Radioimmunoprecipitation assay protein extracts were fractionated by SDS-PAGE and transferred to nitrocellulose. After blocking with 5% milk powder in phosphate-buffered saline (PBS)-0.1% Tween 20, the membrane was probed with anti-CXCR3 (1/1,000 dilution; Abcam) and anti-β-actin (1/2,500, AC-15; Sigma). The secondary antibodies were horseradish peroxidase-conjugated anti-rabbit (1/2,000; Dako) or anti-mouse (1/10,000; Sigma), and bound immunocomplexes were detected by enhanced chemiluminescence (GE Healthcare).
EBER expression plasmids.
The plasmids pBSJJJ1 and pBSJJJ2 contain the B95-8 EBER1 or EBER2 gene, respectively, in pBluescript and can express the EBER RNAs from their own promoters (32). The EBER1 gene was PCR amplified with the primers CCAGATCTCCAGGACCTACGCTGCCCT and CCAAGCTTGGATGCATAAATCCTAA, and the EBER2 gene was PCR amplified with the primers CCAGATCTCCAGGACAGCCGTTGCCCT and CCAAGCTTGGGTGCAAAACTAGCCA, in each case introducing a BglII site at the 5′ end and a HindIII site at the 3′ end. The digested products were cloned between the BglII and HindIII sites of pSUPER (2), the resulting expression plasmids being named pEBER1 and pEBER2, respectively. The region of pSUPER containing the H1 promoter and EBER gene was also cloned as a BamHI-to-XhoI fragment between the BamHI and SalI sites of pHEBo (54) to create pHEBo-EBER1 or pHEBo-EBER2, giving EBER expression plasmids carrying EBV oriP and a hygromycin resistance gene.
32P labeling of cells.
At 16 h prior to radiolabeling, the cells were seeded into wells at a density of 2 × 105 cells/ml in 4 ml of complete medium. Whole-cell RNA was radiolabeled by the addition of 1 mCi of [α-32P]sodium orthophosphate to normal growth medium, followed by incubation of the cells at 37°C for 36 h.
L22 immunofluorescence microscopy.
The cells were transfected with FLAG-L22 plasmid (expressing N-terminal FLAG-tagged L22 from pBK2-CMV) using Lipofectamine 2000. Coverslips were pretreated with gelatin or poly-l-lysine in six-well plates, and 5 × 105 cells/well were added and grown overnight. The cells were washed twice in PBS, fixed with 4% (vol/vol) paraformaldehyde for 45 min at room temperature, and washed three times in PBS before quenching them with 50 mM ammonium chloride for 10 min at room temperature. After three PBS washes, the cells were permeabilized with 0.2% Triton X-100 in PBS for 7 min, washed four times in PBS, and blocked for 30 min in 1% or 5% (wt/vol) bovine serum albumin in PBS. The cells were then stained with α-rpL22 rabbit polyclonal (kindly provided by J. Steitz) or α-Flag M2 mouse monoclonal antibodies (Sigma) and then incubated with secondary antibody (α-rabbit IgG goat-TRITC [Sigma] or α-mouse IgG goat-TRITC [Sigma]). After a washing step, the cells were counterstained with 2 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml and mounted in Mowiol (Calbiochem) on a carrier slide. Slides were analyzed with a Zeiss LSM Pascal or 510 Meta confocal microscope. To quantify the L22 localization, the staining patterns of 100 cells/slide were evaluated in two to four randomly chosen fields and categorized into cytoplasmic, nuclear, or nucleolar L22 localization according to the strongest L22 staining visible in individual cells. Some cells showed equally strong staining in the cytoplasm and nucleolus and were scored in both categories, as indicated where the totals exceed 100% in Table 1.
TABLE 1.
Distribution of endogenous L22 staining in cell linesa
| Cell line | Cytoplasm (%) | Nucleoplasm (%) | Nucleolus (%) |
|---|---|---|---|
| LCL wt EBV | 99.5 | 0.5 | 0 |
| LCL delEBER1 EBV | 99.5 | 0 | 0.5 |
| BJAB + H1 vector | 95 | 0 | 5 |
| BJAB + EBER1 | 98 | 0.5 | 2 |
| BJAB + EBER2 | 97 | 0 | 6 |
| 293 | 95 | 0 | 5 |
| 293 + EBV | 60 | 40 | 0 |
| AGS | 100 | 0 | 5 |
| AGS + EBV | 90 | 10 | 0 |
| HONE1 | 100 | 0 | 25 |
| HONE1 + EBV | 98 | 2 | 0 |
The numbers indicate the percentages of cells in which the predominant staining observed was in the cytoplasm, nucleoplasm, or nucleolus. Some cells showed equally strong staining in the cytoplasm and nucleolus and were scored in both categories; this is indicated where the totals exceed 100%. wt, wild type.
RESULTS
Recombinant EBV BAC lacking EBER1 or EBER2 genes.
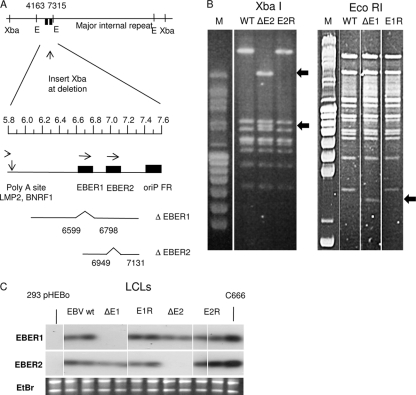
EBV BAC strains with deletion of EBER1 or EBER2 were created by recombination in E. coli of the B95-8 EBV BAC (5) with specific targeting vectors. For deletion of EBER1, both EBER1 transcript and promoter sequences were removed (Fig. 1 A) to avoid the EBER1 promoter upstream sequences potentially disturbing the EBER2 promoter in the recombinant. The EBER2 deletion removed only the EBER2 transcript sequences (Fig. 1A). The EBER mutant BACs that were selected for further study were then mutated back to wt EBER sequence to create revertants as controls. BACs were screened by restriction digestion using AgeI, EcoRI, or XbaI to ensure there were no rearrangements or unwanted deletions in the EBV genomes. An XbaI site was inserted at the point of EBER deletion to facilitate identification of the recombinants, and examples of the pulsed-field electrophoresis analysis of restriction digests are shown in Fig. 1B. The highly characterized BAC DNA (containing a hygromycin resistance gene) was transfected into 293 cells, and cloned cell lines resistant to hygromycin were isolated. These were further characterized by rescuing the BAC plasmid into E. coli and examining again by restriction digestion and PCR across the deletion points (data not shown) to ensure the correct genome modifications were present in the EBV mutants and were returned to the wild type in the revertants. Infectious EBV was recovered and LCLs were produced by infection of human B lymphocytes from peripheral blood or tonsils. EBER expression was determined in the LCLs by Northern blotting total cell RNA with probes for EBER1 or EBER2. The results (Fig. 1C) show the predicted pattern of EBER expression and confirm normal levels of expression of the remaining EBER RNA when one of the EBER genes was deleted. Protein extracts from the LCLs were also tested for the expression of EBNA and LMP1 proteins, and all those tested showed the characteristic levels of EBNA and LMP1 protein expression found in LCLs (data not shown).
FIG. 1.
(A) Summary of EBER deletions introduced into EBV BAC. The relative positions of XbaI and EcoRI (E) restriction sites in the EBV genome are shown; the EcoRI sites at bases 4163 and 7315 flank the EBER genes, represented by the filled boxes. The part of the EBV genome between bases 5800 and 7600 is expanded below under a scale in kilobases. Below that, the EBV content of the targeting plasmids is shown; the deletion of EBER1 (bases 6599 to 6798) includes the EBER1 promoter and the EBER1 sequence. The EBER2 deletion (bases 6949 to 7131) just removes the EBER2 transcript sequence. In both mutants an XbaI site was introduced at the point of deletion to facilitate the analysis. (B) Restriction digestion of BAC DNA analyzed by pulsed-field gel electrophoresis. In the left panels, the XbaI site introduced at the EBER deletion point splits the largest XbaI fragment into two fragments (arrowed) in the XbaI digest; this example is EBER2 mutant and revertant BAC DNA recovered from transfected 293 cells. In the right panel, the EcoRI fragment (bases 4163 to 7315) containing the EBER genes is reduced in length in the EcoRI digest of the EBER1 deletion mutant (arrowed). The largest EcoRI and XbaI restriction fragments span the major internal repeat of EBV, and the similar sizes of these fragments in the parental (WT) and revertant (E1R and E2R) indicate that no loss of internal repeat sequences has occurred in the recombinations. (C) Northern blots of total RNA extracted from the indicated LCLs confirm the expected patterns of EBER expression. Negative control (293pHEBo) and positive control (C666.1) samples are indicated, and the corresponding ethidium bromide stain of rRNA is shown below to confirm equal RNA loading on the gels. Probe specific activities and hybridization efficiencies gave approximately equal signals for EBER1 and EBER2 on these blots.
No significant effect of EBER deletion on transformation efficiency or growth of LCLs.
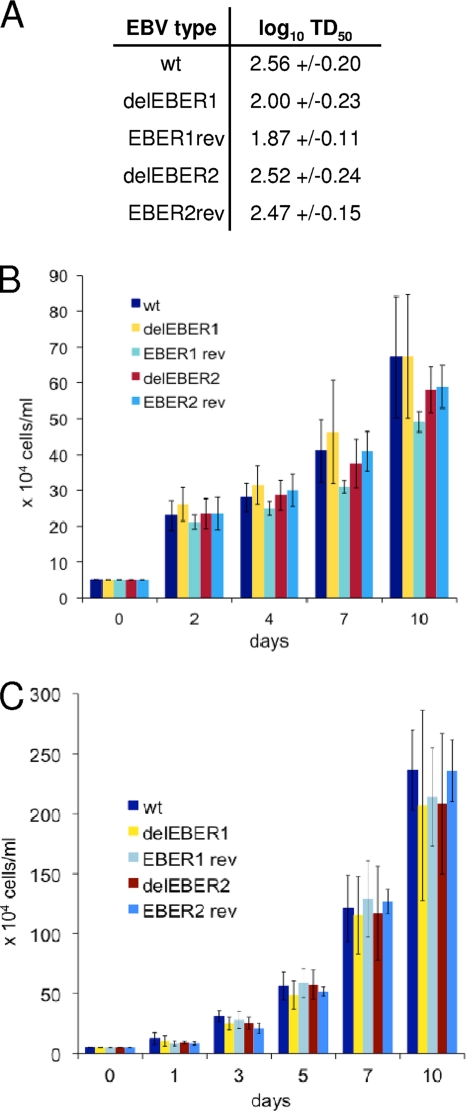
The EBV BAC contains a GFP gene. EBV produced from 293 cells containing parental, EBER deletion or revertant BACs was carefully titered by infection of Raji cells and counting green cells to give a titer in GRU. Equal amounts of GRU of the different viruses were then tested in dilution assays for the ability to transform human B cells into LCLs. The results were expressed as TD50, the transforming dose required to give 50% of wells transformed (Fig. 2 A); there was little effect of either EBER deletion on the transformation efficiency. This contrasts with the results of a previous study where deletion of EBER2 caused a 50-fold reduction of transformation efficiency (50); it should be noted that a different EBV strain background was used in the two experiments.
FIG. 2.
(A) Comparison of the transformation abilities (TD50) of different types of recombinant EBV. Two independent EBVs were used per EBV type and each infection was done in triplicate. Values are given as log10 TD50 with the standard deviation. (B and C) LCL growth assay. LCLs established with the parental EBV or different EBV mutants (delEBER1, EBER1rev, delEBER2, and EBER2rev) were compared for growth rates by plating them at 5 × 104 cells/ml in 24-well plates. The number of cells was counted every 1 to 3 days in a 10-day period, and the results represent the mean values with standard deviations based on six (B) or three (C) experiments. Two independent sets of LCLs (B and C), each derived from B cells isolated from a different blood donor, were used in the assay. For each EBV mutant and the wt EBV, two independent LCLs were analyzed.
Consistent with the lack of effect on transformation, the growth of LCLs did not vary significantly according to EBER gene expression (Fig. 2B and C). There was also little effect (data not shown) of EBER expression on cell growth rate when starting from very low cell densities (starting cell densities between 250 and 4,000 cells/ml were additionally tested). A similar lack of effect of EBER deletion on growth rate or transformation efficiency was reported previously (44) in the B95-8 EBV background, although in that study the mutation deleted both EBER genes. The lack of effect on cell growth was helpful for our subsequent microarray expression profiling since the expression profiling would not simply be identifying genes generically involved in cell proliferation.
Effects of EBER deletion on microarray expression profiles of LCLs.
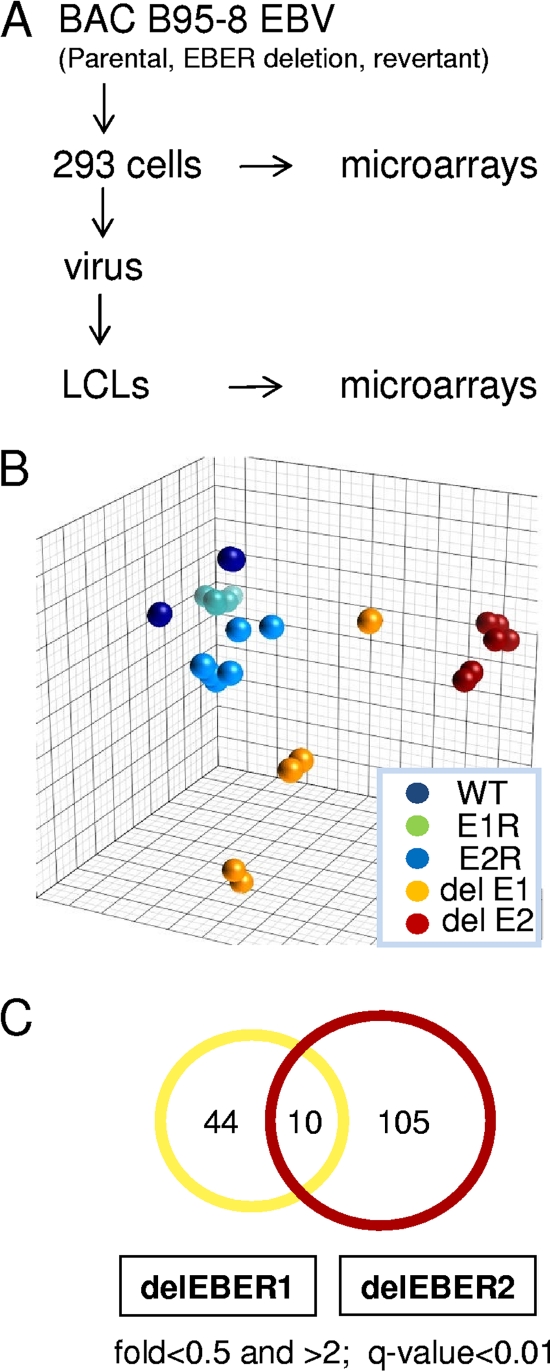
Cytoplasmic RNA from LCLs representing parental EBV, EBER1 deletion, EBER2 deletion, and their respective revertants was assayed for cell gene expression by using Agilent microarrays. These LCLs had all been made using B cells from the same blood donor and RNA from four to six LCLs for each condition was used separately for the microarray analysis (2010 experiment). The microarray strategy is summarized in Fig. 3A. The expression profiles were analyzed using both Genedata and Partek software to identify systematic differences in expression patterns according to EBER status. Principal component analysis (Fig. 3B) showed that EBER1 and EBER2 deletion mutant LCLs segregate away from the wt samples. EBER2 deletion had a more significant effect on gene expression than did EBER1. Most previous investigations of EBER function have focused on EBER1 because it is expressed at higher levels than EBER2, but this result suggests that EBER2 is more likely to have specific effects on LCL gene expression. The LCLs made with revertant EBVs behaved similarly to those with parental EBV, as should be expected.
FIG. 3.
(A) Experimental design for microarrays. (B) Principal component analysis (Partek software) for differences in cell gene expression pattern comparing microarray data from LCLs containing parental EBV (WT), EBV with deletion of EBER1 (del E1), deletion of EBER2 (del E2), revertant of EBER1 deletion (E1R), or revertant of EBER2 deletion (E2R). (C) Venn diagram showing the number of EBER1 or EBER2 regulated genes.
Genes whose expression in the LCLs differed according to EBER expression with high statistical significance were defined by using ANOVA with both Genedata and Partek software. Differentially regulated genes were identified by comparing the wt or revertant samples with deletion samples using an arbitrary fold change threshold of 2 with a P value of <0.001. Lists of differentially regulated genes identified by both analyses are shown in Fig. S2 (for EBER1 deletion) and S3 (for EBER2 deletion) in the supplemental material. A summary of the number of genes and degree to which the regulated genes were unique to EBER1 or EBER2 is shown in the Venn diagram in Fig. 3C. Because of the greater number of differences in cell gene expression, we focused first on EBER2.
EBER2 in LCLs.
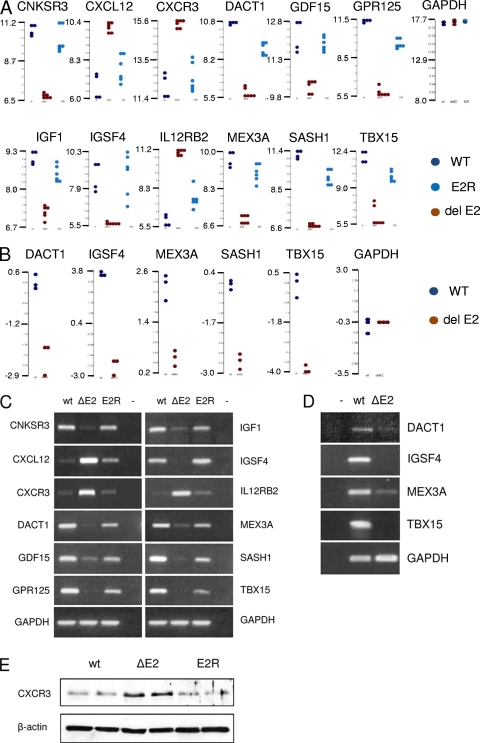
We identified 115 genes that were significantly and consistently regulated with EBER2 expression in the LCLs. To look more specifically at some of the cell genes whose expression correlated with EBER2 expression, the expression values from individual cell lines are shown as dot plots in Fig. 4 A. In each case, the expression level in parental and revertant is similar, but the expression in the EBER2 deletion is consistently different. Some additional data from an earlier comparison of ΔEBER2 and parental LCLs (2008 experiment, Fig. 4B) are shown and are consistent. That independent experiment involved making separate 293 producer cells and separate multiple LCLs from another B-cell donor and was conducted 2 years prior to the main experiment, confirming the reliability of the differences observed. The differences in expression level were also tested by RT-PCR, which confirmed the microarray results (Fig. 4C and D) for all of the 23 targets tested, 12 of which are shown in Fig. 4C. The effects on gene expression may in many cases be indirect; this analysis gives no information on the mechanism of action of EBER2, but it does indicate for the first time that LCL gene expression is modified according to the presence of EBER2. The examples shown in Fig. 4A include genes involved in receptor function and signaling (CNKRS3, CXCL12, CXCR3, DACT1, GDF15, GPR125, IGF1, and IL12RB2A), cellular adhesion (IGSF4), a transcription factor (TBX15), an RNA binding protein (MEX3A), and a proposed tumor suppressor gene (SASH1). Some of the differences in gene expression observed will probably be secondary consequences of the effects of EBER deletion, but an example of altered protein expression correlating with the deletion of EBER2 is shown for CXCR3 in Fig. 4E. As expected from the RNA data, the level of CXCR3 protein was higher in LCLs containing EBV with a deletion of EBER2 than in LCLs containing the parental or revertant virus.
FIG. 4.
(A) Dot plots of EBER2 regulated genes in LCLs (2010 experiment with revertants). Each point represents the level of expression in a separate LCL, plotted on a log base 2 scale. (B) Dot plots of EBER2 regulated genes in LCLs from a 2008 experiment common with a 2010 experiment on a log base 2 scale. (C) RT-PCR of selected genes shown in the dot plots from the 2010 experiment. (D) RT-PCR of selected genes shown in the dot plots from the 2008 experiment. (E) Western blot of CXCR3 showing dependence on EBER2. Protein extracts from two independent LCLs for each EBV variant are shown; actin served as a loading control.
EBER1 in LCLs and comparison with 293 cell expression profiling for EBER1 or EBER2 deletion.
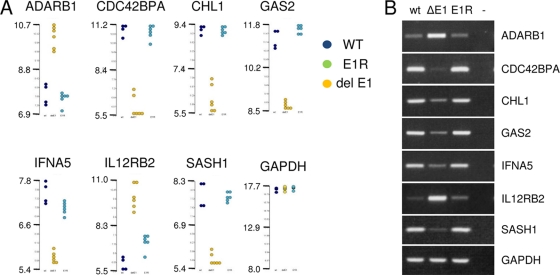
For EBER1, the differences in principal component analysis in LCLs were less pronounced, but clear differences were identified by using ANOVA (see Fig. S3 in the supplemental material). A total of 54 genes were significantly differentially expressed when the wt or revertant LCLs were compared to EBER1 deletion LCLs. The relative expression values of selected genes again indicated clear effects in the dot plot representation of expression values of individual cell lines (Fig. 5 A). A total of 11 of 12 of the genes tested by RT-PCR were confirmed (of which 7 are shown in Fig. 5B). Functions of the examples shown in Fig. 5 include deaminase (ADARB1), protein kinase (CDC42BPA), cell adhesion (CHL1), regulation of apoptosis (GAS2), and receptor signaling (IFNA5 and IL12RB2).
FIG. 5.
(A) Dot plots of EBER1 regulated genes in LCLs (2010 experiment with revertants) on a log base 2 scale. (B) RT-PCR of selected genes shown in the dot plots from 2010 experiment.
The HEK293 cell lines carrying the EBV BAC plasmid that were created as intermediates in the production of LCLs also express the EBER RNAs in the predicted patterns according to the deletion of EBER1 or EBER2 (data not shown). Cytoplasmic RNA from the multiple 293 cell lines (parental, EBER deletion, and revertant; three to four lines of each) used for the production of viruses was therefore also tested by a similar expression profiling technique on Agilent microarrays, and the data were analyzed by using Partek and Genedata software. Using the same high-stringency criteria in the ANOVA of at least 2-fold regulation with a Benjamini-Hochberg q value of <0.01 (1% false discovery rate in a t test), we did not identify any genes that were consistently regulated at the level of cytoplasmic RNA according to EBER status in EBV-infected 293 cells.
EBER1 relocates L22 away from nucleoli in some cell lines, but L22 in LCLs is mainly cytoplasmic and its location is not detectably changed by EBER1.
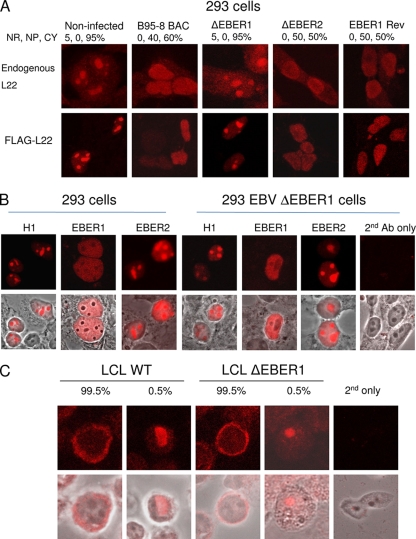
Several previous studies have shown that EBER1 can specifically bind to ribosomal protein L22 and cause a relocation of L22 from a nucleolar location in the nucleus into the nucleoplasm but this has not previously been demonstrated for endogenous L22 by the use of recombinant EBV strains and revertants. Confocal microscopy was used to visualize either endogenous L22 (Fig. 6 A, upper panels) or epitope-tagged L22 (Fig. 6A, lower panels) in 293 cell lines containing the parental BAC EBV, EBER1, or EBER2 deletion mutant EBV or the corresponding revertant EBV. Quantitation of the results confirmed the clear effects of EBER1 but not EBER2 on the L22 location in a proportion of the cells in the culture (Table 1).
FIG. 6.
(A) 293 cells infected as indicated with B95-8 BAC EBV (WT) or BAC EBV with deletion of EBER1, EBER2, or the revertant of the EBER1 deletion were stained for expression of endogenous L22 (upper panel) or transfected FLAG-L22 (lower panel). Staining of the endogenous L22 was classified as predominantly nucleolar (NR), nucleoplasmic (NP), or cytoplasmic (CP), and the percentage of cells with each type of staining, averaged from 100 fields of view, is shown above the image. (B) 293 cells or 293 cells containing the BAC EBV with EBER1 deleted were transfected with the indicated plasmids and with a FLAG-L22 expression plasmid. FLAG-L22 immunofluorescence and phase-contrast overlay images are shown. (C) LCLs stained for endogenous L22 show a predominantly cytoplasmic distribution.
Further confirmation of the specificity of the effect of EBER1 on L22 in 293 cells was provided by transfection of novel expression plasmids for EBER1 and EBER2 into the cells containing EBV with deletion of EBER1 or EBER2. These plasmids use the human H1 promoter in combination with the internal polymerase III promoter elements in the EBER gene to achieve wild-type expression levels of EBER RNA in transfected cells (see Fig. S4A in the supplemental material). The EBER1 RNA produced by this type of plasmid has exactly the same length as the endogenous EBER1 RNA in B95-8 cells (see Fig. S4B in the supplemental material) and was able to cause the relocation of L22 in the 293 cell line containing EBER1-deleted BAC EBV (Fig. 6B). EBER2 produced from the plasmid was also the same length as endogenous EBER2 of B95-8 cells (see Fig. S4B in the supplemental material). The results in Fig. 6B show that EBER1 is able to relocate L22 in 293 cells in the presence or absence of the remainder of the EBV genome, and the activity of EBER1 in this assay demonstrates that EBER1 produced from the H1 plasmids is functional.
Some relocation of L22 was also observed in response to EBV infection in two carcinoma cell lines derived from gastric carcinoma (AGS) or nasopharyngeal carcinoma (HONE1) and in the BJAB lymphoma line in response to EBER expression (Table 1 and see Fig. S4 in the supplemental material). These results support the relevance of this phenomenon to EBV-associated cancers, but the fraction of cells acquiring nucleolar L22 was much lower than in 293 cells (Table 1).
In contrast to the 293 cell results, in LCLs the L22 ribosomal protein staining pattern was mostly cytoplasmic and not detectably changed according to EBER1 expression (Fig. 6C). Several feedback and stress pathways coordinate ribosome assembly in the nucleolus with supply of ribosomal proteins, and it is possible that the lack of nucleolar staining for L22 in the LCLs reflects more normal coordination of ribosome assembly than is present in the tumor cell lines and in 293 cells. The LCL infection is more physiologically relevant to normal EBV infection and gave clear effects of EBER deletion on gene expression, but the 293 results are useful for relating our results to previous studies and demonstrate that the EBER1 made from our novel EBER expression vectors is functional.
DISCUSSION
In the EBV-associated cancers BL, NPC, HL, and PTLD virtually all of the malignant cells are thought to express EBER RNAs. The EBER RNAs are also some of the most abundant EBV transcripts in infected cells. This makes the EBERs a potentially important target for novel therapies that might attempt to use the presence of EBV in tumor cells to target the cancer. Published data on the ability of EBERs to prevent apoptosis in some tumor cell lines support this approach, but much more understanding of the effects of EBER RNAs on cells is likely to be required before this strategy could be explored in detail. The consistent expression of EBER RNAs in latent infection and reduced expression in the lytic cycle implies that they function in latent persistence, but the wide variety of effects has made interpretation of their mechanisms difficult.
Some of the most important published data has come from studies with EBER deletion mutants of the Akata EBV strain (22, 29, 40-42), and these have led to a large number of phenotypic effects described in cells, but only a few cell genes have been reported to have their expression affected by EBERs. We have used mutants of the B95-8 EBV strain lacking EBER1 or EBER2 to identify cell genes whose expression correlates with EBER expression in LCLs. This has resulted in lists of genes (see Fig. S2 and S3 in the supplemental material) whose expression correlates with deletion of either EBER1 or EBER2 from the EBV genome; by applying strict criteria of reproducibility, the final lists of genes are relatively short: 54 for EBER1 deletion and 115 for EBER2 deletion. It is likely that many of the changes in expression observed will be secondary consequences of the specific changes due to EBER status, but these genes should still be useful markers for further investigation of EBER function. The fact that similar genes were not found to be altered when the same EBV mutants were tested in 293 cells suggests that the results may depend on cell type, but it is also possible that slightly smaller changes in 293 cells did not achieve the high degree of statistical significance that was imposed in the present study. Previous work using transient transfection of an EBER2 expression vector into 293 cells and microarray expression profiling described lists of genes that were regulated (8), but we did not see a similar pattern of EBER2-dependent alteration in cell gene regulation in 293 cells containing recombinant EBV.
The clearer effects of EBER2 on LCL gene regulation, the ability of EBER1 to be secreted from cells, and the association of EBER1 with stress conditions might suggest that EBER2 would be a better cell intrinsic therapeutic target in EBV-associated cancers. In fact, one of the genes we found to be regulated with EBER2 in the present study is IGF-1 (Fig. 4A), which was already shown to be regulated by EBER in previous work (16, 17), a consistent result. Regulation of gene expression at the protein level in the LCLs according to EBER status can also be observed, as illustrated for CXCR3 in Fig. 4E, but we do not yet know which of the many regulated genes listed will prove to be the proximal targets of EBER regulation. Future work will allow investigation of this and testing functional consequences of the alterations in RNA levels shown in the present study. Functional classification using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) program (version 6.7; http://david.abcc.ncifcrf.gov/) of the lists of genes regulated in response to EBER1 showed biosynthetic processes and interferon/antiviral responses to be the two most enriched groups in the list. Similar analysis of the EBER2 list showed genes regulating apoptosis and membrane signaling molecules to be the two most enriched groups. These results usefully correspond with previous emphasis in the literature on connection of innate immune responses for EBER1 and effects of EBER genes on apoptosis.
In some respects, our results differ from those published with EBER deletion mutants in the Akata EBV strain (50, 51). We saw no significant effect on transformation efficiency or growth of LCLs in response to the deletion of EBER2, whereas a 50-fold reduction in transformation efficiency and reduced growth from low starting cell density were observed with Akata EBV. This might be a consequence of a high intrinsic transforming ability of the B95-8 strain, or there might be unrecognized procedural differences in the transformation assays used. B95-8 is the most widely used strain in EBV research, but in some respects Akata is a more wild-type genome, and the different results might be a valuable clue to EBER2 function that can be explored in the future. The other published study on the deletion of EBER genes also did not detect any difference in transformation efficiency but was not exactly comparable since it was done by P3HR1 recombination with B95-8 sequence spanning the EBER locus to EBNA2 and both EBER genes were deleted in that experiment (44). These recombinants would be expected to be derived from P3HR1 EBV around the B95-8 deletion region, so the difference between our results and the Akata mutagenesis should not simply be ascribed to the absence of the B95-8 deletion region (and the miRNAs encoded there) in our recombinants.
Several publications using tumor cell lines or overexpression of L22 from transfected expression plasmids have shown EBER1 to cause a relocation of L22 protein in cells, but the mechanism by which this binding of EBER1 to L22 may affect the cell is uncertain. The fact that the L22 gene can be deleted in mice without any gross effect on development or breeding of the mice (1) calls into question the importance of L22 in protein synthesis: L22 is clearly not essential, but it might have quantitative effects on translation efficiency. The remarkable phenotype of the L22 knockout mice, which almost completely lack αβ-T lymphocytes but have otherwise normal blood counts (1), suggests a link between L22 and apoptosis in specific circumstances. The dependence of that phenotype on p53 implies a role for p53 in the apoptosis of precursors to αβ-T lymphocytes and may indicate a function for L22 separate from translation, a function that EBER1 might potentially affect. A recent study in EBV-positive Akata BL cells (13) found that growth-promoting properties of EBER correlated with L22 binding and showed that ribosomes in polysomes lacked L22. In the present study, the nuclear redistribution of endogenous L22 protein away from the nucleolus by EBER1 in EBV-infected 293 cells was confirmed using the recombinant EBV strains, and there was some evidence for EBV-dependent L22 relocation in carcinoma cell lines. Novel EBER expression vectors were capable of achieving high levels of EBER expression, and EBER1 made in this way was also functional in causing L22 relocation. However, in LCLs we could find no significant evidence for a relocation of L22 from nucleoli by EBER1 because the L22 staining was mostly cytoplasmic in LCLs. This might simply be a consequence of slightly different kinetics of ribosome assembly in LCLs not resulting in the accumulation of L22 in the nucleoli, but all of the observations of nucleolar L22 and its relocation by EBER1 have been made in tumor lines or highly transformed cell lines. The relationship of the effects on gene expression that we observed in LCLs to L22 binding thus remains to be established.
We hope that it may be possible in future work to use the novel EBER expression vectors to identify the more direct cell target genes of EBER1 and EBER2 by identifying those in our lists that respond more rapidly or strongly to the re-expression of EBER in LCLs containing EBV with a deletion of EBER1 or EBER2. If clear mechanisms can be identified, it may then be possible to develop novel therapeutic approaches to EBV-associated cancers based on their EBER expression.
Supplementary Material
Acknowledgments
We thank Joan Steitz for the L22 antibody, Wolfgang Hammerschmidt for the B95-8 EBV BAC, and Sankar Swaminathan for helpful discussions.
This study was supported in part by the Sixth Research Framework Programme of the European Union, project INCA (LSHC-CT-2005-018704).
Footnotes
Published ahead of print on 19 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderson, S. J., et al. 2007. Ablation of ribosomal protein L22 selectively impairs αβ T cell development by activation of a p53-dependent checkpoint. Immunity 26:759-772. [DOI] [PubMed] [Google Scholar]
- 2.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 3.Cai, X., et al. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke, P. A., M. Schwemmle, J. Schickinger, K. Hilse, and M. J. Clemens. 1991. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 19:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dirmeier, U., et al. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 63:2982-2989. [PubMed] [Google Scholar]
- 7.Dobbelstein, M., and T. Shenk. 1995. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J. Virol. 69:8027-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eilebrecht, S., et al. 2008. EBER2 RNA-induced transcriptome changes identify cellular processes likely targeted during Epstein-Barr virus infection. BMC Res. Notes 1:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elia, A., J. Vyas, K. Laing, and M. Clemens. 2004. Ribosomal protein L22 inhibits regulation of cellular activities by the Epstein-Barr virus small RNA EBER-1. Eur. J. Biochem. 271:1895-1905. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, P. J., et al. 1997. Direct demonstration of persistent EBV gene expression in peripheral blood of infected common marmosets and analysis of virus-infected tissues in vivo. J. Gen. Virol. 78:1417-1424. [DOI] [PubMed] [Google Scholar]
- 11.Fok, V., K. Friend, and J. A. Steitz. 2006. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell Biol. 173:319-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glickman, J. N., J. G. Howe, and J. A. Steitz. 1988. Structural analyzes of EBER1 and EBER2 ribonucleoprotein particles present in Epstein-Barr virus-infected cells. J. Virol. 62:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houmani, J. L., C. I. Davis, and I. K. Ruf. 2009. Growth-promoting properties of Epstein-Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. J. Virol. 83:9844-9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe, J. G., and J. A. Steitz. 1986. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc. Natl. Acad. Sci. U. S. A. 83:9006-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutzinger, R., et al. 2009. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 5:e1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwakiri, D., Y. Eizuru, M. Tokunaga, and K. Takada. 2003. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 63:7062-7067. [PubMed] [Google Scholar]
- 17.Iwakiri, D., T. S. Sheen, J. Y. Chen, D. P. Huang, and K. Takada. 2005. Epstein-Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene 24:1767-1773. [DOI] [PubMed] [Google Scholar]
- 18.Iwakiri, D., et al. 2009. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 206:2091-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieff, E., and A. Rickinson. 2007. Epstein-Barr virus, p. 2603-2654. In D. Knipe and P. Howley (ed.), Fields virology, 5th ed. Raven Press, Philadelphia, PA.
- 20.Kim, S. K., K. A. Buczynski, G. B. Caughman, and D. J. O'Callaghan. 2001. The equine herpesvirus 1 immediate-early protein interacts with EAP, a nucleolar-ribosomal protein. Virology 279:173-184. [DOI] [PubMed] [Google Scholar]
- 21.Kitagawa, N., et al. 2000. Epstein-Barr virus-encoded poly(A)− RNA supports Burkitt's lymphoma growth through interleukin-10 induction. EMBO J. 19:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komano, J., S. Maruo, K. Kurozumi, T. Oda, and K. Takada. 1999. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J. Virol. 73:9827-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavergne, J. P., F. Conquet, J. P. Reboud, and A. M. Reboud. 1987. Role of acidic phosphoproteins in the partial reconstitution of the active 60S ribosomal subunit. FEBS Lett. 216:83-88. [DOI] [PubMed] [Google Scholar]
- 24.Le, S., R. Sternglanz, and C. W. Greider. 2000. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell 11:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leopardi, R., and B. Roizman. 1996. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein ICP4 with EAP, a nucleolar-ribosomal protein. Proc. Natl. Acad. Sci. U. S. A. 93:4572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the U(L)13 protein kinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerner, M. R., N. C. Andrews, G. Miller, and J. A. Steitz. 1981. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. U. S. A. 78:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna, S. A., D. A. Lindhout, T. Shimoike, C. E. Aitken, and J. D. Puglisi. 2007. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J. Mol. Biol. 372:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanbo, A., K. Inoue, K. Adachi-Takasawa, and K. Takada. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 21:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuhierl, B., R. Feederle, W. Hammerschmidt, and H. J. Delecluse. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. U. S. A. 99:15036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni, J. Q., L. P. Liu, D. Hess, J. Rietdorf, and F. L. Sun. 2006. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 20:1959-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niedobitek, G., et al. 1991. Epstein-Barr virus infection in oral hairy leukoplakia: virus replication in the absence of a detectable latent phase. J. Gen. Virol. 72:3035-3046. [DOI] [PubMed] [Google Scholar]
- 33.Pfeffer, S., et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 34.Pfeffer, S., et al. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 35.Reed, L., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 36.Repellin, C. E., P. M. Tsimbouri, A. W. Philbey, and J. B. Wilson. 2010. Lymphoid hyperplasia and lymphoma in transgenic mice expressing the small non-coding RNA, EBER1 of Epstein-Barr virus. PLoS One 5:e9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosa, M. D., E. Gottlieb, M. R. Lerner, and J. A. Steitz. 1981. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol. 1:785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruf, I. K., K. A. Lackey, S. Warudkar, and J. T. Sample. 2005. Protection from interferon-induced apoptosis by Epstein-Barr virus small RNAs is not mediated by inhibition of PKR. J. Virol. 79:14562-14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rymo, L. 1979. Identification of transcribed regions of Epstein-Barr virus DNA in Burkitt lymphoma-derived cells. J. Virol. 32:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samanta, M., D. Iwakiri, T. Kanda, T. Imaizumi, and K. Takada. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samanta, M., D. Iwakiri, and K. Takada. 2008. Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 27:4150-4160. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu, N., A. Tanabe-Tochikura, Y. Kuroiwa, and K. Takada. 1994. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J. Virol. 68:6069-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swaminathan, S. 2008. Noncoding RNAs produced by oncogenic human herpesviruses. J. Cell Physiol. 216:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swaminathan, S., B. Tomkinson, and E. Kieff. 1991. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc. Natl. Acad. Sci. U. S. A. 88:1546-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toczyski, D. P., A. G. Matera, D. C. Ward, and J. A. Steitz. 1994. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 91:3463-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toczyski, D. P., and J. A. Steitz. 1993. The cellular RNA-binding protein EAP recognizes a conserved stem-loop in the Epstein-Barr virus small RNA EBER 1. Mol. Cell. Biol. 13:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toczyski, D. P., and J. A. Steitz. 1991. EAP, a highly conserved cellular protein associated with Epstein-Barr virus small RNAs (EBERs). EMBO J. 10:459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White, R. E., M. A. Calderwood, and A. Whitehouse. 2003. Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence. J. Gen. Virol. 84:3393-3403. [DOI] [PubMed] [Google Scholar]
- 49.Wood, J., R. M. Frederickson, S. Fields, and A. H. Patel. 2001. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 75:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, Y., S. Maruo, M. Yajima, T. Kanda, and K. Takada. 2007. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J. Virol. 81:11236-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yajima, M., T. Kanda, and K. Takada. 2005. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J. Virol. 79:4298-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto, N., T. Takizawa, Y. Iwanaga, and N. Shimizu. 2000. Malignant transformation of B lymphoma cell line BJAB by Epstein-Barr virus-encoded small RNAs. FEBS Lett. 484:153-158. [DOI] [PubMed] [Google Scholar]
- 53.Yang, L., K. Aozasa, K. Oshimi, and K. Takada. 2004. Epstein-Barr virus (EBV)-encoded RNA promotes growth of EBV-infected T cells through interleukin-9 induction. Cancer Res. 64:5332-5337. [DOI] [PubMed] [Google Scholar]
- 54.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.